Abstract
Background/Aim: Radiation therapy (RT) represents a therapeutic option in breast cancer (BC). Even if a great number of BC patients receive RT, not all of them report benefits, due to radioresistance that gets activated through several factors, such as the hormone receptor status. Herein, we analyzed the gene expression profiles (GEP) induced by RT in triple-negative BC (TNBC) MDA-MB-231, to study signalling networks involved in radioresistance. Materials and Methods: GEP of MDA-MB-231 BC cells treated with a high dose of radiation, went through cDNA microarray analysis. In addition, to examine the cellular effects induced by RT, analyses of morphology and clonogenic evaluation were also conducted. Results: A descriptive report of GEP and pathways induced by IR is reported from our microarray data. Moreover, the MDA-MB-231 Radioresistent Cell Fraction (RCF) selected, included specific molecules able to drive radioresistance. Conclusion: In summary, our data highlight, the RT response of TNBC MDA-MB-231 cell line at a transcriptional level, in terms of activating radioresistance in these cells, as a model of late-stage BC.
Keywords: Ionizing radiation (IR), MDA-MB-231 cells, gene expression profile (GEP)
Radiation therapy (RT) represents one of the most frequently used therapeutic options in cancer clinical practice, including breast cancer (BC), able to control localized tumor (1). As known, BC represents a highly heterogeneous group of tumors at both the clinical and molecular level associated with different outcomes for the patients (2).
Today, a great amount of cancer patients receive RT through several modalities, such internal or external RT, Brachytherapy, intraoperative electron radiation therapy (IOERT), and others (3-5). Although technological advances in radiation delivery have strongly enhanced the tumor killing capacity, the current clinical practice still offers standard RT protocols for patients with cancer in the same anatomical position, without taking into account the molecular characteristics of the tumor histotype.
As described by several groups including our own, IOERT is a RT technique that can destroy residual cancer cells that may be left at the tumor site, immediately following surgical removal of the tumor (6). According to specific eligibility criteria, IOERT may be exclusive (a single radiation dose of 21-23 Gy) or an anticipated boost of 9-12 Gy, followed by conventional RT (7-13).
As recently described by Yadava and Shankara, even if a great number of cancer patients receive RT, not all of them report a therapeutic benefit due to distant metastatic spread and local recurrence, a processes able to induce radioresistance activation (14,15). Indeed, tumor radiosensitivity depends on many factors, some of which are linked to the clinical state of the patient. For example, the fate of BC disease following ionizing radiation (IR), depends on many factors, such as hormone receptor (HR) status of estrogen (ER), progesterone (PR) and human epidermal growth factor (HER2). Moreover, the balance between radiosensitivity and radioresistance is regulated by different and complex factors including the tumor stem cells which are able to repopulate during the course of the RT schedule, tumor microenvironment such as hypoxia, stromal interaction and variations in the intrinsic sensitivity of cells to radiation, modulation of DNA repair or other cell survival pathways (14-19).
To our knowledge, limited data are reported regarding the IR-induced gene expression changes in relation to BC tumor grading (20-24). In this sense, we recently reported IR-induced low grade- and high grade-specific signaling in BC cells (5). This study described the IR-induced genomic biomarkers and gene-signatures of specific breast tumor subtypes, grouped according to HR status. In particular regarding this radiobiological issue, more advanced studies on this topic are needed in order to help personalized biological-driven RT treatment planning alone or in combination with other therapies.
Several authors have reported controversies with respect to the use of RT for patients with triple-negative BC (TNBC) (ER–/PR–/HER2–) (25,26). TNBC (heterogeneous disease which represents 15%-20% of BC incidences), is associated with an early age at presentation, larger tumor sizes, higher rates of recurrence, more aggressive biology, and poorer prognosis (25,26). In addition, the absence of hormonal or targeted therapy against TNBC makes it a clinical challenge for oncologists in terms of patient management. However, Yao and colleagues have recently reported a survival advantage of adjuvant RT for TNBC patients, underling the necessity to study and clarify molecular mechanisms active in these cells and how these are involved in regulating cell survival/death balance (24).
Summarizing, as described in the literature, a major obstacle for effective cancer treatment by RT is the development of radioresistance. Thus, a great focus of research should aim to help clinicians understand the molecular features involved in the activation of this process, including the contribution of the specific tumor subtypes in this process. Such studies could better define the most successful treatment plan, including the choice of the best RT modality and schedule in the clinical practice against cancer.
In this sense, the principal aim of this work was to study gene expression profiles (GEP) by cDNA microarray analyses, induced by a high dose of IR used during IOERT schedules in triple negative MDA-MB-231 BC cells and to identify key factors and networks principally involved in the radioresistance process. Here, we report a list of genes and cellular process involved in cellular and molecular responses to high IR doses. We trust that these data could be used to better clarify mechanisms involved in the radioresistance of patients, particularly those with triple negative BC cells.
Materials and Methods
Radiation treatments. Cellular irradiation setup and dose distribution were conducted as previously reported and were performed using the NOVAC7 IOERT system (Sortina IOERT Technologies, Vicenza, Italy) (5,7,10,27). MDA-MB-231 BC cells were irradiated using the following two doses: i) 9 Gy (according to the boost scheme), and ii) 23 Gy (exclusive modality), to the 100% isodose at a rate of 3.2 cGy/pulse.
Cell cultures and clonogenic survival evaluation. The human breast adenocarcinoma MDA-MB-231 cell line, was purchased from the American Type Culture Collection (ATCC, Manassas, VA, Canada) and cells were cultured according to manufacturer’s guidelines, as previously described (5). To detect the cell radiation effect, clonogenic survival assays, according to the method by Franken et al., and morphological evaluations were conducted as previously reported (10,27).
