Abstract
Background/Aim: Vitamin D receptor (VDR) is present in numerous cellular pathways and it has been suggested that VDR genetic variants influence individual susceptibility to prostate cancer. Also, analyses of single nucleotide polymorphisms (SNPs) in VDR revealed ethnicity-associated polymorphisms. The aim of this study was to identify VDR SNPs in African American men with and without prostate cancer. Materials and Methods: The entire VDR gene was screened for germline mutations in a case-control study by denaturing high performance liquid chromatography and DNA sequencing. Logistic regression was used to estimate the association of SNPs, age, family history, and Gleason score with prostate cancer risk. Results: Six SNPs in the non-coding regions, and one SNP in the coding region, were detected. SNP 1 (c.278-69G>A) and SNP 4 (c.907+75C>T) have not been previously reported. SNP 4 had a significant protective effect (β=–0.6, p<0.05); whereas, SNP 7 (rs7975232) showed an increase association with prostate cancer risk and high Gleason score (β=0.32, p<0.05). SNP 4, SNP 7 and age were better predictors of prostate cancer risk than family history with a high degree of sensitivity (74.7%) and specificity (92.4%). Conclusion: SNP 4 and SNP 7 could be promising markers for prediction of reduced or increased prostate cancer risk, respectively.
Keywords: Prostate cancer, vitamin D, vitamin D receptor gene, African American men, single nucleotide polymorphism
Vitamin D (VD) has been reported as an important candidate implicated in the risk of prostate cancer (1-5). Although the exact mechanism of action is uncertain, various investigators have shown that serum 1,25-Dihydroxyvitamin D [1,25(OH)2 VD] levels can affect tumor cell proliferation and differentiation (6-12). Normal and malignant prostate cells contain VD receptors (VDRs) facilitate the anti-proliferative action of 1,25(OH)2 VD (13,14).
The environmental and physiological factors which mediate the source of cutaneous VD are levels of ultraviolet radiation (UVR) exposure, skin-color, and genes involved in VD synthesis and metabolism (15-18). In our previous work, we described a marginally significant association between skin tanning and risk of prostate cancer. Whereas, no association between prostate cancer development and total UVR exposure or low VD status was noted (19). Analysis from our earlier study on cumulative UVR exposure per year and adult sunbathing scores derived from a validated questionnaire revealed a significant difference in cumulative sun exposure between prostate cancer cases and controls (20). Furthermore, outdoor and recreation UVR exposures were significantly higher in controls when compared to cases. The conditional logistic regression analysis indicated that there was no association between total UVR exposure and risk of prostate cancer after adjusting for age. Outdoor UVR exposure, however, was related with decreased prostate cancer risk. Additionally, we reported a propensity for reduced prostate cancer risk among men with early-life high sun exposure during childhood ages 0-5 years and 6-11 years. This inverse association between risk of prostate cancer and high early-life sun exposure intensity was also observed among young men at ages 12-17 years although not statistically significant. These results show that UVR exposure in early life may decrease prostate cancer risk. In addition to UVR exposure and skin-color, many publications reported that sequence variations within certain genes effect VD synthesis, action, and metabolism. These genes include 1-alpha-hydroxylase, VD binding protein (VDBP) and VDR which are highly polymorphic among different human populations (21-24). Previously, we reported an association of VDR single nucleotide polymorphisms with prostate specific antigen (PSA) level, Gleason score, and prostate cancer risk in African-American men (21). There are various genetic studies that have investigated the relationship between VDR polymorphisms and prostate cancer risk (23,25-33), many of which have suggested statistically significant associations (34-37), weaker associations (38,39), and no associations (40-43) between common VDR variants and prostate cancer. However, those studies lack a consensus on how significantly, if at all, the VDR gene variants contribute to prostate cancer (44). Little is known about the association of VDR variants in high-risk populations, including African-American men with and without a family history of prostate cancer.
The present study was designed to evaluate the possibility that VDR genetic variants may contribute to prostate cancer risk. Therefore, the entire coding region and flanking introns of VDR were screened for germline mutations in a case-control study by denaturing high-performance liquid chromatography (DHPLC) followed by DNA sequencing.
Materials and Methods
Study population. Ninety-one African American men with histologically diagnosed adenocarcinoma of the prostate and 92 ethnicity matched (African American) controls were recruited through the ongoing free prostate cancer screening program at Howard University Cancer Center and from Howard University Hospital between 2005 and 2008. The Howard University Institutional Review Board (IRB-02-MED-42) approved the study protocol, and an informed consent was obtained from the study patients. Demographics and medical history details were previously described (19,20).
Polymerase chain reaction (PCR). QIAmp DNA Blood Maxi Kit (Qiagen Inc., Valencia, CA, USA) was used to extract genomic DNA according to manufacturer’s instructions. List of primers and PCR conditions were followed as previously described (45).
DHPLC. DHPLC instrument (WAVE® DNA Fragment Analysis System, Transgenomics, Omaha, NE, USA) equipped with a DNASep column (Transgenomic Inc., San Jose, CA, USA) was used to detect mutations and SNPs in VDR gene as previously described (45).
DNA sequencing and SNPs identification. Genomic DNA of samples demonstrating two or more heteroduplex peaks was amplified using GeneAmp 9700 thermal cycler as previously described (45) and purified by Qiagen column (QIAquick PCR purification Kit 50; Qiagen, Inc., Valencia, CA, USA). PCR was performed using 8 μl of Terminator ready reaction mix (Applied Biosystems, Foster City, CA, USA ) in a 20 μl reaction volume containing 100 ng of PCR product as a template and 3.2 pmole PCR primers under the following conditions: denaturation at 94˚C for 4 min, 25 cycles of 30 s at 94˚C, 30 s at 50˚C, and 4 min at 60˚C, and a final extension step for 7 min at 60˚C. The PCR product was purified using Centriflex gel filtration cartridge (Edge Biosystems Inc., Gaithersburg, MD, USA); and the DNA pellet was washed with 70% ethanol, and suspended in 25 μl of Template Suppression Reagent (PE; P/N 401674), and denatured at 95˚C for 2 min. Sequencing was performed using ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA, USA), and fluorescent labelled Big-dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) (46). Samples were also sequenced commercially by ACGT Incorporation (Wheeling, IL, USA) for confirmation. The generated data was analysed using Sequencher Version 4.8 software (Gene Codes Corporation), and SNPs were identified using International Hapmap project (http//: www.hapmap.org; https://www.ncbi.nlm.nih.gov/projects/SNP/). Nomenclature for the identified SNPs was assigned according to JT den Dunnen et al. (47), and the Reference SNP accession ID (rs ID) assigned for all previously reported SNPs using BLAST SNP (http://www.ncbi.nlm.nih.gov/SNP/snp_blastByOrg.cgi).
Statistical analyses. Cohen’s d test was used to analyse the age distribution between prostate cancer cases and controls. Logistic regression analysis was used to assess the protective and high-risk SNPs on prostate cancer using prostate cancer as the outcome in relation to each individual SNP, and the effect of each individual SNP on prostate cancer while adjusting for the other SNPs. Logistic regression with Backward substitution was employed to determine the dominant SNPs that are associated with prostate cancer.
