Abstract
Background/Aim: Pseudomyogenic hemangioendo-thelioma is a rare endothelial tumor. Previous genetic investigations have shown that the tumors carry either a SERPINE1-FOSB or an ACTB-FOSB fusion gene. The aim of the study was to identify FOSB fusions linked with pseudomyogenic hemangioendothelioma. Materials and Methods: RNA sequencing, reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing analyses were performed on a pseudomyogenic hemangioendothelioma. Results: An in-frame fusion was found between exon 4 of WWTR1 from 3q25 and exon 2 of FOSB from 19q13. The fusion gene not only places FOSB under the control of the WWTR1 promoter, but is predicted to encode a chimeric WWTR1-FOSB transcription factor. Conclusion: FOSB may be fused with SERPINE1, ACTB, or WWTR1 in pseudomyogenic hemangioendotheliomas. The resulting overexpression of FOSB fusion is a potentially useful marker that could be helpful in the diagnosis of these tumors.
Keywords: Pseudomyogenic hemangioendothelioma, WWTR1, FOSB, WWTR1-FOSB fusion gene, RNA sequencing
Pseudomyogenic hemangioendothelioma is a rare endothelial neoplasm that is multifocal in two-thirds of cases and often occurs within a limited region (1,2). It is more frequently found in young males and primarily in soft tissue, with only 20% presenting with secondary manifestations in bone. About 60% of the lesions occur in the lower limbs. Half of the patients experience pain. Histologically, the tumors are ill-defined, composed of sheets and cords of spindled cells with abundant, eosinophilic cytoplasm. The nucleus is vesicular, with small nucleoli. There are few mitotic figures and little atypia. Immunohistochemistry (IHC) shows diffuse positive expression of AE1/AE3 and ERG and, in 30% of cases, positive staining for CD31 and smooth muscle actin (SMA). The tumors are negative for Desmin, S100 and CD34 (1,2). Cytogenetic information on pseudomyogenic hemangioendothelioma is restricted to 4 tumors all of which carried a t(7;19)(q22;q13) chromosomal translocation (3,4). The translocation resulted in the fusion of the SERPINE1 gene from 7q22 with FOSB gene from 19q13 resulting in overexpression of FOSB (4). In two recent studies, fusion of ACTB from 7p22 with FOSB was described in pseudomyogenic hemangioendotheliomas lacking SERPINE1 rearrangement (5,6). Agaram et al. (5) have studied 15 pseudomyogenic hemangioendotheliomas and, by using fluorescence in situ hybridization and ARCHER FusionPlex analysis, found that seven of the tumors had an ACTB-FOSB fusion whereas eight of them had a SERPINE1-FOSB fusion. Zhu et al. have detected ACTB-FOSB fusion in two pseudomyogenic hemangioendo-theliomas using target RNA sequencing (6).
We here present another pseudomyogenic hemangioendo-thelioma that carried none of the above-mentioned fusions, but instead a novel WWTR1-FOSB fusion gene.
Materials and Methods
Ethics statement. The study was approved by the regional ethics committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no). Written informed consent was obtained from the patient for publication of the case details. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
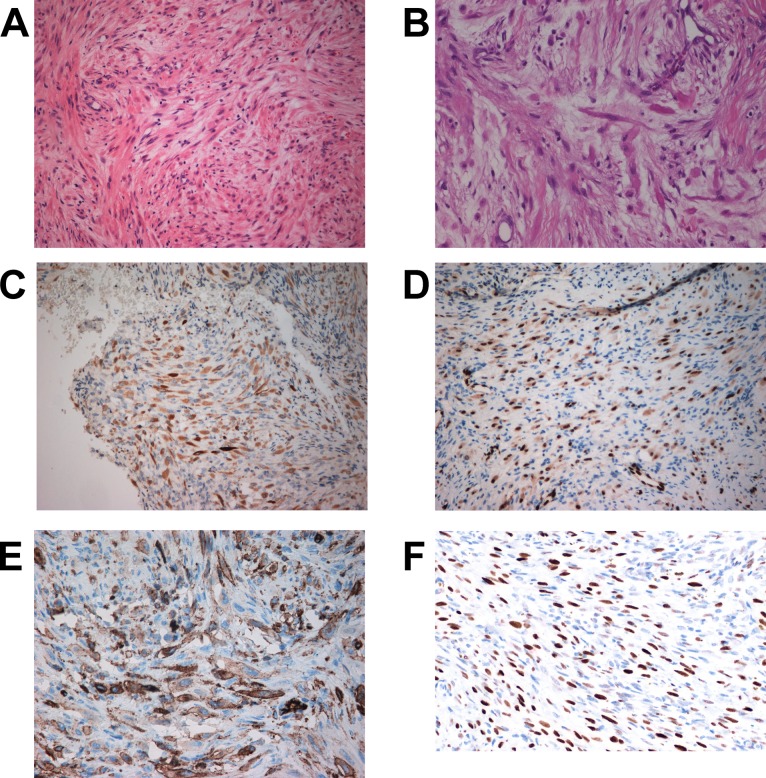
Case description. The patient was a 33-year-old woman who had a history of back pain for over 6 years. A sudden worsening of her symptoms led to radiological examination which revealed a lytic lesion in the upper lateral region of the sacral bone, presumably of a benign, fibrous nature, in addition to a prolapse in the lumbar spine. The lesion in the sacrum had presumably brought about the longstanding dumb, continuous pain. Magnetic resonance imaging (MRI) and positron emission tomography (PET) examinations showed also a second lytic lesion in the lumbar spine. Examination of an initial core needle biopsy showed a spindle cell lesion without evidence of atypia leading to a tentative diagnosis of benign fibrous histiocytoma. In a second