Whole-genome cDNA microarray expression analysis. GEP of the MDA-MB-231 BC cell line treated with 9 and 23 Gy of IR doses was performed. In addition, we also conducted microarray experiments on the surviving fraction of MDA-MB-231 cells collected 7 days after IOERT treatment, using 9 Gy of IR dose (hereafter named Radioresistent Cell Fraction, RCF). The total RNA was extracted from cells and was evaluated for its concentration and purity, as previously reported (5). cRNA synthesis and labeling were conducted according to the Agilent Two-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Santa Clara, CA, USA), as previously described by our group (5). For this purpose we used Whole Human Genome 4x44K microarrays (Agilent Technologies) that contain all known human genes. Array hybridization, microarray scanning and feature detection, as well as statistical data analysis, background correction, normalization and summary of expression measures were conducted with the Feature Extraction and GeneSpring GX 10.0.2 software (Agilent Technologies), as previously described in detail (7). In addition, the GEPs obtained in this work were also analyzed by pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) network building tool (https://david.ncifcrf.gov/tools.jsp). The data discussed here are deposited in the NCBI Gene Expression Omnibus (28) and are accessible through the GEO Series accession number (GSE127789). Microarray data are available in compliance with Minimum Information about a Microarray Experiment standards (29).
Results
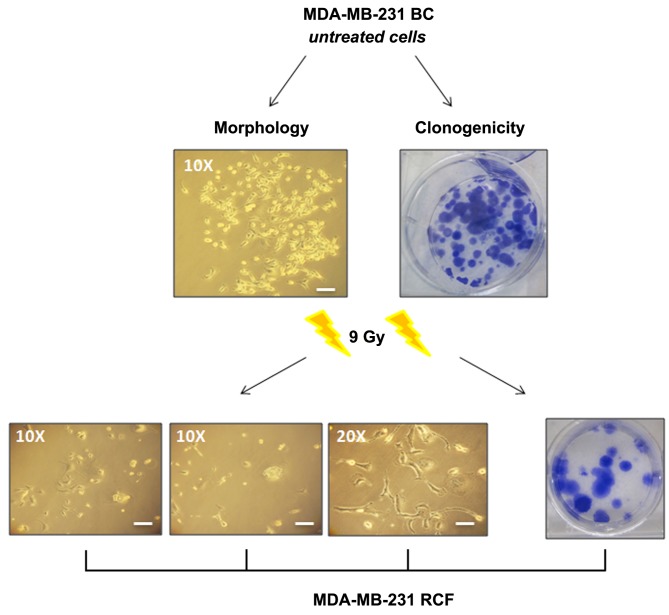
Cell survival and morphology. To evaluate MDA-MB-231 cell viability in terms of reproductive capacity, we performed a clonogenic survival assay (10,27). 23 Gy of IR exposure inhibited the colony-forming ability of cells. On the other hand, 3 weeks following the 9 Gy exposure an RCF was observed (SF=6.4%) (Figure 1). Cell morphology modifications induced by IR, such as membranous and cytoplasmic damages, were starting from 72 hours post-treatment with either dose of 9 or 23 Gy.
Figure 1. Micrographs of MDA-MB-231 RCF cell line following 9 Gy for morphological and clonogenic survival evaluation. Scale bars: 30 μm.
Moreover, irradiated MDA-MB-231 cells displayed a large flat shape with evident macroscopic plasma membrane and nucleus alterations, suggesting a typical senescent phenotype (the well-known ‘fried egg’), confirmed by SA-b-Gal activity, as previously described (30).
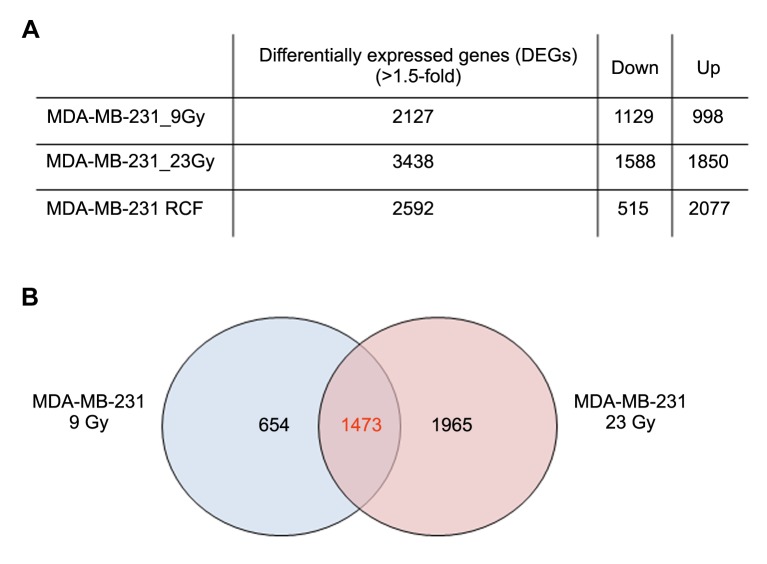
Overview of cDNA microarray gene expression analyses after 9Gy and 23Gy irradiations. Whole human genome Microarray-Based Gene Expression Analyses (Agilent Technologies) were conducted on MDA-MB-231 cells treated with 9 Gy and 23 Gy, delivered by IOERT. MDA-MB-231 untreated cells were used as reference sample for comparing the differential gene expression analyses (Figure 2A).
Figure 2. (A) Genes whose expression was significantly altered by 1.5-fold or greater following different irradiation conditions compared to the untreated reference group. (B) Venn diagrams showing the number of unique and shared differentially expressed genes (DEG) of the MDA-MB-231 BC cells, following exposure to doses of 9 and 23 Gy.
