Emerging antibacterial resistance is a consequence of the continued use of our current antibacterial therapies, and it is limiting their utility, especially for infections caused by multidrug-resistant isolates. β-Lactams have enjoyed extensive clinical success, but their broad usage is linked to perhaps the most extensive and progressive example of resistance development for any antibacterial scaffold. In Gram-negative pathogens, this largely involves constant evolution of new β-lactamases able to degrade successive generations of this scaffold. In addition, more recently, alterations in the targets of these compounds, penicillin-binding proteins (PBPs), are being described in clinical isolates, which often also have multiple β-lactamases. This study underscores the multifactorial nature of β-lactam resistance by uncovering alterations of PBP2 that reduce susceptibility to carbapenems in E. coli clinical isolates that also have alterations of PBP3 and express the NDM-1 β-lactamase. The changes in PBP2 also reduced susceptibility to the intrinsic antibacterial activity of some diazabicyclooctane (DBO) compounds that can target PBP2. This may have implications for the development and use of the members of this relatively newer scaffold that are inhibitors of PBP2 in addition to their inhibition of serine-β-lactamases.
KEYWORDS: NDM-1, carbapenems, diazabicyclooctane, penicillin-binding proteins
ABSTRACT
Penicillin-binding proteins (PBPs) are essential for bacterial cell wall biosynthesis, and several are clinically validated antibacterial targets of β-lactam antibiotics. We identified mutations in the mrdA gene encoding the PBP2 protein in two Escherichia coli blaNDM-1 clinical isolates that reduce susceptibility to carbapenems and to the intrinsic antibacterial activity of a diazabicyclooctane (DBO) PBP2 and β-lactamase inhibitor. These mutations coexisted with previously described mutations in ftsI (encoding PBP3) that reduce susceptibility to monobactams, penicillins, and cephalosporins. Clinical exposure to β-lactams is driving the emergence of multifactorial resistance that may impact the therapeutic usefulness of existing antibacterials and novel compounds that target PBPs.
IMPORTANCE Emerging antibacterial resistance is a consequence of the continued use of our current antibacterial therapies, and it is limiting their utility, especially for infections caused by multidrug-resistant isolates. β-Lactams have enjoyed extensive clinical success, but their broad usage is linked to perhaps the most extensive and progressive example of resistance development for any antibacterial scaffold. In Gram-negative pathogens, this largely involves constant evolution of new β-lactamases able to degrade successive generations of this scaffold. In addition, more recently, alterations in the targets of these compounds, penicillin-binding proteins (PBPs), are being described in clinical isolates, which often also have multiple β-lactamases. This study underscores the multifactorial nature of β-lactam resistance by uncovering alterations of PBP2 that reduce susceptibility to carbapenems in E. coli clinical isolates that also have alterations of PBP3 and express the NDM-1 β-lactamase. The changes in PBP2 also reduced susceptibility to the intrinsic antibacterial activity of some diazabicyclooctane (DBO) compounds that can target PBP2. This may have implications for the development and use of the members of this relatively newer scaffold that are inhibitors of PBP2 in addition to their inhibition of serine-β-lactamases.