core needle biopsy, areas with giant cells were observed. Based on the evidence that a variant of giant cell tumor was present, the patient treated with Denosumab. After initial alleviation of symptoms, the pain came back after four injections of Denosumab. A new PET scan showed minimal reduction in signal intensity and no reduction in tumor size. The treatment was discontinued and an open biopsy was performed, which again revealed a spindle cell lesion without atypia (Figure 1A-B). Immunohistochemistry (IHC) showed positivity for SMA, high molecular weight cytokeratins AE1/AE3 (Figure 1C), ERG (Figure 1D), and CD31 (Figure 1E). After consulting Professor Nielsen at Massachusetts General Hospital, the patient was diagnosed with a pseudomyogenic hemangioendothelioma. IHC for FOSB was then performed which showed strong nuclear positivity (Figure 1F).
Figure 1. Microscopic examination of the pseudomyogenic hemangioendothelioma. A) H&E-stained section showing a tumor with a spindle cell lesion without atypia, ×20. B) H&E-stained section, ×40. C) Immunoexpression of cytokeratin AE1/AE3, ×20. D) Immunoexpression of ERG, ×20, E) Immunoexpression of CD31, ×20. F) Immunoexpression of FOSB showing strong nuclear positivity, ×20.
Chromosome banding analysis. Fresh tissue from a representative area of the tumor was received and isolated cells were short-term cultured and analysed cytogenetically, as part of our diagnostic routine, as described elsewhere (7).
RNA sequencing. Total RNA was extracted from frozen (–80˚C) tumor tissue using miRNeasy Mini Kit (Qiagen Nordic, Oslo, Norway) and one μg of total RNA was sent to the Genomics Core Facility at the Norwegian Radium Hospital, Oslo University Hospital (http://genomics.no/oslo/) for high-throughput paired-end RNA-sequencing according to the Illumina TruSeq Stranded mRNA protocol. The “grep” command was used to search the fastq files of the sequence data for FOSB sequence. The principle of this approach has been described in detail elsewhere (7,8). The search term was the 20-nucleotide-sequence (nt) “GAGTGCGCCGGTC TCGGGGA” which is the first 20 nt in exon 2 of FOSB corresponding to nt 719-738 in the FOSB reference sequence with accession number NM_006732.2.
Reverse transcription (RT) and genomic PCR analyses. One μg of total RNA was reverse-transcribed in a 20 μl reaction volume using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad, Oslo, Norway). One μl of the synthesized cDNA was used as template in subsequent PCR assays. PCR amplifications were performed in a 25 μl reaction volume which contained 12.5 μl Premix Ex Taq™ DNA Polymerase Hot Start Version (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France), template (1 μl cDNA), and 0.4 μM of each of the forward and reverse primers. The forward primer was WWTR1-996F1: TGA GTA TGC CCA ATG CGC TGA CCA corresponding to position 996-1019 in the WWTR1 reference sequence with accession number NM_015472.4. The reverse primer was FOSB-817R1: TGG GAC TGG GCC ATG GAA GAG ATG corresponding to position 840-817 in the FOSB reference sequence with accession number NM_006732.2. PCR amplifications were run on a C-1000 Thermal cycler (Bio-Rad) and the cycling was: an initial denaturation at 94˚C for 30 sec, followed by 35 cycles of 7 sec at 98˚C and 2 min at 68˚C, and a final extension for 5 min at 72˚C. Three μl of the PCR products were stained with GelRed (Biotium, VWR International, Oslo, Norway), analyzed by electrophoresis through 1.0 % agarose gel, and photographed. The remaining PCR products were purified using the MinElute PCR Purification Kit (Qiagen) and analysed by direct sequencing using the dideoxy procedure with the BigDye terminator v1.1 cycle sequencing kit (ThermoFisher Scientific, Waltman, MA, USA) on the Applied Biosystems Model 3500 Genetic Analyzer sequencing system. The BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for computer analysis of sequence data.