A total of 2,127 genes changed their expression levels by 1.5-fold or more (1129 genes were down-regulated and 998 up-regulated) compared to the untreated reference group of MDA-MB-231 cells irradiated with 9 Gy.
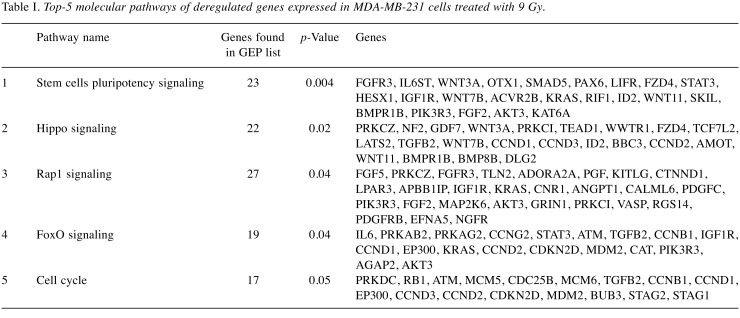
Deregulated genes (DEGs) were grouped according to their involvement in specific pathways using DAVID and REACTOME tools (https://david.ncifcrf.gov/tools.jsp; https://reactome.org). The result of this mapping, shown in Table I highlights the involvement of a set of factors, principally involved in stem cell pluripotency pathway: Hippo, Rap1 and FoxO signaling as well as in cell cycle control.
Table I. Top-5 molecular pathways of deregulated genes expressed in MDA-MB-231 cells treated with 9 Gy.
From the MDA-MB-231 cells irradiated with 23 Gy, 3438 DEGs were selected a difference in their GEP by 1.5-fold or greater compared to the control: 1588 genes were down-regulated and 1850 up-regulated (Figure 2A).
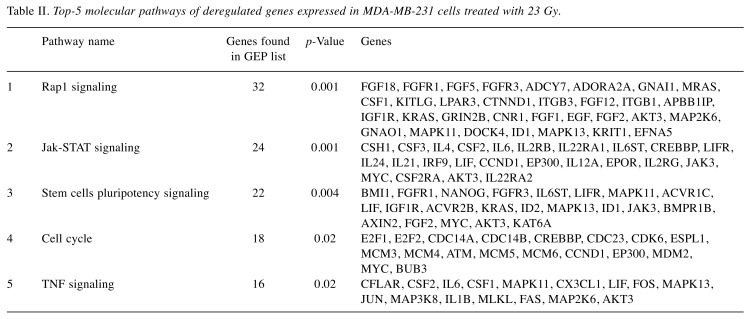
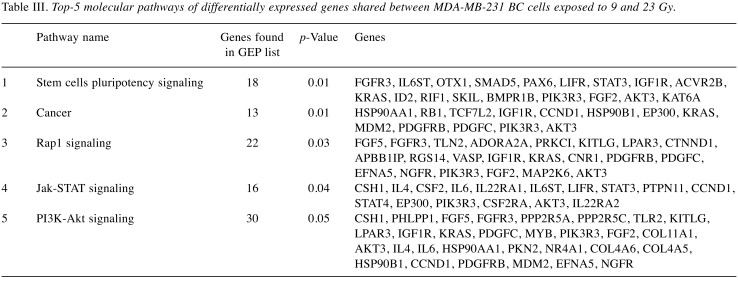
The top 5 statistically and biologically relevant pathways, deregulated in MDA-MB-231 following 23 Gy, were analyzed by using the DAVID tool, underlining the involvement of the following listed pathways: i) Rap1 signaling, ii) Jak-STAT network, iii) stem cell pluripotency pathways, iv) cell cycle control and v) Tumor Necrosis Factor (TNF) network (Table II). Moreover, we performed Venn diagrams in order to study the number of unique and shared differentially expressed genes between the cells exposed to IR doses of 9 and 23 Gy (Figure 2B). 1473 genes were found to be common to MDA-MB-231 BC cells exposed to either 9 or 23 Gy (848 were down- and 625 were up-regulated). As shown in Table III, the stem cells pluripotency pathway, Rap1, Jak-STAT and PI3K-Akt signaling, were the top-5 statistically relevant pathways selected using the DAVID tool.
Table II. Top-5 molecular pathways of deregulated genes expressed in MDA-MB-231 cells treated with 23 Gy.
Table III. Top-5 molecular pathways of differentially expressed genes shared between MDA-MB-231 BC cells exposed to 9 and 23 Gy.
Interestingly, the analysis GO biological processes of all the configurations analyzed in this work, underlined the involvement of multiple cellular processes associated with chromatin assembly, such as nucleosome and telomere organization.
Overview of cDNA microarray gene expression analyses of RCF. As described above, following 9 Gy treatment a surviving MDA-MB-231 RCF (6.4%), was isolated and maintained up to 3-weeks post-IOERT (Figure 1). In order to analyze key genes involved in radioresistant cell response, we performed GEP analysis of MDA-MB-231 RCF samples collected 7 days post IOERT treatment. We used a comparative reference sample (MDA-MB-231 9 Gy-treated cells collected 24 hours post irradiation) in order to study genes specifically and temporally involved in the activation of the radioresistant process in the post irradiation time window analyzed. Precisely, 2592 genes were found to be deregulated in the MDA-MB-231 RCF pool by 2-fold or more compared to control (515 genes were down-regulated and 2,077 were up-regulated) (Figure 2A).
