Abstract
Background/Aim: Chloride intracellular channel 1 (CLIC1) represents a promising target for personalized therapy. Our aim was to assess CLIC1 expression in clear cell renal cell carcinoma (cc RCC) and identify its possible prognostic role. Materials and Methods: Fifty cases of cc RCC were evaluated and selected for immunohistochemistry. CLIC1 expression was correlated with tumor grade, invasion and heterogeneity. Results: A total of 87.5% of the cases were CLIC1 positive, with either a homogeneous (31.42%) or a heterogeneous (68.57%) pattern. Low, mild and strong CLIC1 expressing tumors were defined based on nuclear (N), cytoplasmic (C), membrane (M) or combinations of them (NC, NM, CM, NCM) in terms of CLIC1 distribution. A significant correlation was found between tumor grade and percent of positive tumor cells (p=0.017). For G3 tumors, CLIC1 cytoplasmic expression was strongly correlated with high expression status (p=0.025) and tumor heterogeneity (p=0.004). CLIC1 expression was also correlated with metastasis (p=0.046). Conclusion: We defined four cc RCC groups depending on G, CLIC1 expression and pattern: i) G3/NM/low CLIC1+, ii) G2/CM/mild CLIC1+ iii) G1 or G2/NM or CM /high CLIC1+, and iv) G2/M /high CLIC1+.
Keywords: Chloride Intracellular Channel 1 (CLIC1), clear cell renal cell carcinoma (cc RCC)
CLIC1 (chloride intracellular channel 1) is a protein that belongs to the family of ion channels of chlorine. This protein is naturally expressed in the human body and is involved in many cellular processes, such as cell volume regulation, regulation of membrane potential, cell cycle regulation, cell proliferation and cell differentiation (1). CLIC1 normally exists as a soluble globular protein, however, in response to oxidative stress, it translocates from the cytoplasm to the cell membrane where it selectively acts as a chloride channel (2,3). A potential involvement of CLIC1 in tumor development is suspected both for its role in cell cycle regulation as well as for its functional expression during oxidative stress.
Amongst the six members of intracellular chloride ion channel protein family (CLICs), CLIC1 and CLIC4 have been extensively studied regarding their involvement in tumor development (4). Indeed, CLIC1 is overexpressed in several tumor types, such as gastric cancer (5), oral squamous cell carcinomas (6) or glioblastoma (7). Recently, Barbieri et al. reported that CLIC1 inhibition induces glioblastoma growth inhibition, mainly by acting on CLIC1-rich glioblastoma stem cells, decreasing their proliferation and subsequently tumor progression (8). Although CLIC1 expression in cancer stem cells (CSC) has been certified only for glioblastoma (7,8), indirect evidence suggests that CLIC1 is also expressed in tumor stem cells from other aggressive malignancies, such as pancreatic (9), ovarian (10) and esophageal cancer (11).
CLIC1 is characterized by a high versatility regarding its ability to translocate from cytosol to the nucleus and/or the plasma membrane during malignant transformation (12), based on its ability to spontaneously convert from soluble to an integral membrane-bound form. This changeable state is most probably regulated by the membrane lipid composition, especially by cholesterol, together with external factors, such as oxidation and pH (13,14), however, a precise mechanism is currently unknown. Once CLIC1 is incorporated in the plasma membrane, tumor cells become highly active, with a rapid proliferation rate (15), increased invasiveness (16) and metastatic potential (17).