Results
The G-banding analysis revealed a normal karyotype 46, XX in all examined 25 metaphases (data not shown).
Using the “grep” command and a search term corresponding to the first 20 nt in exon 2 of FOSB on the raw RNA sequence data, which were in the text-based fastq format, 23 unique sequences were extracted (Table I). Alignment of each of them with the human genome using BLAT on the genome browser (https://genome-euro.ucsc.edu/index.html) and BLAST on the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi ) showed that 19 sequences were from FOSB whereas 4 sequences were hybrids containing sequences from exon 4 of WWTR1 (sequence with accession number NM_015472) and exon 2 of FOSB (sequence with accession number NM_006732) (Table I).
Table I. Detection of WWTR1-FOSB fusion in the RNA sequencing data of pseudomyogenic hemangioendothelioma. The retrieved sequences from the fastq file of the RNA sequencing using the “grep” command and the search term “GAGTGCGCCGGTCTCGGGGA” which is the first 20 nt in the exon 2 of FOSB corresponding to nt 719-738 in the FOSB reference sequence with the accession number NM_006732.2.
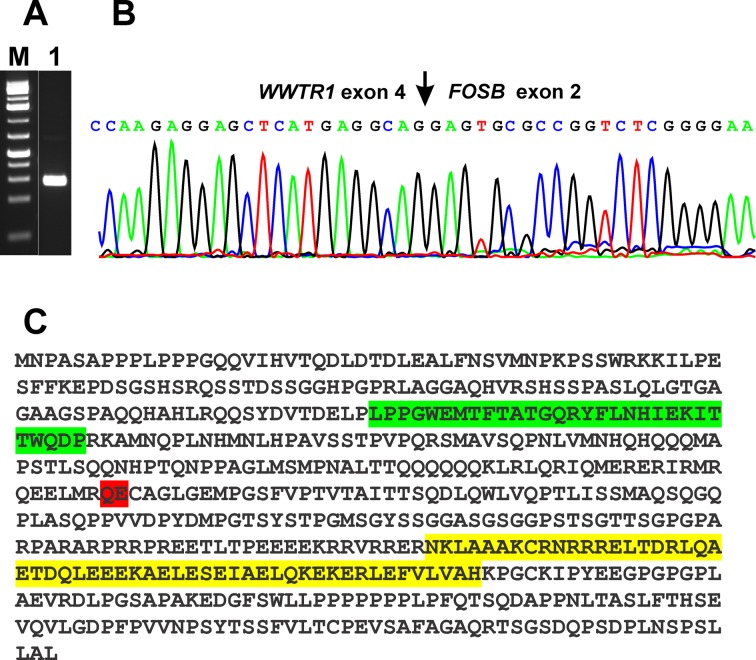
RT-PCR with the primer set WWTR1-996F1/FOSB-817R2 amplified a cDNA fragment strongly suggesting the presence of a WWTR1-FOSB fusion transcript in the examined tumor (Figure 2A, lane 1). Sequencing of the amplified cDNA fragment showed that it consisted of a WWTR1-FOSB chimeric fragment in which exon 4 of WWTR1 (nucleotide 1322 accession number NM_012330 version 3) was fused in-frame to exon 2 of FOSB (Figure 2B). Thus, taking into account the relevant reference sequences NM_015472/NP_056287 for WWTR1 and NM_006732/NP_006723 for FOSB, the fusion WWTR1-FOSB appears to code for a 553 amino acid residue chimeric WWTR1-FOSB protein consisting of the first 257 and the last 296 (position 43-338) amino acid residues of the WWTR1 and FOSB proteins, respectively (Figure 2C).
Figure 2. RT-PCR and Sanger sequencing of the PCR products in pseudomyogenic hemangioendothelioma. A) Gel electrophoresis showing the amplified WWTR1-FOSB cDNA fragment. M, GeneRuler 1 Kb DNA ladder (Thermo Scientific). Lane 1, amplification using the forward primer WWTR1-996F1 and the reverse primer FOSB-817R1. B) Partial sequence chromatograms of the cDNA amplified fragment showing the fusion (arrow) of WWTR1 with FOSB. C) The putative WWTR1-FOSB fusion protein. The WW domain from WWTR1 is in green, the bZIP region from FOSB is in yellow, the WWTR1-FOSB junction is in red.