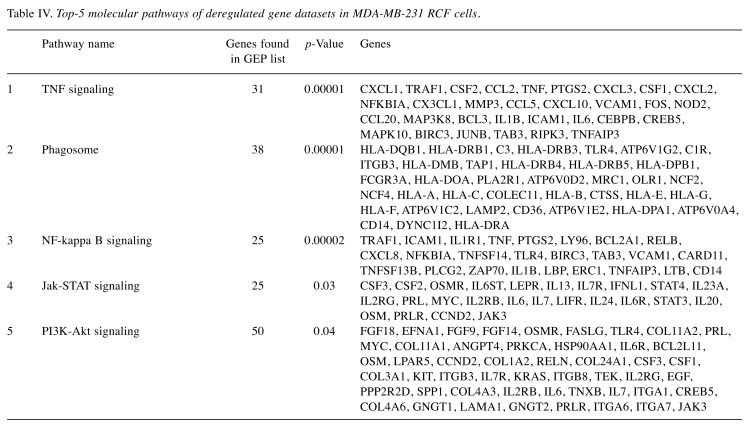
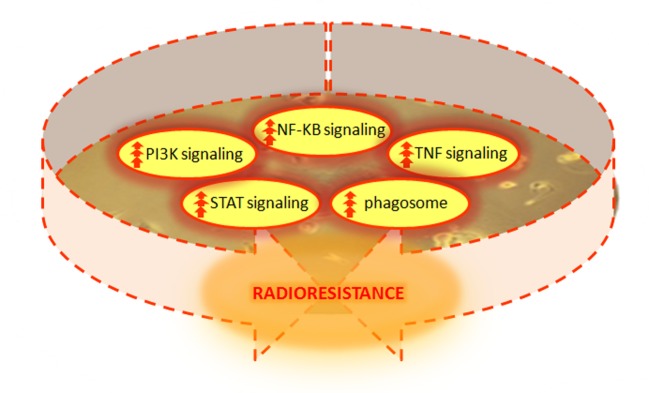
Also in this case, we performed DAVID analysis in order to analyze the top-5 cellular pathways regulated by the selected gene list, as shown in Table IV. This analysis confirmed once again the involvement of networks, such as TNF, NF-kappa B, Jak-STAT and PI3K-Akt signaling and phagosome pathway. Interestingly, the analysis of GO biological processes performed using DAVID tool, underlined once again the involvement of multiple processes associated with chromatin assembly, possibly driving the activation of radioresistance processes in MDA-MB--231 RCF cells (Figure 3).
Table IV. Top-5 molecular pathways of deregulated gene datasets in MDA-MB-231 RCF cells.
Figure 3. Schematic of the master regulators and signaling pathways strictly involved in the development of the radioresistance phenotype observed in MDA-MB-231 BC cells.
Discussion
As recently described by Sosin et al., IOERT demonstrates favorable patient-reported outcomes and low rates of toxicity with adequate local disease control at 2-year follow-up when implemented in early BC (6). However, the radiosensitivity of BC depends on many factors, including a positive or negative HR status (5). In particular, controversial data are available in the literature regarding the radiation responses of TNBC cells. In this work we analyzed genetic and cellular pathways deregulated after 9 Gy (IOERT BC ‘boost’ scheme) and 23 Gy (BC exclusive treatment modality) of IR doses, conventionally delivered during the IOERT plans.
Cell viability showed that 9 Gy treatment did not totally inhibit cell growth and proliferation of the MDA-MB-231 BC cell line, which generated RCF (SF=6.4%). On the contrary, radiation treatment using 23 Gy of IR dose was able to inhibit cell clonogenic activity (undetectable colonies) (30). IR-induced morphological changes, that were similar at either dose. Irradiated MDA-MB-231 cells displayed a large flat cell shape with evident macroscopic plasma membrane and nuclear alterations, suggesting a typical senescent phenotype, confirmed by SA-b-Gal activity and linked with a senescence-associated secretory phenotype (SASP), as previously reported (30).
As suggested by GO analyses, both irradiation conditions created conspicuous changes in DNA structure assembly and in nucleosome and telomere organization, and consequently in gene expression regulation.
In particular, following 9 Gy IR exposure, MDA-MB-231 BC cells changed the expression levels in a large set of genes controlling the stem cell pluripotency pathway: Hippo, Rap1 and FoxO signaling as well as cell cycle regulators. In particular, DEGs belonging to the selected pathways, are listed in Table I.
The Hippo signaling is a novel tumor suppressor pathway that regulates cell proliferation and stem cell state. Hippo also controls organ size by inhibiting cell proliferation and promoting apoptosis (31). In turn, as described by Saeg and Anbalagan, the deregulation of Hippo signaling is believed to be responsible for the formation of Cancer Stem Cells (CSCs) in various types of cancer, including BC (32-33). The proliferation of CSCs is known to be involved with activation of radioresistance. In MDA-MB-231 9 Gy cells, Hippo signaling was inhibited, as highlighted by the down-regulation of several factors that are part of this network, such as neurofibromin 2 (NF2 also known as Merlin), large tumor suppressor kinase 2 (LATS2), WW domain containing transcription regulator 1 (WWTR1) and TEA domain family member 1 (TEAD1) (32,34-35). Despite extensive studies, the mechanisms by which breast tumors become radioresistant are not fully understood. Tumor heterogeneity is a key product of CSCs and a key feature of therapy resistance. In particular, following irradiation CSCs up-regulate IGF (insulin like growth factor) type 1 receptor (up-regulated also in MDA-MB-231 9 Gy) and increase the IGF1 secretion. In the resting G0 phase of the cell cycle, this expression pattern inhibits PI3K-AKT signaling and activates Fox0, resulting in cell cycle deregulation and stimulation of self-renewal (32,34-36). Our data here confirm these findings as FoxO signaling and cell cycle key regulators (such as RB1, ATM, CDC25B, CCNB1, CCND1, EP300, CCND3, CCND2, CDKN2D genes and others listed in Table I), were deregulated in MDA-MB-231 9 Gy (Table I). Finally, Rap1 pathway was included in the top 5 biologically relevant pathways activated in MDA-MB-231 BC cells following either irradiation dose. Interestingly, Rap1 gene encodes a protein that is part of a complex involved in the regulation of telomere length and possibly in the activation of senescence (37).