We further investigated the association of the best predictor SNPs, 4 and 7, with prostate cancer as an outcome and adjusting for the other risk factors (e.g., age, family history). Particularly, 4 models of risk factors were assessed: Model 1 [SNP4, SNP7 and family history], Model 2 [SNP4, SNP7 and age], Model 3 [SNP4, SNP7, family history and age], and Model 4 [age and family history (as a substitute for SNPs)].
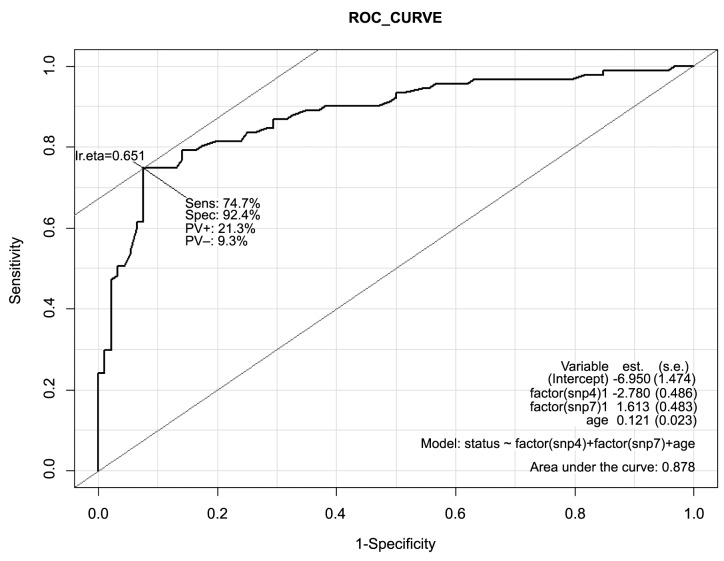
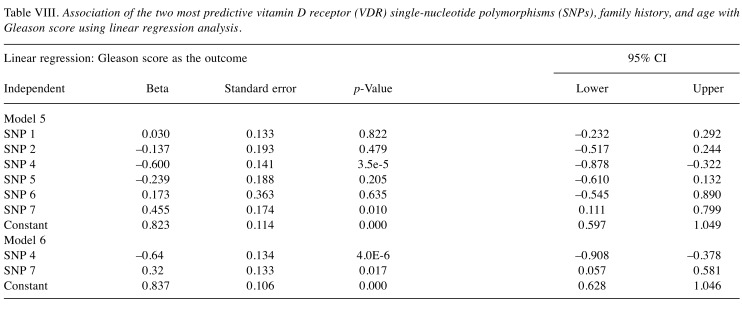
The association of the SNPs, age, and family history with prostate cancer risk was also demonstrated using the area under the receiver-operating-characteristic curve (ROC; AUC) as a measure of prediction performance (Figure 1). The ROC was also used to determine the best model among the 4 models for predicting the risk of prostate cancer.
Figure 1. Area under the curve (AUC) for the receiver-operatingcharacteristic curve analysis for sensitivity and specificity of the models: (a) Prostate cancer (PCa) versus SNP4, SNP7, age; (b) PCa versus age, family history; (c) PCa versus SNP4, SNP7, family history, age; (d) PCa versus SNP4, SNP7, family history.
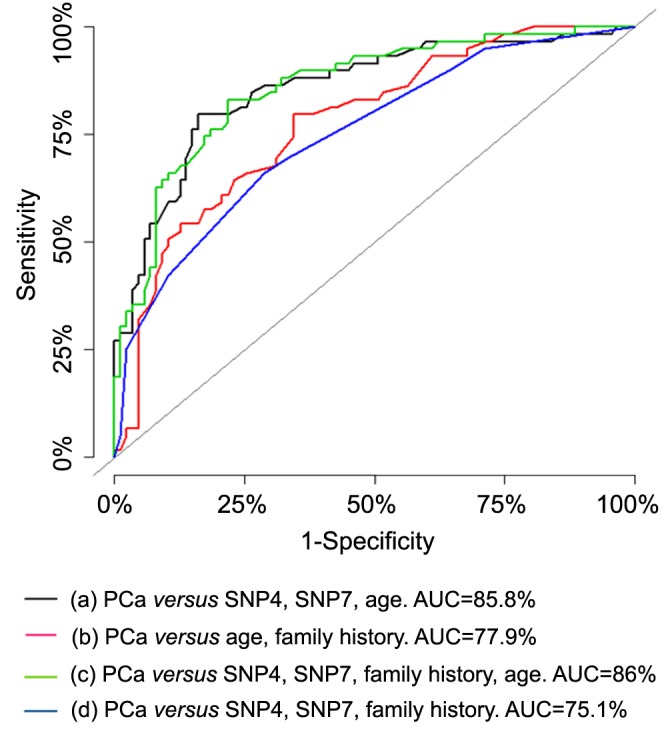
Each SNP was stratified by family history of prostate cancer and Gleason score as one of the composite variables that reflect prostate cancer aggressiveness. Gleason scores less than 7 and higher than 7 were considered as less and more advanced, respectively. Linear regression analysis was used to assess the association of VDR SNPs with Gleason score as the outcome using Model 5 (SNP 1, SNP 2, SNP 4, SNP 5, SNP 6, SNP 7, and Gleason score) and Model 6 (SNP 4, SNP 7, and Gleason score). All p-values less than 0.05 were considered statistically significant.
Results
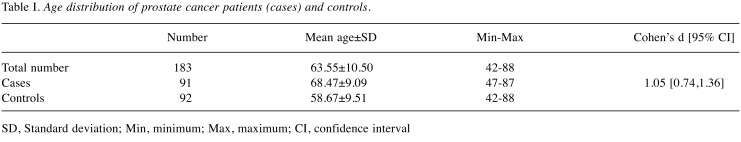
Ninety-one African American men with prostate cancer and 92 control subjects were analyzed to identify SNPs in VDR gene. The mean age of prostate cancer and control subjects was 68.47 and 58.67 years; respectively. Cohen’s d test of 1.05 indicated large effect size for average age difference between prostate cancer and control individuals (Table I).
Table I. Age distribution of prostate cancer patients (cases) and controls.