Discussion
In the present study, a WWTR1-FOSB fusion gene was identified in a pseudomyogenic hemangioendothelioma, the third fusion gene to be detected in tumors of this type. One consequence of the fusion is that it places coding regions of the FOSB gene under the control of the WWTR1 promoter. Replacement of the native FOSB promoter with the promoter of the fusion partner has been a theme common to all reported FOSB fusions described (4-6,9). For example, in the first reported SERPINE1-FOSB fusion gene in pseudomyogenic hemangioendothelioma, the breakpoint in SERPINE1 was in the non-coding region of exon 1. Therefore, SERPINE1 provided a strong promoter for the expression of its fusion partner, FOSB (4). Similarly, in the ZFP36-FOSB fusion gene, exon 1 of ZFP36 was fused in-frame to exon 2 of FOSB and contributed only eight amino acid residues to the ZFP36-FOSB chimeric protein (9). Thus, the role of ZFP36 in the fusion with FOSB was to provide a promoter for regulating the expression of FOSB.
In the present case, however, the fusion WWTR1-FOSB appears to code for a chimeric transcription factor containing the N-terminal 14-3-3 binding region and the WW domain from WWTR1 (10,11). It also contains the region required for transformation activity, the basic-leucine zipper (bZIP) domain, and the C-terminal transcriptional activation domain from FOSB (10,11). A WWTR1-FOSB fusion gene was also reported previously, in an epithelioid hemangioma (9) where exon 3 of WWTR1 was fused in-frame with exon 1 of FOSB, and the coding chimeric WWTR1-FOSB protein contained the above-mentioned functional regions.
The recently described ACTB-FOSB fusion gene in pseudomyogenic hemangioendothelioma also codes for a chimeric transcription factor (5,6). The ACTB-FOSB protein contains the nucleotide-binding domain of the sugar kinase/HSP70/actin superfamily from ACTB and all the above-mentioned regions of FOSB (12). However, there seemed to be no clinicopathological differences between pseudomyogenic hemangioendothelioma carrying an ACTB-FOSB fusion and the tumors with SERPINE1-FOSB, except that ACTB-FOSB positive tumors were often solitary (5).
At the genomic level, WWTR1 (on chromosome band 3q25) is transcribed in the direction from telomere to centromere whereas the transcription of FOSB (on chromosome band 19q13) proceeds in the opposite direction, from centromere to telomere. Hence, formation of a WWTR1-FOSB fusion should not be possible from a simple and balanced t(3;19)(q25;q13) translocation. An additional genomic aberration would be required, for example an inversion or an insertion in one of the derivative chromosomes, der(3) and der(19). The orientation of WWTR1 relative to FOSB may also explain the possibly low frequency of WWTR1-FOSB fusions. The SERPINE1-FOSB fusion gene is the result of a balanced t(7;19)(q22;q13) chromosomal aberration because SERPINE1 and FOSB are transcribed in the direction from centromere to telomere (3,4). The ACTB-FOSB fusion gene could also be the result of a balanced t(7;19)(p22;q13) since ACTB on 7p22 is transcribed from centromere to telomere, although such a translocation has not been reported yet in pseudomyogenic hemangioendotheliomas (5,6).
The SERPINE1-FOSB, ACTB-FOSB, and WWTR1-FOSB fusion genes and the taking over of FOSB expression control by SERPINE1, ACTB, and WWTR1 strong promoters makes FOSB a useful immunohistochemical diagnostic marker for pseudomyogenic hemangioendotheliomas as it was the case here (5,13,14). However, genomic analyses are needed to clarify which gene recombinations occur at which frequencies in these tumors, and also whether as yet unknown variants of the FOSB-activation theme exist.
Conflicts of Interest
The Authors declare that they have no potential conflicts of interest exist.
Authors’ Contributions
IP designed the research, performed the molecular genetic analyses, interpreted the data, and wrote the manuscript. IL did the pathological evaluations. LG performed cytogenetic experiments and interpreted the data. SH evaluated the cytogenetics wrote the manuscript.
Acknowledgements
The Authors thank Professor G. Petur Nielsen, M.D., Department of Pathology, Massachusetts General Hospital, Boston for help with diagnosis, performing FOSB IHC analysis and providing Figure 1F. This work was supported by grants from Radiumhospitalets Legater.