The analysis of the top-5 pathway deregulated genes in MDA-MB-231 treated with 23 Gy, confirmed the deregulation of Rap1, and of pathways affecting stem cells pluripotency and cell cycle signaling following radiation exposure. Interestingly, two out of the five pathways selected were specifically deregulated following 23 Gy exposure: Jak-STAT and TNF networks. JAK-STAT signaling plays a key role in regulating the immune responses to IR. The STAT proteins (STAT1 and STAT3 were up-regulated in MDA-MB-231 23Gy) are considered to be important for cell viability in response to different stimuli, including IR (38-40), and play a significant role in tumor development. Interestingly, Khodarev NN et al., have shown that a radiation-resistant phenotype could be induced by ectopic STAT over-expression, while its down-regulation could significantly increase the radiosensitivity of renal carcinoma cell lines (38,41,42). Thus, the role of JAK-STAT in acquired radioresistance seems to be based not only on its transcriptional regulation, but also on its expression.
Several groups have highlighted the key role of TNF signaling in cancer, including BC. Tumour Necrosis Factor alpha (TNF-α), is a well-known pro-inflammatory cytokine often up-regulated following radiation and has been implicated in the development of fibrosis induced by IR (38,43). In addition, TNF-α is both an inducer and a target of NF-kB, one of the most important transcription factors involved in immunological responses induced by radiation, and thus a master regulator of radiosensitivity/radioresistance cell balance (38). In summary, Jak-STAT and TNF pathways are strictly involved in immunological responses to radiation, as reported by the literature and by our research group (30,38,44). Here, we confirmed their key role in cell radiation response to high IR doses, such as those delivered by IOERT with 23 Gy, according to the exclusive BC RT schedule.
Finally, in order to analyze the number of unique and shared differentially expressed genes of the MDA-MB-231 cells exposed to IR doses of 9 and 23 Gy, we performed Venn diagrams showing a conspicuous amount of genes deregulated following either type of high-dose irradiation. From these, we then listed the top-5 biologically relevant pathways regulated by the 473 genes common to MDA-MB-231 BC cells exposed to either 9 or 23 Gy, underling once again the relevant role in cell radiation response of some of the above mentioned cellular signaling.
Finally, in order to analyze key genes involved in the radioresistant cell responses observed in the MDA-MB-231 RCF samples, we performed GEP analysis. Precisely, 2592 genes were found to be deregulated in MDA-MB-231 RCF by 2-fold or greater compared to control. Even in this case, the top GO biological process were involved in chromatin assembly able to regulate gene expression changes and probably able to drive the activation of radioresistance process in MDA-MB-231 RCF cells. In addition, the top five biological relevant pathways, confirm the involvement of the networks above described and following listed: TNF, NF-kappa B, Jak-STAT, PI3K-Akt signaling and phagosome regulations. In MDA-MB-231 RCF cells, the regulation of phagosome could be involved in radiation-induced autophagy known to be able to enhance radioresistance and leads to activation of the survival pathway, as recently observed (45). In particular, the master regulators of these pathways were up-regulated as displayed in Figure 3, and to our opinion strictly involved in the development of radioresistance phenotype observed in MDA-MB-231 RCF cells, after high radiation dose exposure.
In our opinion, this RCF gene list, needs more investigation in order to clarify the key roles of these factors in driving cell survival after the exposure of high IR doses and potentially used in targeted therapy intervention in tandem with RT schedules.
To date, targeted therapies against these molecules is under investigation and is supported by encouraging data in the literature (also in TNBC patients), however, these need to be further supported and clarificed by radiobiology approach to evaluate their synergistic behavior in tandem with RT schedules in order to optimize BC care (46-48).
IOERT demonstrates favorable BC patient outcomes and low rates of toxicity with adequate control of local disease (6). However, a limited number of studies describe the molecular basis of radioresistance activation following RT, especially in the triple negative BC subtype (7,10,27,49). Herein, we report a list of genes and cellular processes involved in cellular and molecular responses to high IR doses, delivered during IOERT schedules, and highlighting key factors involved in radioresistance activation. We trust that these data could be used to better clarify mechanisms involved in the radioresistance process, particularly in triple-negative BC cells in order to optimize RT successful plans in tandem with targeted therapies.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
Authors’ Contributions
All Authors participated in the conception and elaboration of the stud. In particular, GR performed the irradiation setups, and, together with VB, LM, FPC and GIF performed the IR cell treatments. LM, RM and GP carried out the cell cultures and the cell survival assays. VB performed the microarray experiments and the gene expression analyses. All authors were involved in the interpretation of the findings and approved the final content of the manuscript.
Acknowledgements
This work was carried out thanks to the support of the following project grants: FIRB/MERIT (RBNE089KHH) and GeSeTON (funded by Italian MISE grant n. 489 of 21/02/2018).