SD, Standard deviation; Min, minimum; Max, maximum; CI, confidence interval
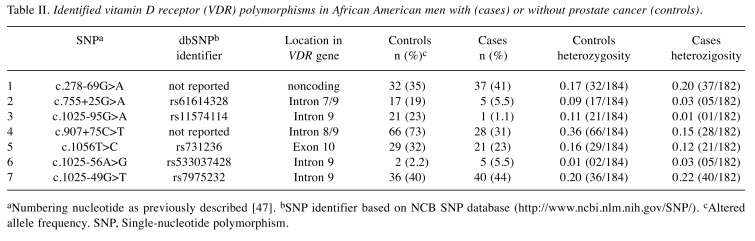
Six distinct polymorphisms [SNP 1 (c.278-69G>A), SNP 2 (rs61614328), SNP 3 (rs11574114), SNP 4 (c.907+75 C>T), SNP 6 (rs533037428), and SNP 7 (rs7975232, ApaI)] in non-coding regions; and one distinct polymorphism in the coding region (SNP 5: rs731236, TaqI) have been detected in VDR (Table II). None of the SNPs were located in the conserved regions or splicing sites, or in the VDR promoter region. Only one nonsense polymorphism SNP 5 (a synonymous Taq1 RFLP) in exon 9 was detected in the 3’ coding region. Two of the detected polymorphisms, SNP 1 and SNP 4 have not been previously reported in the Entrez database SNP (dpSNP) and may be unique to African Americans. In our study, the prevalence of the variant alleles within SNP 1, SNP 2, SNP 3, SNP 4, SNP 5, SNP 6 and SNP 7 among cases was 35%, 19%, 23%, 73%, 32%, 2.2%, and 40%, respectively; and 41%, 5.5%, 1.1%, 31%, 23%, 5.5%, and 44% among controls, respectively (Table II).
Table II. Identified vitamin D receptor (VDR) polymorphisms in African American men with (cases) or without prostate cancer (controls).
aNumbering nucleotide as previously described [47]. bSNP identifier based on NCB SNP database (http://www.ncbi.nlm.nih.gov/SNP/). cAltered allele frequency. SNP, Single-nucleotide polymorphism.
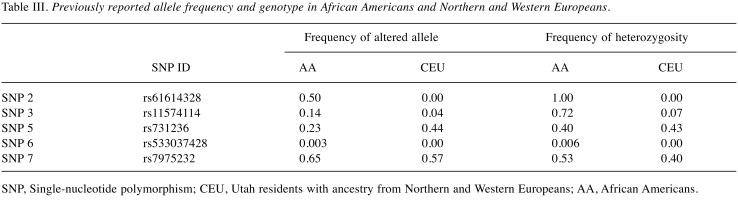
Genotype and allele frequencies of SNP 2, SNP 3, SNP 5, SNP 6 and SNP 7 were previously reported in Hapmap database (http://hapmap.ncbi.nlm.nih.gov/index.html.en) and were different in African Americans than in Northern and Western Europeans (Table III). More specifically, frequencies of the altered alleles for SNP 2, SNP 3, SNP 5, SNP 6 and SNP 7 (0.50, 0.14, 0.23, 0.003 and 0.65; respectively) in African Americans were lower than in Europeans (0.0, 0.04, 0.44, 0.00 and 0.57; respectively).
Table III. Previously reported allele frequency and genotype in African Americans and Northern and Western Europeans.
SNP, Single-nucleotide polymorphism; CEU, Utah residents with ancestry from Northern and Western Europeans; AA, African Americans.
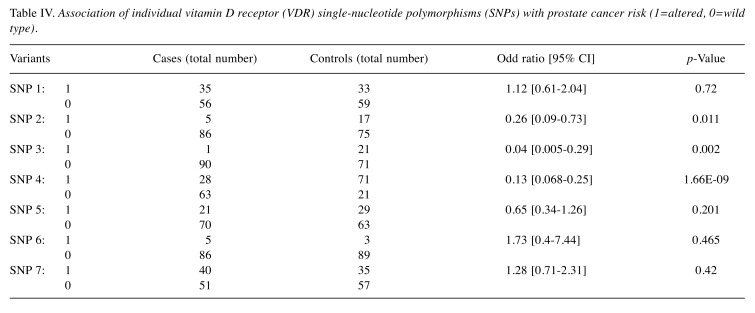
A significant association of prostate cancer with SNP 2 (OR=0.26, 95% CI=0.09-0.73; p=0.011), SNP 3 (OR=0.04, 95% CI=0.005-0.29; p=0.002), and SNP 4 (OR=0.13, 95% CI=0.068-0.25; p<0.005); and a non-significant association of prostate cancer with SNP 1, SNP 5, SNP 6, and SNP 7 were found (p>0.005) (Table IV). SNP 1, SNP 6 and SNP 7 were associated with an increasing risk of prostate cancer (OR=1.12, OR=1.73, OR=1.28; respectively), although the association was not statistically significant (p>0.05). A statistically significant protective effect from prostate cancer risk was found for SNP 2 (OR=0.26, 95% CI=0.09-0.73; p=0.011), SNP 3 (OR=0.04, 95% CI=0.005-0.29; p=0.002), and SNP 4 (OR=0.13, 95% CI=0.068-0.25; p=0.000) (Table IV). SNP 3 was detected in only one prostate cancer case; therefore, it was excluded from further analysis.
Table IV. Association of individual vitamin D receptor (VDR) single-nucleotide polymorphisms (SNPs) with prostate cancer risk (1=altered, 0=wild type).
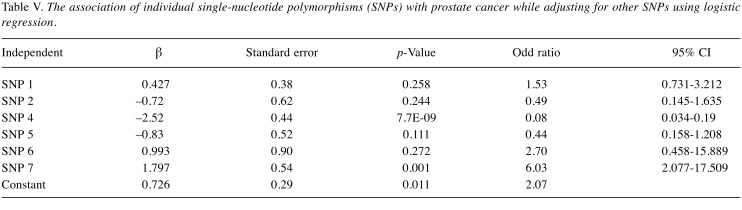
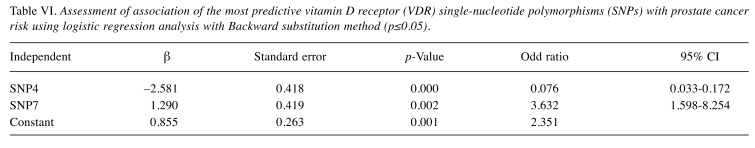
When we used logistic regression analysis to study the effect of individual VDR SNPs on prostate cancer risk, we found that SNP 4 had a highly significant protective effect by reducing the risk of prostate cancer by 92% (OR=0.08, 95% CI=0.034-0.19; β=–2.52, p<0.05), while SNP 7 had a significant direct association by increasing the risk of prostate cancer by 6-fold (OR=6.03, 95% CI=2.077-17.509; β=1.80, p=0.001) when controlling for SNPs 1, 2, 5, and 6. Whereas, no significant associations were found between prostate cancer risk and SNP 1, SNP 5, and SNP 6 with (p>0.05) (Table V). When the 6 SNPs (SNP 1, SNP 2, SNP 3, SNP 4, SNP 5, SNP 6) were further analyzed using logistic regression with Backward substitution method, only SNPs 4 and 7 were the dominant SNPs associated with prostate cancer; and thus, the rest of the SNPs were dropped out from any further analysis. SNP 4 reduced the risk of prostate cancer by 92.4% (OR=0.076, 95% CI=0.033-0.172; β=-2.581, p=0.000) when adjusting for SNP 7. Moreover, SNP 7 increased the risk by 3.6-fold (OR=3.632, 95% CI=1.598-8.254; β=1.290, p=0.002) when adjusting for SNP 4 (Table VI).