OBSERVATION
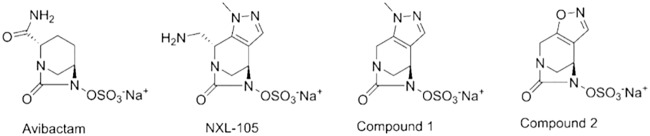
β-Lactams are a broad class of antibacterial agents that inhibit penicillin-binding proteins (PBPs) essential for transglycosylation and transpeptidation of peptidoglycan strands during bacterial cell wall biosynthesis (1). Many β-lactam antibiotics have been developed and extensively used in the clinic over the past several decades. These antibiotics have a wide range of PBP affinities and specificities, with most inhibiting multiple PBPs (2). Unfortunately, the utility of this class of antibiotics is being undermined by continuing resistance development. In particular, resistance to carbapenems, a group that has been widely used as an agent of last resort to treat severe infections caused by extended-spectrum-β-lactamase (ESBL)-expressing Enterobacteriaceae, is a growing concern (3). Clinical resistance to β-lactams in Gram-negative bacteria had primarily been attributed to the expression of plasmid-carried or chromosomal β-lactamase genes (4), with reports of specific target mutations being rare in Gram-negative pathogens. Recently, however, mutations in ftsI encoding insertions in PBP3 that reduce susceptibility to certain β-lactams have been identified in Escherichia coli clinical isolates (5, 6). In addition, clinical isolates and in vitro-selected E. coli mutants with reduced susceptibility to carbapenems showed changes in the gene encoding PBP2 (7–9). Given the continuing emergence and spread of new β-lactamases, there is interest in the development of novel β-lactamase inhibitors (BLIs) (4) and in the development of novel β-lactam mimetics that are not impacted by β-lactamases (10). One such class of BLIs and β-lactam mimetics is the diazabicyclooctane (DBO) scaffold, which was discovered by chemists at Hoechst Marion Roussel, such as NXL-104 (avibactam), NXL-105 (11) (US2010092443 [12]), and compounds 1 and 2 (WO 02/100860 [13]) (Fig. 1). DBOs are potent inhibitors of β-lactamases but were originally designed as β-lactam mimetics, and some analogs such as NXL-105 also had intrinsic antibacterial activity through inhibition of PBP2 and were viewed as potential antibacterial drugs (11). The notion that the antibacterial activity of such compounds should be considered in drug discovery has also been echoed more recently (14). Antibacterials that act mainly by inhibition of PBP2 can exhibit relatively high frequencies of selecting non-target-based resistance in vitro via a multiplicity of mutations affecting the stringent response, thereby reducing susceptibility to inhibition of PBP2 (15, 16). The potential for target-based resistance to DBOs is currently not well understood. We reported previously (17) that four NDM-1-expressing E. coli clinical isolates (NB27236, NB27330, NB27307, and NB27326 [Table 1]) possessed a previously characterized (5) mutation in ftsI encoding a YRIN insertion in PBP3 that reduces susceptibility to PBP3 inhibitors such as aztreonam (which is not degraded by NDM-1). Intriguingly, one of these isolates, NB27307, was 4-fold less susceptible to the antibacterial activity of the very potent DBO molecule NXL-105 than was NB27236 or the control E. coli strain NB27001 and 2-fold less susceptible than NB27330 (Table 1), based on broth microdilution assay according to CLSI methodology (18). Strain NB27326 was also 2- to 4-fold less susceptible than NB27236 and NB27001 (Table 1). Strains NB27307 and NB27326 were also less susceptible to the less potent DBO compounds 1 and 2 (Fig. 1) than were NB27236 and NB27330 (4- to 16-fold) and were 64-fold (compound 1) and 8-fold (compound 2) less susceptible than the control strain NB27001 (Table 1). However, strains NB27236 and NB27330 were also less susceptible to compound 1 (8- to 16-fold) than the control strain NB27001 whereas they did not show these substantial shifts in susceptibility to compound 2 (Table 1). Since the antibacterial activity of DBOs is mediated by inhibition of PBP2 (14), we asked if NB27307 and NB27326 harbored mutations in mrdA, which encodes PBP2. PCR amplification and sequencing of mrdA from all four isolates, and the control strain NB27001 (ATCC 25922), using the primers listed in Table 2, indicated that NB27307 and NB27326, the two isolates that were the most consistently less susceptible to DBOs, each had unique mutations encoding amino acid substitutions in PBP2 compared to the other isolates or to the E. coli ATCC 25922 or strain MG1655 mrdA reference sequences (19). These were T1718A, encoding a unique L573Q substitution for strain NB27307, and G1564A, encoding a unique V522I substitution for strain NB27326 (Table 1). NB27326 also had a V217M substitution in addition to V522I, while the two more-susceptible isolates, NB27236 and NB27330, harbored only the V217M substitution.
FIG 1.
Chemical structures of diazabicyclooctane (DBO) molecules. NXL-105 is described in reference 11 (US2010092443 [12]), and compounds 1 and 2 are described in WO 02/100860 (13).
TABLE 1.