References
- 1.Schwartzberg BS, Chin DT, Dorn PL, Herron DS, Howell KT, Kemmis TJ, Miller SA, Moore JA, Paul D. Application of 21-gene recurrence score results and ASTRO suitability criteria in breast cancer patients treated with intraoperative radiation therapy (IORT) Am J Surg. 2018;216(4):689–693. doi: 10.1016/j.amjsurg.2018.07.022. PMID: 30041733. DOI: 10.1016/j.amjsurg.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 2.Bravatà V, Cammarata FP, Forte GI, Minafra L. "Omics" of HER2-positive breast cancer. OMICS. 2013;17(3):119–129. doi: 10.1089/omi.2012.0099. PMID: 23421906. DOI: 10.1089/omi.2012.0099. [DOI] [PubMed] [Google Scholar]
- 3.Formenti SC, Demaria S. Combining radiotherapy and cancer immunotherapy: A paradigm shift. J Natl Cancer Inst. 2013;105:256–265. doi: 10.1093/jnci/djs629. PMID: 23291374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskar R, Dai J, Wenlong N, Yeo R, Yeoh KW. Biological response of cancer cells to radiation treatment. Front Mol Biosci. 2014;1:24. doi: 10.3389/fmolb.2014.00024. PMID: 25988165. DOI: 10.3389/fmolb.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravatà V, Cava C, Minafra L, Cammarata FP, Russo G, Gilardi MC, Castiglioni I, Forte GI. Radiation-induced gene expression changes in high and low grade breast cancer cell types. Int J Mol Sci. 2018;19(4):pii: E1084. doi: 10.3390/ijms19041084. PMID: 29617354. DOI: 10.3390/ijms19041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sosin M, Gupta SS, Wang JS, Costellic CD, Gulla A, Bartholomew AJ, O’Neill SC, Hechenbleikner EM, Collins BT, Rudra S, Collins SP, Chaldekas KM, Seevaratnam S, Langan RC, Willey SC, Tousimis EA. A prospective analysis of quality of life and toxicity outcomes in treating early breast cancer with breast conservation therapy and intraoperative radiation therapy. Front Oncol. 2018;8:545. doi: 10.3389/fonc.2018.00545. PMID: 30560085. DOI: 10.3389/fonc.2018.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minafra L, Bravatà V, Cammarata FP, Russo G, Gilardi MC, Forte GI. Radiation gene-expression signatures in primary breast cancer cells. Anticancer Res. 2018;38(5):2707–2715. doi: 10.21873/anticanres.12512. PMID: 29715090. DOI: 10.21873/anticanres.12512. [DOI] [PubMed] [Google Scholar]
- 8.Wallner P, Arthur D, Bartelink H Connolly J, Edmundson G, Giuliano A, Goldstein N, Hevezi J, Julian T, Kuske R, Lichter A, McCormick B, Orecchia R, Pierce L, Powell S, Solin L, Vicini F, Whelan T, Wong J, Coleman CN and Workshop Participants. Workshop on partial breast irradiation: state of the art and the science. J Natl Cancer Inst. 2004;96:175–184. doi: 10.1093/jnci/djh023. PMID: 14759984. [DOI] [PubMed] [Google Scholar]
- 9.Smith BD, Arthur DW, Buchholz TA, Haffty BG, Hahn CA, Hardenbergh PH, Julian TB, Marks LB, Todor DA, Vicini FA, Whelan TJ, White J, Wo JY, Harris JR. Accelerated partial breast irradiation consensus statement from the American Society for Radiation Oncology (ASTRO) Int J Radiat Oncol Biol Phys. 2009;74:987–1001. doi: 10.1016/j.ijrobp.2009.02.031. PMID: 19545784. DOI: 10.1016/j.ijrobp.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 10.Bravatà V, Minafra L, Russo G, Forte GI, Cammarata FP, Ripamonti M, Casarino C, Augello G, Costantini F, Barbieri G, Messa C, Gilardi MC. High-dose ionizing radiation regulates gene expression changes in the MCF7 breast cancer cell line. Anticancer Res. 2015;35(5):2577–2591. PMID: 2596453. [PubMed] [Google Scholar]
- 11.Harris EER, Small W Jr. Intraoperative radiotherapy for breast cancer. Front Oncol. 2017;7:317. doi: 10.3389/fonc.2017.00317. PMID: 29312887. DOI: 10.3389/fonc.2017.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedlmayer F, Reitsamer R, Wenz F, Sperk E, Fussl C, Kaiser J, Ziegler I, Zehentmayr F, Deutschmann H, Kopp P, Fastner G. Intraoperative radiotherapy (IORT) as boost in breast cancer. Radiat Oncol. 2017;12(1):23. doi: 10.1186/s13014-016-0749-9. PMID: 28103903. DOI: 10.1186/s13014-016-0749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veronesi U, Orecchia R, Luini A, Galimberti V, Zurrida S, Intra M, Veronesi P, Arnone P, Leonardi MC, Ciocca M, Lazzari R, Caldarella P, Rotmensz N, Sangalli C, Sances D, Maisonneuve P. Intraoperative radiotherapy during breast conserving surgery: a study on 1, 822 cases treated with electrons. Breast Cancer Res Treat. 2010;124:141–151. doi: 10.1007/s10549-010-1115-5. PMID: 20711810. DOI: 10.1007/s10549-010-1115-5. [DOI] [PubMed] [Google Scholar]
- 14.Yadav P, Shankar BS. Radio resistance in breast cancer cells is mediated through TGF-β signalling, hybrid epithelial-mesenchymal phenotype and cancer stem cells. Biomed Pharmacother. 2018;111:119–130. doi: 10.1016/j.biopha.2018.12.055. PMID: 30579251. DOI: 10.1016/j.biopha.2018.12.055. [DOI] [PubMed] [Google Scholar]