Kidney cancer accounts for 5% and 3% of all malignanciens in adult men and women, respectively (18). The most frequent histological type (around 80%) is represented by clear cell renal cell carcinoma (cc RCC). Cc RCC is a kidney malignancy with an unpredictable behaviour, especially in terms of response to treatment (19). Although in most cases kidney cancer is diagnosed when already localized, approximately 20% of patients will develop metastasis following nephrectomy with curative intent (20). In other cases, cc RCC is already invasive at the time of diagnosis and, despite of new targeted therapies, patients present a high mortality rate (16). This suggests that the spectrum of cc RCC is not fully characterized regarding its molecular profile. Unpredictable response to treatment represents one of the most controversial issues in cc RCC. Resistance to therapy is based on an incomplete mechanism of action (21), and there is no effective definitive therapeutic tool available (22). Lichner et al. recently reported that Sunitinib therapy for advanced RCC induces selection of a tumor cells subpopulation able to survive and to grow as spheroids in tumor models xenografts (23). The group also showed that these cells express E cadherin as a key contributor to the survival of RCC cells under Sunitinib treatment. The Memorial Sloan Kettering Cancer Centre RCC classification (Motzer criteria) focuses on clinical features of RCC to predict survival. This system evaluates: i) performance status, ii) LDH value, iii) haemoglobin value, iv) serum calcium concentration and v) time from initial diagnosis to systemic treatment (24); however, no pathological variables are taken into consideration. A previous attempt to integrate the clinical data with tissue markers so as to predict disease mortality in metastatic RCC was done by Kim et al. (25), which, unfortunately, highlighted the limitations. Taken together, it is clear that there is the strong need for novel prognostic biological markers reliably assessing tumor invasion as well as its metastatic potential in cc RCC. These tools are also needed for the identification of patients at higher risk of recurrence following local treatment.
CLIC1 is expressed in the normal kidney in the glomerular structures and also on the apical domain of proximal tubules’ epithelial cells (26), however, its role of in renal cancer is yet to be elucidated. The Human Atlas Gene Protein mentions CLIC1 as an unfavorable prognostic marker in renal cancer, with no adequate details available on it (https://www. proteinatlas.org/ENSG00000213719-CLIC1/t).
Evidence regarding the interrelation and crosstalk between CLIC1 and RCC tumor cells derive mostly from in vitro and experimental tumor xenograft studies using different renal cancer cell lines or tumor cells isolated from patients with localized and metastatic RCC (15). CLIC1 inhibition seems to decrease tumor invasion and metastasis due to a blockage of the myosin light chain kinase (MYLK) and of β3 integrin, suggesting an important role for CLIC1 in integrin-mediated actomyosin dynamics in cells with invasive and metastatic potential.
Despite of clear experimental evidence about the role of CLIC1 in RCC progression, no data are available about its expression on human cc RCC specimens at present. Thus, we aimed to describe CLIC1 expression pattern using immunohistochemistry on human tumor samples of cc RCC and to correlate the pattern of reaction with tumor grade, invasion and tumor heterogeneity.
Materials and Methods
Tumor samples. Fifty cases of cc RCC were collected during open surgery performed for kidney tumor mass of suspicious or proven malignant origin. Initial diagnosis was made by imaging (e.g. CT scan, MRI). Both localized and metastatic diseases were studied. Informed consent was obtained from all patients before surgery and all procedures respected ethical principles regarding the use of human tissue specimens for research purposes, according with WMA Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
Tissue primary processing. Tumor samples fixed for 24 h in 10% buffered formalin were included in a routine automated workflow of paraffin embedding by using automated ThermoShandon carousel for histopathology (ThermoScientific Fischer, Cambridge, UK) including detailed protocols for each type of tissue. Automatic workflow follows the routine steps widely known in histopathology and has as a final product the parraffin block ready for sectioning Three micrometer-thick serial sections were cut from each specimen and were stained with haematoxylin and eosin for histopathological evaluation.