Table V. The association of individual single-nucleotide polymorphisms (SNPs) with prostate cancer while adjusting for other SNPs using logistic regression.
Table VI. Assessment of association of the most predictive vitamin D receptor (VDR) single-nucleotide polymorphisms (SNPs) with prostate cancer risk using logistic regression analysis with Backward substitution method (p≤0.05).
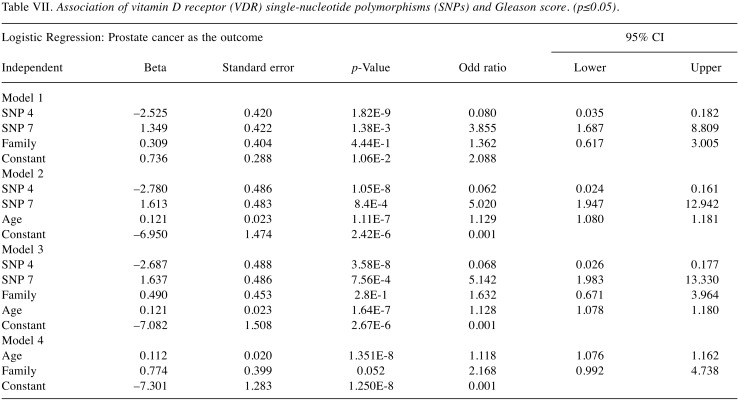
Furthermore, logistic regression analysis was used to examine whether the effect of the best predicted SNPs 4 and 7 is modulated by another factor (such as age, family history, Gleason score) (Table VII). In particular, we assessed “SNP 4, SNP 7, family history”; “SNP 4, SNP 7, age”; “SNP 4, SNP 7, family history, age”; “Age, family history”; in 4 models (Model, 1, Model 2, Model 3, and Model 4; respectively). Models 1, 2, and 3 showed that SNP 4 and SNP 7 maintained their protective effect and risk association with prostate cancer; respectively. Model 4 showed a marginal significant association of family history with prostate cancer risk when adjusting for age. As expected, an increased significant association of age and family history with prostate cancer risk was found (p<0.05).
Table VII. Association of vitamin D receptor (VDR) single-nucleotide polymorphisms (SNPs) and Gleason score. (p≤0.05).
The statistically significant association of the SNPs, age, and family history with prostate cancer risk was also demonstrated using the AUC as a measure of prediction performance. The overall accuracy expressed by the AUC value increased from 77.9% for age and family history (Model 4) to 86% when the SNPs 4 and 7 were included in the prediction model (Model 3) for prostate cancer risk. Similarly, an increase from 75.1% for SNP 4, SNP 7 and family history (Model 1) to 86% was observed when the age was included (Model 3).
The ROC curves and AUC values (Figure 1 and Figure 2) showed that SNP 4, SNP 7, and age were better predictors (AUC=85.8%) than family history in predicting prostate cancer risk with a high degree of sensitivity (74.7%), and specificity (92.4%) for the optimal cut-off point for Model 2. The combination of SNP 4, SNP 7, family history and age had a higher AUC value (86%) than the age and family history (AUC=77.9%), or SNP 4, SNP 7 and family history (AUC=75.1%).
Figure 2. Validation of the best predictive model for prostate cancer using the area under the receiver-operating-characteristic curve as a measure of prediction performance.
The association between VDR SNPs and Gleason score was also determined using linear regression in two Models 5 and 6 (Table VIII). Gleason score as the response variable and SNPs 1, 2, 3, 4, 5, 6 and 7 as the predictors of prostate cancer. Model 5, the effect of SNP 7 after adjusting for all other SNPs (SNP 1, SNP 2, SNP 4, SNP 5, SNP 6, SNP 7, and Gleason score); and Model 6, the effect of each SNP at the level of the other SNPs (SNP 4, SNP 7, and Gleason score). The significant results of Models 5 and 6 indicate that SNP 4 (β=–0.64, p<0.05) had a significant protective effect against prostate cancer and was associated with a low Gleason score. On the other hand, SNP 7 showed a statistically significant association with increased prostate cancer risk and high Gleason score (β=0.32, p<0.05).
Table VIII. Association of the two most predictive vitamin D receptor (VDR) single-nucleotide polymorphisms (SNPs), family history, and age with Gleason score using linear regression analysis.
Discussion
Common genetic variants in VDR have been related to prostate cancer risk in several studies, but the overall results remain contradictory. Two initial studies showed a 3 to 4-fold increase risk of prostate cancer associated with VDR polymorphisms in the 3’ end of the gene (23,25). However, a meta-analysis involving 17 studies that assessed the TaqI (rs731236), BsmI, and poly-A repeat polymorphisms, as well as the FokI polymorphism in exon 2, concluded that none of these variants was likely to be a major determinant of prostate cancer risk (48). Further, it has been suggested that VDR SNPs may be more related to advanced disease (23,26,49-51). Earlier investigations including cases with localized and advanced disease revealed a reduced risk of prostate cancer related with the TaqI t allele or an allele in linkage disequilibrium (LD) with TaqI t (52-58). Among Japanese populations, ApaI (rs7975232) was not significantly associated with either familial prostate cancer (59) or sporadic prostate cancer and benign prostatic hyperplasia (60). Prostate cancer has been inversely associated with the TaqI tt genotype in White and Black men in North Carolina (25). Similarly, it was reported by Ma et al. (49) that reduced prostate cancer risk is associated with the TaqI tt genotype; however, only among men with low serum 1,25 (OH)2 VD levels. In a recent meta-analysis, it was reported that VDR TaqI polymorphism may be associated with prostate cancer risk in the Asian population (37), and particularly in the Japanese population (24). Although, VDR SNPs have been evaluated as markers of prostate cancer risk; their impact remains uncertain especially in African-Americans. In the current study, we identified and evaluated seven SNPs in the VDR gene which had a significant association or no association with the risk of prostate cancer. Two of the SNPs, 1 and 4, were not reported in the SNPs database. Thus, we postulate that these two SNPs are novel. Moreover, the present study is the first to show SNP 4 as a determinant of prostate cancer risk.
Two of the detected SNPs, SNP 5 (Taql) and SNP 7 (Apal), have been previously reported (48,61). In our previous study (21), SNP 5 was found to be significantly associated with prostate cancer risk in African American men after adjusting for age using the codominant, dominant, and the log-additive models; whereas, SNP 7 was significantly associated with an increased prostate cancer risk using the recessive model. There was no evidence that these associations were modified by family history of prostate cancer, which is inconsistent with the present study. Moreover, we have previously investigated whether DHPLC elution patterns of VDR PCR products can serve as indicators of susceptibility to prostate cancer risk (45). A similar elution pattern of exons 1, 6, 7 and 9 along with the higher prevalence of heteroduplex DNA in prostate cancer cases than in controls was observed. Moreover, exons 4 and 8 had highly significant protective effects against prostate cancer. On the other hand, exons 5, 7, and 9 were positively correlated with prostate cancer risk, thus they could be better predictors of susceptibility to prostate cancer. This data is also inconsistent with our present study in which SNP 4 (Intron 8/9) showed a protective effect for prostate cancer development, while SNP 7 (Intron 9) was associated with increased risk of prostate cancer.