Mutations identified in E. coli clinical isolates and antibiotic susceptibilities of engineered E. coli mrdA mutantsa
| E. coli strain | Source | PBP2 alteration | MIC (μg/ml) of drug: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| NXL-105 | Comp 1 | Comp 2 | IPM | MEM | ATM | CAZ | |||
| NB27001 | ATCC 25922 | None | 0.008 | 0.5 | 2 | 0.25 | ≤0.06 | 0.125 | 0.25 |
| NB27236 (FtsIYRIN) | JMI | V217M | 0.008 | 4 | 1–2 | 16 | >64 | >64 | >64 |
| NB27330 (blaCTX-M-15 blaCMY-2 blaNDM-1, FtsIYRIN) | IHMA | V217M | 0.016 | 8 | 4 | 32 | >64 | >64 | >64 |
| NB27307 (blaNDM-1 FtsIYRIN) | ATCC BAA-2471 (27) | L573Q | 0.032 | 32 | 16 | 16 | 32 | >64 | >64 |
| NB27326 (blaSHV-12 blaCMY-2 blaNDM-1 FtsIYRIN) | IHMA | V217M, V522I | 0.016–0.032 | 32 | 16 | 4 | 16 | >64 | >64 |
| BW25113 | None | 0.004 | 1 | 2 | 0.25 | 0.032 | 0.125 | 0.25 | |
| BW25113-CDK0001 | This study | L573Q | 0.016 | 4 | 16 | 0.5 | 0.125 | 0.125 | 0.25 |
| BW25113-CDK0004 | This study | V522I | 0.004 | 4 | 16 | 0.5 | 0.06 | 0.125 | 0.25 |
Abbreviations: ATM, aztreonam; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; ATCC, American Type Culture Collection; IHMA, International Health Management Associates; Comp, compound. IPM, MEM, ATM, and CAZ were obtained from commercial sources. DBOs were synthesized at Novartis. MIC data were generated according to CLSI methodology (18). Unique amino acid substitutions in PBP2 are indicated in bold. Strain numbers are Novartis internal numbering. Strain NB27330 was isolated from the colon of a patient in India. NB27307 is a respiratory isolate from Pakistan, and NB27326 is an isolate from peritoneal fluid in India. No additional information is available for strain NB27236.
TABLE 2.
Oligonucleotide primers used in this study
| Primer purpose and name | Sequence (5′–3′) |
|---|---|
| mrdA (PBP2) sequencinga | |
| SR176 | CATCACCACCAACCATCCTT |
| SR177 | CCGTGCAGCACATCTTCATA |
| SR178 | TGACGATATTGCTGCATTCC |
| SR179 | GGTTCACCAGCGGTGTATTC |
| SR180 | TGGTTTCCACGCCTAGTTATG |
| SR181 | AGGTTTCGTTCGCTTTCAGA |
| SR182 | CCGAATGGATGGGTAAATTC |
| SR183 | TGTGGGATCGAGATGGACTT |
| Gene manipulations and diagnostic PCR | |
| SR200 | TTGACGGTATCTCCAGCAAA |
| SR201 | GCTAAGGCCAGAGAGGAACA |
| SR202 | ACACCATTCCGGTTGGTATC |
| SR203 | TACGCTCCATCATGCCAATA |
The mrdA gene was amplified from clinical isolates for sequencing in segments using primer pairs SR176/SR177, SR178/SR179, SR180/SR181, SR182/SR183, and SR176/SR183.
To establish whether the two unique alterations reduced susceptibility to DBOs or β-lactams that act in part by inhibition of PBP2, each alteration was introduced individually into the susceptible E. coli laboratory strain BW25113 by recombination as previously described (20). Briefly, linear DNA fragments encompassing the appropriate region of mrdA were amplified from E. coli clinical isolates NB27307 and NB27326 using primers SR200 and SR201 (Table 2). There were three silent mrdA mutations in NB27326, and these were present on the PCR fragment from this isolate. The fragments were then individually transformed into electrocompetent E. coli BW25113 cells harboring the pKD46 helper plasmid (20). Mutants were selected on individual LB agar plates (tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 10 g/liter; agar, 1.5%) containing 0.25% (wt/vol) arabinose and 0.25 μg/ml imipenem at 37°C. The introduced missense mutations were confirmed on the genome for BW25113-CDK0001 and BW25113-CDK0004 (Table 1) by PCR and sequencing of the mrdA gene using primers SR202 and SR203 (Table 2). The three silent mutations within mrdA from clinical isolate NB27326 were also introduced into mrdA of BW25113-CDK0004. Whole-genome sequencing of BW25113-CDK0001 and BW25113-CDK0004 using previously described methodology (21) confirmed that only the intended mutations were introduced on the genome. These alterations caused a modest 2- to 4-fold decrease in susceptibility to imipenem and meropenem, which act in part by inhibition of PBP2 (7) (BW25113-CDK0001 [L573Q] and BW25113-CDK0004 [V522I] [Table 1]). Susceptibility to aztreonam and ceftazidime, which do not significantly engage PBP2 (2, 22), was not affected by either alteration.