References
- 1.Hornick JL, Fletcher CD. Pseudomyogenic hemangioendothelioma: a distinctive, often multicentric tumor with indolent behavior. Am J Surg Pathol. 2011;35:190–201. doi: 10.1097/PAS.0b013e3181ff0901. PMID: 21263239. DOI: 10.1097/PAS.0b013e3181ff0901. [DOI] [PubMed] [Google Scholar]
- 2.Al-Qaderi A, Mansour AT. Pseudomyogenic Hemangioendothelioma. Arch Pathol Lab Med. 2018 doi: 10.5858/arpa.2017-0430-RS. PMID: 30576238. DOI: 10.5858/arpa.2017-0430-RS. [DOI] [PubMed] [Google Scholar]
- 3.Trombetta D, Magnusson L, von Steyern FV, Hornick JL, Fletcher CD, Mertens F. Translocation t(7;19)(q22;q13)-a recurrent chromosome aberration in pseudomyogenic hemangio-endothelioma. Cancer Genet. 2011;204:211–215. doi: 10.1016/j.cancergen.2011.01.002. PMID: 21536240. DOI: 10.1016/j.cancergen.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Walther C, Tayebwa J, Lilljebjorn H, Magnusson L, Nilsson J, von Steyern FV, Ora I, Domanski HA, Fioretos T, Nord KH, Fletcher CD, Mertens F. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J Pathol. 2014;232:534–540. doi: 10.1002/path.4322. PMID: 24374978. DOI: 10.1002/path.4322. [DOI] [PubMed] [Google Scholar]
- 5.Agaram NP, Zhang L, Cotzia P, Antonescu CR. Expanding the spectrum of genetic alterations in pseudomyogenic hemangioendothelioma with recurrent Novel ACTB-FOSB gene fusions. Am J Surg Pathol. 2018;42:1653–1661. doi: 10.1097/PAS.0000000000001147. PMID: 30256258. DOI: 10.1097/PAS.0000000000001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu G, Benayed R, Ho C, Mullaney K, Sukhadia P, Rios K, Berry R, Rubin BP, Nafa K, Wang L, Klimstra DS, Ladanyi M, Hameed MR. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol, 2018 doi: 10.1038/s41379-018-0175-7. PMID: 30459475. DOI: 10.1038/s41379-018-0175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The "grep" command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13) PLoS One. 2014;9:e99439. doi: 10.1371/journal.pone.0099439. PMID: 24950227. DOI: 10.1371/journal.pone.0099439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. Novel KAT6B-KANSL1 fusion gene identified by RNA sequencing in retroperitoneal leiomyoma with t(10;17)(q22;q21) PLoS One. 2015;10:e0117010. doi: 10.1371/journal.pone.0117010. PMID: 25621995. DOI: 10.1371/ journal.pone.0117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonescu CR, Chen HW, Zhang L, Sung YS, Panicek D, Agaram NP, Dickson BC, Krausz T, Fletcher CD. ZFP36-FOSB fusion defines a subset of epithelioid hemangioma with atypical features. Genes Chromosomes Cancer. 2014;53:951–959. doi: 10.1002/gcc.22206. PMID: 25043949. DOI: 10.1002/gcc.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. PMID: 11118213. DOI: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wisdom R, Verma IM. Proto-oncogene FosB: the amino terminus encodes a regulatory function required for transformation. Mol Cell Biol. 1993;13:2635–2643. doi: 10.1128/mcb.13.5.2635. PMID: 8474434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley JH. The sugar kinase/heat shock protein 70/actin superfamily: implications of conserved structure for mechanism. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. PMID: 8800467. DOI: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 13.Hung YP, Fletcher CD, Hornick JL. FOSB is a Useful Diagnostic Marker for Pseudomyogenic Hemangioendothelioma. Am J Surg Pathol. 2017;41:596–606. doi: 10.1097/PAS.0000000000000795. PMID: 28009608. DOI: 10.1097/PAS.0000000000000795. [DOI] [PubMed] [Google Scholar]
- 14.Sugita S, Hirano H, Kikuchi N, Kubo T, Asanuma H, Aoyama T, Emori M, Hasegawa T. Diagnostic utility of FOSB immunohistochemistry in pseudomyogenic hemangioendo-thelioma and its histological mimics. Diagn Pathol. 2016;11:75. doi: 10.1186/s13000-016-0530-2. PMID: 27515856. DOI: 10.1186/s13000-016-0530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]