Both Gleason score and TNM stage are accepted as valid and reliable measures for assessing the aggressiveness of prostate cancer (62). However, Gleason score was found to be a more sensitive measure of aggressive prostate cancer than TNM stage (59,61,63), and the associations of VDR variants with Gleason grade were identified (64). In the present study, we found that carriers of the variant SNP 4 had a significant protective effect and low Gleason score of prostate cancer (β=–0.6, p<0.05); whereas, carriers of SNP 7 had higher risk and poorly differentiated prostate cancer (β=0.32, p<0.05). Further large studies that replicate these results are warranted to confirm the association of SNPs 4 and 7 with prostate cancer.
It is speculated that a substitute for long-term serum VD levels is the level of UVR exposure. Prostate cancer and control patients classified into low- and high- exposure group based on cumulative UVR exposure may mask any effect of VDR variants in men with different VD levels (23). Several publications reported that the pathogenesis of prostate cancer in men with low levels of UVR exposure is different from that in men with higher levels (8,65-67). However, if the functional differences between the VDR genotypes are small relative to the consequences of low vitamin levels, the effect of the polymorphisms may be concealed. In contrast, men with UVR exposure above the median would be expected to synthesize adequate amounts of VD. The functional consequences of the polymorphisms may be sufficiently great in the presence of an adequate concentration of the VD to influence prostate cancer risk. Thus, the VD pathway may have an etiologic role in the development of prostate cancer.
Conclusion
In African American men, SNP 4, SNP 7, and age were better predictors of prostate cancer risk than family history. Moreover, our results suggested that SNP 4 and 7 could be promising predicting markers of reduced or increased prostate cancer risk, respectively. Further, larger studies are warranted to confirm the association of these markers with prostate cancer.
Authors ’ Contributions
MD and DB conducted the experiments. VA performed the statistical analyses. TN collected the clinical data and contributed to data analyses. OOK contributed to the data analyses and manuscript writing. RLC and YK were responsible for the experimental design and contributed to the data analyses and manuscript writing
Conflicts of Interest
The Authors have no personal or financial conflicts of interest.
Acknowledgements
This work was supported by US Army Medical Research and Materiel Command (USAMRMC) [DAMD17-03-1-0069].
References
- 1.Studzinski GP, Moore DC. Sunlight - Can it prevent as well as cause cancer. Cancer Res. 1995;55(18):4014–4022. PMID: 7664274. [PubMed] [Google Scholar]
- 2.Selby PL, Mawer EB. Sunlight and health. Exposure to sunlight may reduce cancer risk. BMJ. 1999;319:1067–1068. PMID: 10576851. [PubMed] [Google Scholar]
- 3.McCarty MF. Parathyroid hormone may be a cancer promoter - an explanation for the decrease in cancer risk associated with ultraviolet light, calcium, and vitamin D. Med Hypotheses. 2000;54(3):475–482. doi: 10.1054/mehy.1999.0880. PMID: 10783492. DOI: 10.1054/mehy.1999. 0880. [DOI] [PubMed] [Google Scholar]
- 4.Travis RC, Perez-Cornago A, Appleby PN, Albanes D, Joshu CE, Lutsey PL, Mondul AM, Platz EA, Weinstein SJ, Layne TM, Helzlsouer KJ, Visvanathan K, Palli D, Peeters PH, Bueno-de-Mesquita B, Trichopoulou A, Gunter MJ, Tsilidis KK, Sánchez MJ, Olsen A, Brenner H, Schöttker B, Perna L, Holleczek B, Knekt P, Rissanen H, Yeap BB, Flicker L, Almeida OP, Wong YYE, Chan JM, Giovannucci EL, Stampfer MJ, Ursin G, Gislefoss RE, Bjørge T, Meyer HE, Blomhoff R, Tsugane S, Sawada N, English DR, Eyles DW, Heath AK, Williamson EJ, Manjer J, Malm J, Almquist M, Marchand LL, Haiman CA, Wilkens LR, Schenk JM, Tangen CM, Black A, Cook MB, Huang WY, Ziegler RG, Martin RM, Hamdy FC, Donovan JL, Neal DE, Touvier M, Hercberg S, Galan P, Deschasaux M, Key TJ, Allen NE. A collaborative analysis of individual participant data from 19 prospective studies assesses circulating vitamin D and prostate cancer risk. Cancer Res. 2018;79(1):274–285. doi: 10.1158/0008-5472.CAN-18-2318. PMID: 30425058. DOI: 10.1158/0008-5472.CAN-18-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan C, Shui IM, Wilson KM, Stampfer MJ, Mucci LA, Giovannucci EL. Circulating 25-hydroxyvitamin D, vitamin D binding protein and risk of advanced and lethal prostate cancer. Int J Cancer. 2018;144(10):240–2407. doi: 10.1002/ijc.31966. PMID: 30411792. DOI: 10.1002/ijc.31966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haussler MR, Whitfield GK, Haussler CA, Hsieh JC, Thompson PD, Selznick SH, Dominguez CE, Jurutka PW. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13(3):325–349. doi: 10.1359/jbmr.1998.13.3.325. PMID: 9525333. DOI: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 7.Holick MF. Sunlight and Vitamin D: The Bone and Cancer Connections. Radiat Prot Dosimetry. 2000;91(1-3):65–71. DOI: 10.1093/oxfordjournals.rpd.a006404. [Google Scholar]
- 8.John EM, Koo J, Schwartz GG. Sun exposure and prostate cancer risk: evidence for a protective effect of early-life exposure. Cancer Epidemiol Biomarkers Prev. 2007;16(6):1283–1286. doi: 10.1158/1055-9965.EPI-06-1053. PMID: 17548698. DOI: 10.1158/1055-9965.EPI-06-1053. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz GG. Vitamin D and the epidemiology of prostate cancer. Semin Dial. 2005;18(4):276–289. doi: 10.1111/j.1525-139X.2005.18403.x. PMID: 16076349. DOI: 10.1111/j.1525-139X.2005.18403.x. [DOI] [PubMed] [Google Scholar]