BW25113-CDK0001 also exhibited a 4-fold decrease in susceptibility to NXL-105 (11) (Table 1). No shift in NXL-105 MIC was mediated by the V522I alteration alone (BW25113-CDK0004); however, the possibility that V522I might cause a more subtle shift (lower than 2-fold) in this strain background was not tested by growth curve analysis. We speculated that less potent but structurally related DBOs such as compounds 1 and 2 (Fig. 1) might reveal a more consistent effect of both the L573Q and V522I alterations. Indeed, the activity of both compounds was decreased (4- to 8-fold, respectively) for BW25113-CDK0001 (L573Q) and BW25113-CDK0004 (V522I) (Table 1). As mentioned above, isolates NB27236 and NB27330, which harbored only the V217M alteration, were less susceptible to compound 1 than NB27001, suggesting that this change may differentially affect susceptibility to different DBO molecules, but this remains to be confirmed. It should be noted that we did not determine the PBP binding profiles for compounds 1 and 2 and therefore do not know if they differ. Nonetheless, it is possible that an additional contribution of the V217M alteration that was not tested here together with V522I in mutant BW25113-CDK0004 could be necessary to see a shift in NXL-105 susceptibility in this mutant. Lastly, both BW25113-CDK0001 and BW25113-CDK0004 were 4-fold less susceptible to the non-DBO PBP2 inhibitor amdinocillin (MIC of 1 μg/ml for both compared to 0.25 μg/ml for BW25113 or NB27001). The L573Q and related L573V substitutions were previously implicated in reducing susceptibility to amdinocillin but were only tested along with other PBP2 alterations (9, 23). Overall, our data suggest that the amino acid substitutions identified in this study affect the intrinsic antibacterial activity of these DBO molecules, but it is unlikely that they would interfere with inhibition of β-lactamases by DBOs.
The PBP2 L573Q substitution identified here in the blaNDM-1-containing E. coli clinical isolate NB27307 occurs at the same position as a previously described L573V substitution associated with decreased susceptibility to imipenem (9) and is immediately adjacent to an M574I substitution previously associated with reduced susceptibility to carbapenems (7, 23). Furthermore, an L573Q substitution emerged in E. coli during in vitro serial passaging studies using ertapenem (8). However, the V522I substitution identified here in the blaNDM-1-containing E. coli clinical isolate NB27326 has not, to our knowledge, been previously associated with resistance.
A BLAST search of publicly available NCBI sequences using the PBP2 L573Q variant found one E. coli genome (NZ_CP021879.1; strain AR_0137) encoding the identical PBP2 protein. E. coli strain AR_0137 is an antibiotic-resistant carbapenemase-producing strain that is part of the FDA-CDC resistant bacteria panel (https://www.ncbi.nlm.nih.gov/bioproject/294416). A similar BLAST search using only the truncated sequence KIAERLRDHKQMTAFAPYNNPQVA, encompassing L573Q, yielded the same single hit. Searching publicly available sequences using the PBP2 V522I (single) variant sequence yielded 17 E. coli genomes encoding the identical protein (GenBank accession numbers AXN84639.1, TBI06625.1, RKP81685.1, AWR48380.1, OXK11063.1, RWS74779.1, KZJ44915.1, ROK48460.1, AUY30260.1, OCO29799.1, QAY43423.1, QBC12101.1, KDG76004.1, AXP27762.1, AYL12293.1, AWA17983.1, and VCY82512.1). These strains were mainly multidrug-resistant (MDR) and carbapenem-resistant/carbapenemase-producing clinical isolates, with RWS74779.1 having been isolated from water. This suggests that the V5221 substitution in PBP2 could be emerging among MDR isolates. Searching with the PBP2 V217M V522I double variant sequence dropped the number of hits to two (GenBank accession numbers KSY14719.1 and PSF33251.1). One of these, strain 50673720, was a carbapenemase-producing clinical isolate from Norway (24).