- 15.Langlands FE, Horgan K, Dodwell DD, Smith L. Breast cancer subtypes: Response to radiotherapy and potential radiosensitisation. Br J Radiol. 2013;86(1023):20120601. doi: 10.1259/bjr.20120601. PMID: 23392193. DOI: 10.1259/bjr.20120601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19(3):200–207. doi: 10.1097/PPO.0b013e318292e4e3. PMID: 23708066. DOI: 10.1097/PPO.0b013e318292e4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodward WA, Debeb BG, Xu W, Buchholz TA. Overcoming radiation resistance in inflammatory breast cancer. Cancer 116(11. 2010;Suppl):2840–2845. doi: 10.1002/cncr.25173. PMID: 20503417. DOI: 10.1002/cncr.25173. [DOI] [PubMed] [Google Scholar]
- 18.Bravatà V, Stefano A, Cammarata FP, Minafra L, Russo G, Nicolosi S, Pulizzi S, Gelfi C, Gilardi MC, Messa C. Genotyping analysis and 18FDG uptake in breast cancer patients: A preliminary research. J Exp Clin Cancer Res. 2013;32:23. doi: 10.1186/1756-9966-32-23. PMID: 23631762. DOI: 10.1186/1756-9966-32-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camphausen K, Purow B, Sproull M, Scott T, Ozawa T, Deen DF, Tofilon PJ. Orthotopic growth of human glioma cells quantitatively and qualitatively influences radiation-induced changes in gene expression. Cancer Res. 2005;65:10389–10393. doi: 10.1158/0008-5472.CAN-05-1904. PMID: 16288029. DOI: 10.1158/0008-5472.CAN-05-1904. [DOI] [PubMed] [Google Scholar]
- 20.Khodarev NN, Park JO, Yu J, Gupta N, Nodzenski E, Roizman B, Weichselbaum RR. Dose-dependent and independent temporal patterns of gene responses to ionizing radiation in normal and tumor cells and tumor xenografts. Proc Natl Acad Sci. 2001;98:12665–12670. doi: 10.1073/pnas.211443698. PMID: 11675498. DOI: 10.1073/ pnas.211443698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, Trent JM, Bittner ML, Meltzer PS, Fornace AJ Jr. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. PMID: 18199535. DOI: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 22.Lü X, de la Peña L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–1061. doi: 10.1158/0008-5472.CAN-05-3459. PMID: 16424041. DOI: 10.1158/0008-5472.CAN-05-3459. [DOI] [PubMed] [Google Scholar]
- 23.Essmann F, Engels IH, Totzke G, Schulze-Osthoff K, Jänicke RU. Apoptosis resistance of MCF7 breast carcinoma cells to ionizing radiation is independent of p53 and cell cycle control but caused by the lack of caspase-3 and a caffeine-inhibitable event. Cancer Res. 2004;64:7065–7072. doi: 10.1158/0008-5472.CAN-04-1082. PMID: 15466201. DOI: 10.1158/0008-5472.CAN-04-1082. [DOI] [PubMed] [Google Scholar]
- 24.Yao Y, Chu Y, Xu B, Hu Q, Song Q. Radiotherapy after surgery has significant survival benefits for patients with triple-negative breast cancer. Cancer Med. 2019;8(2):554–563. doi: 10.1002/cam4.1954. PMID: 30632300. DOI: 10.1002/cam4.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhani , Parshad R, Kazi M, Seenu V, Mathur S, Dattagupta S, Haresh KP. Triple-negative breast cancers: are they always different from nontriple-negative breast cancers? An experience from a tertiary center in India. Indian J Cancer. 2017;54(4):658–663. doi: 10.4103/ijc.IJC_348_17. PMID: 30082553. DOI: 10.4103/ijc.IJ C_348_17. [DOI] [PubMed] [Google Scholar]
- 26.Gong CC, Ma G, Hu XC, Zhang Y, Wang Z, Zhang J, Zhao Y, Li Y, Xie Y, Yang Z, Wang B. Pretreatment 18F-FDG uptake heterogeneity predicts treatment outcome of first-line chemotherapy in patients with metastatic triple negative breast cancer. Oncologist. 2018;23(10):1144–1152. doi: 10.1634/theoncologist.2018-0001. PMID: 30082489. DOI: 10.1634/theoncologist.2018-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minafra L, Bravatà V, Russo G, Forte GI, Cammarata FP, Ripamonti M, Candiano G, Cervello M, Giallongo A, Perconti G, Messa C, Gilardi MC. Gene-expression profiling of MCF10A breast epithelial cells exposed to IOERT. Anticancer Res. 2015;35(6):3223–3234. PMID: 26026082. [PubMed] [Google Scholar]
- 28.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–995. doi: 10.1093/nar/gks1193. PMID: 23193258. DOI: 10.1093/ nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. PMID: 11726920. DOI: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 30.Bravatà V, Minafra L, Forte GI, Cammarata FP, Russo G, Di Maggio FM, Augello G, Lio D, Gilardi MC. Cytokine profile of breast cell lines after different radiation doses. Int J Radiat Biol. 2017;93(11):1217–1226. doi: 10.1080/09553002.2017.1362504. PMID: 28763256. DOI: 10.1080/09553002.2017.1362504. [DOI] [PubMed] [Google Scholar]
- 31.Johnson R, Halder G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. doi: 10.1038/nrd4161. PMID: 24336504. DOI: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeg F, Anbalagan M. Breast cancer stem cells and the challenges of eradication: a review of novel therapies. Stem Cell Investig. 2018;5:39. doi: 10.21037/sci.2018.10.05. PMID: 30498750. DOI: 10.21037/ sci.2018.10.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei C, Wang Y, Li X. The role of Hippo signal pathway in breast cancer metastasis. Oncol Targets Ther. 2018;11:2185–2193. doi: 10.2147/OTT.S157058. PMID: 29713187. DOI: 10.2147/OTT.S157058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol Ther. 2016;160:145–158. doi: 10.1016/j.pharmthera.2016.02.008. PMID: 26899500. DOI: 10.1016/j.pharmthera.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borst P, Jonkers J, Rottenberg S. What makes tumors multidrug resistant. Cell Cycle. 2007;6:2782–2787. doi: 10.4161/cc.6.22.4936. PMID: 17998803. DOI: 10.4161/cc.6.22.4936. [DOI] [PubMed] [Google Scholar]