- 10.Lambert JR, Young CD, Persons KS, Ray R. Mechanistic and pharmacodynamic studies of a 25-hydroxyvitamin D3 derivative in prostate cancer cells. Biochem Biophys Res Commun. 2007;361(1):189–195. doi: 10.1016/j.bbrc.2007.07.012. PMID: 17658477. DOI: 10.1016/j.bbrc.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Chen TC, Schwartz GG, Burnstein KL, Lokeshwar BL, Holick MF. The in vitro evaluation of 25-hydroxyvitamin D3 and 19-nor-1{{alpha}},25-dihydroxyvitamin D2 as therapeutic agents for prostate cancer. Clin Cancer Res. 2000;6(3):901–908. PMID: 10741714. [PubMed] [Google Scholar]
- 12.Swamy N, Chen TC, Peleg S, Dhawan P, Christakos S, Stewart LV, Weigel NL, Mehta RG, Holick MF, Ray R. Inhibition of proliferation and induction of apoptosis by 25-hydroxyvitamin D3-3beta-(2)-bromoacetate, a nontoxic and vitamin D receptor-alkylating analog of 25-hydroxyvitamin D3 in prostate cancer cells. Clin Cancer Res. 2004;10(23):8018–8027. doi: 10.1158/1078-0432.CCR-04-0881. PMID: 15585637. DOI: 10.1158/1078-0432.CCR-04-0881. [DOI] [PubMed] [Google Scholar]
- 13.Ahonen MH, Tenkanen L, Teppo L, Hakama M, Tuohimaa P. Prostate cancer risk and prediagnostic serum 25-hydroxyvitamin D levels (Finland) Cancer Causes Control. 2000;11(9):847–852. doi: 10.1023/a:1008923802001. PMID: 11075874. [DOI] [PubMed] [Google Scholar]
- 14.Miller GJ, Stapleton GE, Ferrara JA, Lucia MS, Pfister S, Hedlund TE, Upadhya P. The human prostatic carcinoma cell line LNCaP expresses biologically active, specific receptors for 1 alpha,25-dihydroxyvitamin D3. Cancer Res. 1992;52(3):515–520. PMID: 1370648. [PubMed] [Google Scholar]
- 15.Carnevale V, Modoni S, Pileri M, Di Giorgio A, Chiodini I, Minisola S, Vieth R, Scillitani A. Longitudinal evaluation of vitamin D status in healthy subjects from southern Italy: seasonal and gender differences. Osteoporos Int. 2001;12(12):1026–1030. doi: 10.1007/s001980170012. PMID: 11846328. DOI: 10.1007/s001980170012. [DOI] [PubMed] [Google Scholar]
- 16.Shaw NJ, Pal BR. Vitamin D deficiency in UK Asian families: activating a new concern. Arch Dis Child. 2002;86(3):147–149. doi: 10.1136/adc.86.3.147. PMID: 11861227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka LY, Wortsman J, Haddad JG, Kolm P, Hollis BW. Racial pigmentation and the cutaneous synthesis of vitamin D. Arch Dermatol. 1991;127(4):536–538. PMID: 1848745. [PubMed] [Google Scholar]
- 18.Rasmussen LB, Hansen GL, Hansen E, Koch B, Mosekilde L, Molgaard C, Sorensen OH, Ovesen L. Vitamin D: should the supply in the Danish population be increased. Int J Food Sci Nutr. 2000;51(3):209–215. doi: 10.1080/09637480050029719. PMID: 10945117. [DOI] [PubMed] [Google Scholar]
- 19.Beyene D, Daremipouran M, Apprey V, Williams R, Ricks-Santi L, Kassim OO, Naab TJ, Kanaan YM, Copeland RL Jr. Use of tanning potential as a predictor for prostate cancer risk in African-American men. In Vivo. 2014;28(6):1181–1187. PMID: 25398820. [PubMed] [Google Scholar]
- 20.Kanaan YM, Beyene D, Daremipouran M, Mireku-Boateng A, Williams R, Jackson A, Bonney GE, Apprey V, Daniel MG, Wutoh AK, Rohan JP, Ricks-Santi L, Copeland RL Jr. Association of cumulative ultraviolet radiation exposure with prostate cancer risk in a case-control study of African-American men. The Open Prostate Cancer Journal. 2012;5:8–14. DOI: 10.2174/1876822901205010008. [Google Scholar]
- 21.Jingwi EY, Abbas M, Ricks-Santi L, Winchester D, Beyene D, Day A, Naab TJ, Kassim OO, Dunston GM, Copeland RL Jr, Kanaan YM. Vitamin D receptor genetic polymorphisms are associated with PSA level, Gleason score and prostate cancer risk in African-American men. Anticancer Res. 2015;35 (3):1549–1558. PMID: 25750310. [PMC free article] [PubMed] [Google Scholar]
- 22.Braun A, Bichlmaier R, Cleve H. Molecular analysis of the gene for the human vitamin-D-binding protein (group-specific component): allelic differences of the common genetic GC types. Hum Genet. 1992;89(4):401–406. doi: 10.1007/BF00194311. PMID: 1352271. [DOI] [PubMed] [Google Scholar]
- 23.Ingles SA, Ross RK, Yu MC, Irvine RA, La Pera G, Haile RW, Coetzee GA. Association of prostate cancer risk with genetic polymorphisms in vitamin D receptor and androgen receptor. J Natl Cancer Inst. 1997;89(2):166–170. doi: 10.1093/jnci/89.2.166. DOI: 10.1093/jnci/ 89.2.166. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Wei J, Zhang S, Lou Z, Wang X, Ren Y, Qi H, Xie Z, Chen Y, Chen F, Wu Q, Fan X, Huang S, Weng G. Association of VDR gene TaqI polymorphism with the susceptibility to prostate cancer in Asian population evaluated by an updated systematic meta-analysis. OncoTargets and Therapy. 2018;11:3267–3280. doi: 10.2147/OTT.S151002. PMID: 29910622. DOI: 10.2147/OTT.S151002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor JA, Hirvonen A, Watson M, Pittman G, Mohler JL, Bell DA. Association of prostate cancer with vitamin D receptor gene polymorphism. Cancer Res. 1996;56(18):4108–4110. PMID: 8797574. [PubMed] [Google Scholar]
- 26.Ingles SA, Coetzee GA, Ross RK, Henderson BE, Kolonel LN, Crocitto L, Wang W, Haile RW. Association of prostate cancer with vitamin D receptor haplotypes in African-Americans. Cancer Res. 1998;58(8)):1620–1623. http://cancerres.aacrjournals.org/content/56/18/4108 A. [PubMed] [Google Scholar]
- 27.Correa-Cerro L, Berthon P, Häussler J, Bochum S, Drelon E, Mangin P, Fournier G, Paiss T, Cussenot O, Vogel W. Vitamin D receptor polymorphisms as markers in prostate cancer. Hum Genet. 1999;105(3):281–287. doi: 10.1007/s004390051102. PMID: 10987658. [DOI] [PubMed] [Google Scholar]
- 28.Chokkalingam AP, McGlynn KA, Gao YT, Pollak M, Deng J, Sesterhenn IA, Mostofi FK, Fraumeni JF Jr., Hsing AW. Vitamin D receptor gene polymorphisms, insulin-like growth factors, and prostate cancer risk: a population-based case-control study in China. Cancer Res. 2001;61(11):4333–4336. PMID: 11389055. [PubMed] [Google Scholar]
- 29.Holick MF. The vitamin D epidemic and its health consequences. J Nutr. 2005;135(11):2739S–2748S. doi: 10.1093/jn/135.11.2739S. PMID: 16251641. DOI: 10.1093/jn/135.11.2739S. [DOI] [PubMed] [Google Scholar]