As mentioned above, it is well established that Gram-negative bacteria can become less susceptible to PBP2 inhibition at high frequency through a multiplicity of nontarget mutations related to the stringent response (15, 16). PBP2 target mutations affecting DBO activity, being more rare, may be difficult to identify using standard in vitro selection experiments, given the high background of stringent response mutants. Here, we noticed a trend suggesting differential susceptibility of clinical isolates and confirmed the presence of PBP2 alterations affecting DBO susceptibility. To our knowledge, this is the first report of alterations in PBP2 that reduce susceptibility to the intrinsic antibacterial activity of a DBO molecule. Antibacterial DBO molecules are referred to as “enhancers” of the activity of β-lactams that inhibit other PBPs besides PBP2, due to synergy arising during inhibition of multiple PBPs (25, 26). It is reasonable to suspect that the PBP2 variants such as those identified in this study may therefore also reduce this effect, but this remains to be determined. Nonetheless, the presence of these PBP2 variants in clinical isolates that also harbor the YRIN insertion in PBP3 (5) as well as blaNDM-1, provides yet another glimpse into the clinical emergence of multifactorial resistance, not only β-lactamase mediated but also involving multiple target mutations. The latter can clearly impact currently used therapeutics but may also hamper the future potential of novel β-lactam mimetics such as DBOs to the extent that their effectiveness relies on engaging these targets.
ACKNOWLEDGMENTS
We thank JMI Laboratories and International Health Management Associates (IHMA) for clinical isolates and Cindy Li, Katherine Thompson, and Johanne Blais for helpful discussion. We also thank Fergal Casey and David Barkan for bioinformatics assistance.
REFERENCES
- 1.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 2.Davies TA, Shang W, Bush K, Flamm RK. 2008. Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1510–1512. doi: 10.1128/AAC.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Bradford PA. 2016. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alm RA, Johnstone MR, Lahiri SD. 2015. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Kashikar A, Brown CA, Denys G, Bush K. 2017. Unusual Escherichia coli PBP 3 insertion sequence identified from a collection of carbapenem-resistant Enterobacteriaceae tested in vitro with a combination of ceftazidime-, ceftaroline-, or aztreonam-avibactam. Antimicrob Agents Chemother 61:e00389-17. doi: 10.1128/AAC.00389-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamachika S, Sugihara C, Kamai Y, Yamashita M. 2013. Correlation between penicillin-binding protein 2 mutations and carbapenem resistance in Escherichia coli. J Med Microbiol 62:429–436. doi: 10.1099/jmm.0.051631-0. [DOI] [PubMed] [Google Scholar]
- 8.Adler M, Anjum M, Andersson DI, Sandegren L. 2016. Combinations of mutations in envZ, ftsI, mrdA, acrB and acrR can cause high-level carbapenem resistance in Escherichia coli. J Antimicrob Chemother 71:1188–1198. doi: 10.1093/jac/dkv475. [DOI] [PubMed] [Google Scholar]
- 9.Aissa N, Mayer N, Bert F, Labia R, Lozniewski A, Nicolas-Chanoine MH. 2016. A new mechanism to render clinical isolates of Escherichia coli non-susceptible to imipenem: substitutions in the PBP2 penicillin-binding domain. J Antimicrob Chemother 71:76–79. doi: 10.1093/jac/dkv318. [DOI] [PubMed] [Google Scholar]
- 10.Zervosen A, Sauvage E, Frere JM, Charlier P, Luxen A. 2012. Development of new drugs for an old target: the penicillin binding proteins. Molecules 17:12478–12505. doi: 10.3390/molecules171112478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman K. 2011. Diazabicyclooctanes (DBOs): a potent new class of non-β-lactam β-lactamase inhibitors. Curr Opin Microbiol 14:550–555. doi: 10.1016/j.mib.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 12.Levasseur P, Pace JL, Coleman K, Lowther J. 5 August 2009, filing date. Novel combinations of nitrogenated heterocyclic antibacterial compounds with other antibacterial compounds and the use of same as drugs. US patent application 20100092443.