- 36.Osuka S, Sampetrean O, Shimizu T, Saga I, Onishi N, Sugihara E, Okubo J, Fujita S, Takano S, Matsumura A, Saya H. IGF1 receptor signaling regulates adaptive radioprotection in glioma stem cells. Stem Cells. 2013;31:627–640. doi: 10.1002/stem.1328. PMID: 23335250. DOI: 10.1002/stem.1328. [DOI] [PubMed] [Google Scholar]
- 37.Martínez P, Gómez-López G, Pisano DG, Flores JM, Blasco MA. A genetic interaction between RAP1 and telomerase reveals an unanticipated role for RAP1 in telomere maintenance. Aging Cell. 2016;15(6):1113–1125. doi: 10.1111/acel.12517. PMID: 27586969. DOI: 10.1111/ acel.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Maggio FM, Minafra L, Forte GI, Cammarata FP, Lio D, Messa C, Gilardi MC, Bravatà V. Portrait of inflammatory response to ionizing radiation treatment. J Inflamm (Lond) 2015;12:14. doi: 10.1186/s12950-015-0058-3. PMID: 25705130. DOI: 10.1186/s12950-015-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poczeta M, Bednarek I, Nowak E, Galilejczyk A, Gawlik N. Silencing of the STAT3 gene expression activity and cancer cells metastatic potential at in vitro studies. Wiad Lek. 2013;66:226–232. PMID: 24483027. [PubMed] [Google Scholar]
- 40.Murray PJ. The JAK-STAT signaling pathway: Input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. PMID: 17312100. [DOI] [PubMed] [Google Scholar]
- 41.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, Darga TE, Beveridge MG, Sood RF, Sutton HG, Beckett MA, Mauceri HJ, Posner MC, Weichselbaum RR. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. PMID: 19503789. DOI: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hui Z, Tretiakova M, Zhang Z, Li Y, Wang X, Zhu JX, Gao Y, Mai W, Furge K, Qian CN, Amato R, Butler EB, Teh BT, Teh BS. Radiosensitization by inhibiting STAT1 in renal cell carcinoma. Int J Radiat Oncol Biol Phys. 2009;73:288–295. doi: 10.1016/j.ijrobp.2008.08.043. PMID: 19100922. DOI: 10.1016/j.ijrobp.2008.08.043. [DOI] [PubMed] [Google Scholar]
- 43.Veeraraghavan J, Natarajan M, Aravindan S, Herman TS, Aravindan N. Radiation-triggered tumor necrosis factor (TNF) alpha-NFkappaB cross-signaling favors survival advantage in human neuroblastoma cells. J Biol Chem. 2011;286:21588–21600. doi: 10.1074/jbc.M110.193755. PMID: 21527635. DOI: 10.1074/jbc.M110.193755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaue D, Kachikwu EL, McBride WH. Cytokines in radiobiological responses: A review. Radiat Res. 2012;178:505–523. doi: 10.1667/RR3031.1. PMID: 23106210. DOI: 10.1667/RR3031.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin HJ, Liu HH, Lin CD, Kao MC, Chen YA, Chiang-Ni C, Jiang ZP, Huang MZ, Lin CJ, Lo UG, Lin LC, Lai CK, Lin H, Hsieh JT, Chiu CH, Lai CH. Cytolethal distending toxin enhances radiosensitivity in prostate cancer cells by regulating autophagy. Front Cell Infect Microbiol. 2017;7:223. doi: 10.3389/fcimb.2017.00223. PMID: 26774497. DOI: 10.1016/j.clbc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jhaveri K, Teplinsky E, Silvera D, Valeta-Magara A, Arju R, Giashuddin S, Sarfraz Y, Alexander M, Darvishian F, Levine PH, Hashmi S, Zolfaghari L, Hoffman HJ, Singh B, Goldberg JD, Hochman T, Formenti S, Esteva FJ, Moran MS, Schneider RJ. Hyperactivated mTOR and JAK2/STAT3 pathways: Molecular drivers and potential therapeutic targets of inflammatory and invasive ductal breast cancers after neoadjuvant chemotherapy. Clin Breast Cancer. 2016;16(2):113–22.e1. doi: 10.1016/j.clbc.2015.11.006. PMID: 26774497. DOI: 10.1016/j.clbc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mundt F, Rajput S, Li S, Ruggles KV, Mooradian AD, Mertins P, Gillette MA, Krug K, Guo Z, Hoog J, Erdmann-Gilmore P, Primeau T, Huang S, Edwards DP, Wang X, Wang X, Kawaler E, Mani DR, Clauser KR, Gao F, Luo J, Davies SR, Johnson GL, Huang KL, Yoon CJ, Ding L, Fenyö D, Ellis MJ, Townsend RR, Held JM, Carr SA, Ma CX. Mass spectrometry-based proteomics reveals potential roles of NEK9 and MAP2K4 in resistance to PI3K inhibition in triple-negative breast cancers. Cancer Res. 2018;78(10):2732–2746. doi: 10.1158/0008-5472.CAN-17-1990. PMID: 29472518. DOI: 10.1158/0008-5472.CAN-17-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buckley NE, Haddock P, De Matos Simoes R, Parkes E, Irwin G, Emmert-Streib F, McQuaid S, Kennedy R, Mullan P. A BRCA1 deficient, NFĸB driven immune signal predicts good outcome in triple negative breast cancer. Oncotarget. 2016;7(15):19884–19896. doi: 10.18632/oncotarget.7865. PMID: 26943587. DOI: 10.18632/oncotarget.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pucci M, Bravatà V, Forte GI, Cammarata FP, Messa C, Gilardi MC, Minafra L. Caveolin-1, breast cancer and ionizing radiation. Cancer Genomics Proteomics. 2015;12(3):143–152. PMID: 25977173. [PubMed] [Google Scholar]