- 30.Mikhak B, Hunter DJ, Spiegelman D, Platz EA, Hollis BW, Giovannucci E. Vitamin D receptor (VDR) gene polymorphisms and haplotypes, interactions with plasma 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D, and prostate cancer risk. The Prostate. 2007;67(9):911–923. doi: 10.1002/pros.20570. PMID: 17440943. DOI: 10.1002/pros.20570. [DOI] [PubMed] [Google Scholar]
- 31.Patino-Garcia B, Arroyo C, Rangel-Villalobos H, Soto-Vega E, Velarde-Felix JS, Gabilondo F, Sandoval-Ramirez L, Figuera LE. Association between polymorphisms of the androgen and vitamin D receptor genes with prostate cancer risk in a Mexican population. Rev Invest Clin. 2007;59(1):25–31. PMID: 17569297. [PubMed] [Google Scholar]
- 32.Andersson P, Varenhorst E, Soderkvist P. Androgen receptor and vitamin D receptor gene polymorphisms and prostate cancer risk. Eur J Cancer. 2006;42(16):2833–2837. doi: 10.1016/j.ejca.2006.06.030. PMID: 17010601. DOI: 10.1016/j.ejca.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 33.Rukin NJ, Luscombe C, Moon S, Bodiwala D, Liu S, Saxby MF, Fryer AA, Alldersea J, Hoban PR, Strange RC. Prostate cancer susceptibility is mediated by interactions between exposure to ultraviolet radiation and polymorphisms in the 5’ haplotype block of the vitamin D receptor gene. Cancer Lett. 2007;247(2):328–335. doi: 10.1016/j.canlet.2006.05.012. PMID: 16815628. DOI: 10.1016/ j.canlet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Ekman P, Pan Y, Li C, Dich J. Environmental and genetic factors: a possible link with prostate cancer. Br J Urol. 1997;79 Suppl:234–241. doi: 10.1111/j.1464-410x.1997.tb16919.x. PMID: 9126068. [DOI] [PubMed] [Google Scholar]
- 35.Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, Walsh PC. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150(3):797–802. doi: 10.1016/s0022-5347(17)35617-3. PMID: 8345587. [DOI] [PubMed] [Google Scholar]
- 36.Braczkowski RS, Kwiatkowski R, Danikiewicz A, Gorczynska-Kosiorz S, Trautsolt W, Braczkowska B, Grzeszczak W. Vitamin D receptor gene polymorphisms and prostate cancer. J Biol Regul Homeost Agents. 2018;32(5):1245–1248. PMID: 30334420. [PubMed] [Google Scholar]
- 37.Kang S, Zhao Y, Wang L, Liu J, Chen X, Liu X, Shi Z, Gao W, Cao F. Vitamin D receptor Taq I polymorphism and the risk of prostate cancer: a meta-analysis. Oncotarget. 2018;9(6):7136–7147. doi: 10.18632/oncotarget.23606. PMID: 29467956. DOI: 10.18632/oncotarget.23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaram S, S K, Mazumdar S, Shukla A. Overlap syndrome between primary biliary cholangitis and primary sclerosing cholangitis. ACG Case Rep J. 2018;5:e54. doi: 10.14309/crj.2018.54. PMID: 30038926. DOI: 10.14309/crj.2018.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274(5291):1371–1374. doi: 10.1126/science.274.5291.1371. PMID: 8910276. [DOI] [PubMed] [Google Scholar]
- 40.Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, Wilkens E, Bujnovszky P, Bova GS, Walsh P, Isaacs W, Schleutker J, Matikainen M, Tammela T, Visakorpi T, Kallioniemi OP, Berry R, Schaid D, French A, McDonnell S, Schroeder J, Blute M, Thibodeau S, Grönberg H, Emanuelsson M, Damber JE, Bergh A, Jonsson BA, Smith J, Bailey-Wilson J, Carpten J, Stephan D, Gillanders E, Amundson I, Kainu T, Freas-Lutz D, Baffoe-Bonnie A, Van Aucken A, Sood R, Collins F, Brownstein M, Trent J. Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet. 1998;20(2):175–179. doi: 10.1038/2477. PMID: 9771711. DOI: 10.1038/2477. [DOI] [PubMed] [Google Scholar]
- 41.Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wohr G, Latil A, Millasseau P, Mellah I, Cohen N, Blanché H, Bellané-Chantelot C, Demenais F, Teillac P, Le Duc A, de Petriconi R, Hautmann R, Chumakov I, Bachner L, Maitland NJ, Lidereau R, Vogel W, Fournier G, Mangin P, Cussenot O. Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2-43. Am J Hum Genet. 1998;62(6):1416–1424. doi: 10.1086/301879. PMID: 9585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheteri MB, Stanford JL, Friedrichsen DM, Peters MA, Iwasaki L, Langlois MC, Feng Z, Ostrander EA. Vitamin D receptor gene polymorphisms and prostate cancer risk. Prostate. 2004;59(4):409–418. doi: 10.1002/pros.20001. PMID: 15065089. DOI: 10.1002/pros.20001. [DOI] [PubMed] [Google Scholar]
- 43.Kang S, Zhao Y, Wang L, Liu J, Chen X, Liu X, Shi Z, Gao W, Cao F. Lack of association between the risk of prostate cancer and vitamin D receptor Bsm I polymorphism: a meta-analysis of 27 published studies. Cancer management and research. 2018;10:2377–2387. doi: 10.2147/CMAR.S171305. PMID: 30122987. DOI: 10.2147/ CMAR.S171305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gsur A, Madersbacher S, Haidinger G, Schatzl G, Marberger M, Vutuc C, Micksche M. Vitamin D receptor gene polymorphism and prostate cancer risk. Prostate. 2002;51(1):30–34. doi: 10.1002/pros.10064. PMID: 11920955. [DOI] [PubMed] [Google Scholar]
- 45.Copeland RL, Beyene D, Apprey V, Daremipouran MR, Naab TJ, Kassim OO andKanaan YM. DHPLC elution patterns of VDR PCR products can predict prostate cancer susceptibility in african american men. Cancer genomics proteomics. 2017;14(6):461–467. doi: 10.21873/cgp.20056. PMID: 29109096. DOI: 10.21873/cgp.20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stolzenberg-Solomon RZ, Hayes RB, Horst RL, Anderson KE, Hollis BW, Silverman DT. Serum vitamin D and risk of pancreatic cancer in the prostate, lung, colorectal, and ovarian screening trial. Cancer Res. 2009;69(4):1439–1447. doi: 10.1158/0008-5472.CAN-08-2694. PMID: 19208842. DOI: 10.1158/0008-5472.CAN-08-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. PMID: 10612815. DOI: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4> 3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 48.Ntais C, Polycarpou A, Ioannidis JP. Vitamin D receptor gene polymorphisms and risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2003;12(12):1395–1402. PMID: 14693728. [PubMed] [Google Scholar]