- 13.Aszodi J, Lampilas M, Musicki B, Rowlands DA, Collette P. 19 December 2002. Novel heterocyclic compounds, method for preparing same and use thereof as medicines, in particular as anti-bacterial agents. World patent application WO 02/100860.
- 14.King AM, King DT, French S, Brouillette E, Asli A, Alexander JA, Vuckovic M, Maiti SN, Parr TR Jr, Brown ED, Malouin F, Strynadka NC, Wright GD. 2016. Structural and kinetic characterization of diazabicyclooctanes as dual inhibitors of both serine-β-lactamases and penicillin-binding proteins. ACS Chem Biol 11:864–868. doi: 10.1021/acschembio.5b00944. [DOI] [PubMed] [Google Scholar]
- 15.Thulin E, Sundqvist M, Andersson DI. 2015. Amdinocillin (mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia coli. Antimicrob Agents Chemother 59:1718–1727. doi: 10.1128/AAC.04819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore DM, Warner M, Mushtaq S, Woodford N. 2016. Interactions of OP0595, a novel triple-action diazabicyclooctane, with beta-lactams against OP0595-resistant Enterobacteriaceae mutants. Antimicrob Agents Chemother 60:554–560. doi: 10.1128/AAC.02184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blais J, Lopez S, Li C, Ruzin A, Ranjitkar S, Dean CR, Leeds JA, Casarez A, Simmons RL, Reck F. 2018. In vitro activity of LYS228, a novel monobactam antibiotic, against multidrug-resistant Enterobacteriaceae. Antimicrob Agents Chemother 62:e00552-18. doi: 10.1128/AAC.00552-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacterial that grow aerobically. Approved standard—10th ed, suppl M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean CR, Barkan DT, Bermingham A, Blais J, Casey F, Casarez A, Colvin R, Fuller J, Jones AK, Li C, Lopez S, Metzger LET, Mostafavi M, Prathapam R, Rasper D, Reck F, Ruzin A, Shaul J, Shen X, Simmons RL, Skewes-Cox P, Takeoka KT, Tamrakar P, Uehara T, Wei JR. 2018. Mode of action of the monobactam LYS228 and mechanisms decreasing in vitro susceptibility in Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother 62:e01200-18. doi: 10.1128/AAC.01200-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes MV, Orr DC. 1983. Mode of action of ceftazidime: affinity for the penicillin-binding proteins of Escherichia coli K12, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother 12:119–126. doi: 10.1093/jac/12.2.119. [DOI] [PubMed] [Google Scholar]
- 23.Lange F, Pfennigwerth N, Hofken LM, Gatermann SG, Kaase M. 2019. Characterization of mutations in Escherichia coli PBP2 leading to increased carbapenem MICs. J Antimicrob Chemother 74:571–576. doi: 10.1093/jac/dky476. [DOI] [PubMed] [Google Scholar]
- 24.Samuelsen O, Overballe-Petersen S, Bjornholt JV, Brisse S, Doumith M, Woodford N, Hopkins KL, Aasnaes B, Haldorsen B, Sundsfjord A, Norwegian Study Group On CPE. 2017. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS One 12:e0187832. doi: 10.1371/journal.pone.0187832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morinaka A, Tsutsumi Y, Yamada M, Suzuki K, Watanabe T, Abe T, Furuuchi T, Inamura S, Sakamaki Y, Mitsuhashi N, Ida T, Livermore DM. 2015. OP0595, a new diazabicyclooctane: mode of action as a serine β-lactamase inhibitor, antibiotic and β-lactam ‘enhancer’. J Antimicrob Chemother 70:2779–2786. doi: 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- 26.Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. Potent β-lactam enhancer activity of zidebactam and WCK 5153 against Acinetobacter baumannii, including carbapenemase-producing clinical isolates. Antimicrob Agents Chemother 61:e01238-17. doi: 10.1128/AAC.01238-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]