- 49.Ma J, Stampfer MJ, Gann PH, Hough HL, Giovannucci E, Kelsey KT, Hennekens CH, Hunter DJ. Vitamin D receptor polymorphisms, circulating vitamin D metabolites, and risk of prostate cancer in United States physicians. Cancer Epidemiol Biomarkers Prev. 1998;7(5):385–390. PMID: 9610787. [PubMed] [Google Scholar]
- 50.Hamasaki T, Inatomi H, Katoh T, Ikuyama T andMatsumoto T. Significance of vitamin D receptor gene polymorphism for risk and disease severity of prostate cancer and benign prostatic hyperplasia in Japanese. Urol Int. 2002;68(4):226–231. doi: 10.1159/000058440. PMID: 12053022. DOI: 10.1159/000058440. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe M, Fukutome K, Murata M, Uemura H, Kubota Y, Kawamura J, Yatani R. Significance of vitamin D receptor gene polymorphism for prostate cancer risk in Japanese. Anticancer Res. 1999;19(5C):4511–4514. PMID: 10650802. [PubMed] [Google Scholar]
- 52.Ma J, Stampfer MJ, Gann PH, Hough HL, Giovannucci E, Kelsey KT, Hennekens CH, Hunter DJ. Vitamin D receptor polymorphisms, circulating vitamin D metabolites, and risk of prostate cancer in United States physicians. Cancer Epidemiol Biomarkers Prev. 1998;7(5):385–390. PMID: 9610787. [PubMed] [Google Scholar]
- 53.Ingles SA, Ross RK, Yu MC, Irvine RA, La Pera G, Haile RW, Coetzee GA. Association of prostate cancer risk with genetic polymorphisms in vitamin D receptor and androgen receptor. J Natl Cancer Inst. 1997;89(2):166–170. doi: 10.1093/jnci/89.2.166. PMID: 8998186. [DOI] [PubMed] [Google Scholar]
- 54.Ingles SA, Coetzee GA, Ross RK, Henderson BE, Kolonel LN, Crocitto L, Wang W, Haile RW. Association of prostate cancer with vitamin D receptor haplotypes in African-Americans. Cancer Res. 1998;58(8):1620–1623. PMID: 9563471. [PubMed] [Google Scholar]
- 55.Hamasaki T, Inatomi H, Katoh T, Ikuyama T, Matsumoto T. Significance of vitamin D receptor gene polymorphism for risk and disease severity of prostate cancer and benign prostatic hyperplasia in Japanese. Urol Int. 2002;68(4):226–231. doi: 10.1159/000058440. PMID: 12053022. DOI: 10.1159/000058440. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe M, Fukutome K, Murata M, Uemura H, Kubota Y, Kawamura J, Yatani R. Significance of vitamin D receptor gene polymorphism for prostate cancer risk in Japanese. Anticancer Res. 1999;19(5C):4511–4514. PMID: 10650802. [PubMed] [Google Scholar]
- 57.Yamamoto H, Miyamoto K, Li B, Taketani Y, Kitano M, Inoue Y, Morita K, Pike JW, Takeda E. The caudal-related homeodomain protein Cdx-2 regulates vitamin D receptor gene expression in the small intestine. J Bone Miner Res. 1999;14(2):240–247. doi: 10.1359/jbmr.1999.14.2.240. PMID: 9933478. DOI: 10.1359/jbmr.1999.14.2.240. [DOI] [PubMed] [Google Scholar]
- 58.Lock-Andersen J, Knudstorp ND, Wulf HC. Facultative skin pigmentation in Caucasians: an objective biological indicator of lifetime exposure to ultraviolet radiation. Br J Dermatol. 1998;138(5):826–832. doi: 10.1046/j.1365-2133.1998.02220.x. PMID: 9666829. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki K, Matsui H, Ohtake N, Nakata S, Takei T, Koike H, Nakazato H, Okugi H, Hasumi M, Fukabori Y, Kurokawa K, Yamanaka H. Vitamin D receptor gene polymorphism in familial prostate cancer in a Japanese population. Int J Urol. 2003;10(5):261–266. doi: 10.1046/j.1442-2042.2003.00617.x. PMID: 12694466. [DOI] [PubMed] [Google Scholar]
- 60.Habuchi T, Suzuki T, Sasaki R, Wang L, Sato K, Satoh S, Akao T, Tsuchiya N, Shimoda N, Wada Y, Koizumi A, Chihara J, Ogawa O, Kato T. Association of vitamin D receptor gene polymorphism with prostate cancer and benign prostatic hyperplasia in a Japanese population. Cancer Res. 2000;60(2):305–308. PMID: 10667581. [PubMed] [Google Scholar]
- 61.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1990–1999. doi: 10.1158/1055-9965.EPI-07-0487. PMID: 17932346. DOI: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 62.Pannek J, Partin AW. Prostate-specific antigen: what’s new in 1997. Oncology (Williston Park) 1997;11(9):1273–1278. discussion 1279-1282. [PubMed] [Google Scholar]
- 63.Mikhak B, Hunter DJ, Spiegelman D, Platz EA, Hollis BW, Giovannucci E. Vitamin D receptor (VDR) gene polymorphisms and haplotypes, interactions with plasma 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D, and prostate cancer risk. Prostate. 2007;67(9):911–923. doi: 10.1002/pros.20570. PMID: 17440943. DOI: 10.1002/pros. 20570. [DOI] [PubMed] [Google Scholar]
- 64.Nunes SBR, de Matos Oliveira F, Neves AF, Araujo GR, Marangoni K, Goulart LR, Araújo TG. Association of vitamin D receptor variants with clinical parameters in prostate cancer. Springerplus. 2016;5(1):364. doi: 10.1186/s40064-016-2009-8. PMID: 27066374. DOI: 10.1186/s40064-016-2009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bodiwala D, Luscombe CJ, French ME, Liu S, Saxby MF, Jones PW, Fryer AA, Strange RC. Polymorphisms in the vitamin D receptor gene, ultraviolet radiation, and susceptibility to prostate cancer. Environ Mol Mutagen. 2004;43(2):121–127. doi: 10.1002/em.20000. PMID: 14991752. DOI: 10.1002/em.20000. [DOI] [PubMed] [Google Scholar]
- 66.Luscombe CJ, Fryer AA, French ME, Liu S, Saxby MF, Jones PW, Strange RC. Exposure to ultraviolet radiation: association with susceptibility and age at presentation with prostate cancer. Lancet. 2001;358(9282):641–642. doi: 10.1016/S0140-6736(01)05788-9. PMID: 11530156. DOI: 10.1016/S0140-6736(01)05788-9. [DOI] [PubMed] [Google Scholar]
- 67.Bodiwala D, Luscombe CJ, Liu S, Saxby M, French M, Jones PW, Fryer AA, Strange RC. Prostate cancer risk and exposure to ultraviolet radiation: further support for the protective effect of sunlight. Cancer Lett. 2003;192(2):145–149. doi: 10.1016/s0304-3835(02)00710-3. PMID: 12668278. [DOI] [PubMed] [Google Scholar]