Immunohistochemistry. Following initial evaluation, additional paraffin-embedded slides from each RCC case were immunostained using the monoclonal mouse anti human CLIC1 antibody (Clone 356.1, dilution 1:2,000) (Santa Cruz Biotechnology, Heidelberg, Germany), incubated at room temperature for 30 min. Incubation with the primary antibody was followed by the use of Bond Polymer Refine Detection System (Leica Biosystems, Newcastle Upon Tyne, UK) specific for BOND MAX autostainer having well standardized protocol including all steps of routine immunohistochemistry. Automated process selected into BOND MAX Autostainer performs automated dewax for 30 min, incubation with primary antibodies as it was described before, and then incubation with polymer for 30 min followed by DAB chromogen for 10 min. Counterstain was the final step automated immunostaining. Prostate tissue expressing CLIC1 was used as external positive control, and normal kidney tissue adjacent to the tumor as internal positive control. A dark brown color detected by microscopy on stained specimens having nuclear, cytoplasmic and/or membranar pattern was considered as positive for CLIC1. All immunohistochemical steps were fully automated and controlled by the Bond Max autostainer (Leica Biosystems, Newcastle Upon Tyne, UK). The interpretation of immunostained specimens included all three patterns: i) nuclear (N), ii) cytoplasmic (C) and iii) membrane (M), but we also checked combined patterns: i) all three (NMC), ii) nuclear and cytoplasmic (NC), iii) nuclear and membranar (NM), iv) cytoplasmic and membrane (CM). We stratified cases according to the ammounts of CLIC1 positive cells as follows: i) low expressing tumors (10-30% of tumor cells expressed CLIC1), ii) mild expressing tumors (30-50%) and iii) high expressing tumors (>50% tumor cells positive for CLIC1). The intensity of positive reaction was then assessed as: i) weak (1), ii) moderate (2) and iii) strong (3).
Image aquisition and data analysis. All slides were scanned using the Pannoramic Desk slide scanner (3D Histech, Budapest, Hungary). Digital slides were stored in Case Center and were assessed by using the Pannoramic Viewer Platform (3D Histech, Budapest, Hungary). By using these methods we evaluated the whole section of each specimen for CLIC1 expression pattern, percentage of positive cells and CLIC1 signal intensity. Statistical analysis was performedusing SPSS version 17, for evaluating correlations tests (Pearson, Spearman and Kendall). A p-value less than 0.05 being considered statistically significant.
Results
The normal kidney tissue adjacent to the tumor presented a heterogeneously positive reaction for CLIC1. The renal cortex was intensely stained compared to the medulla. We persistently detected CLIC1 expression inside the renal corpuscle with a nuclear pattern restricted to podocytes. The number of positive podocytes was significantly lower compared to the presumed total number of podocytes in the normal glomerulus. The tubular system of the normal kidney had a different expression, the strongest immunostaining was observed inside the proximal renal tubules compared to distal and collecting tubes where a weak CLIC1 expression or negative reaction was found (Figure 1).
Figure 1. CLIC1 expression in the normal human renal parenchyma. Note the CLIC1 nuclear expression inside renal corpuscules (red arrow, podocytes) and its cytoplasmic expression in the proximal renal tubules (red arrowhead). Immunohistochemistry for CLIC1, ×400 magnification.
Within the tumor tissue, CLIC1 was expressed in malignant cells and in the endothelium of small intratumoral and peritumoral blood vessels. CLIC1 showed high accuracy in detecting locoregional and intravascular invasion thanks to its high ability of intensively staining tumor cells, unlike the staining of the surrounding tissues. Endothelial cells lining some tumor capillaries expressed CLIC1 as well.
CLIC1 was detected in 87.5% of all the examined cases of cc RCC. Positive reaction was homogeneously distributed in the whole tumor area in 31.42% of the total positive cases. For the remaining cases (68.57%), positive tumor cells were distributed in a “mosaic-like” fashion, with groups of positive tumor cells intermingled within negative areas (Figure 2A and B). 53.33% out of G2 cases and 91.66% of G3 cases had a heterogeneous distribution of CLIC1 positivity. Heterogeneity of expression increased with tumor grading, the most heterogeneous distribution of positive reaction for CLIC1 observed for G3 tumors (Figure 3). Based on this distribution pattern, we thought appropiate to quantify the percent of positive cells from each case and we found that 8.57% of all positive cases were CLIC1 low-expressing tumors, 17.14% were CLIC1 mild-expression tumors and 71.42% were CLIC1 high-expressing tumors. The WHO/ISUP grading system was used to assess nuclear and nucleolar features; in this classification, tumor grade is assigned according to the highest grade cells present in the tissue analyzed (27).
Figure 3. CLIC1 heterogeneity expression according to tumor grade.
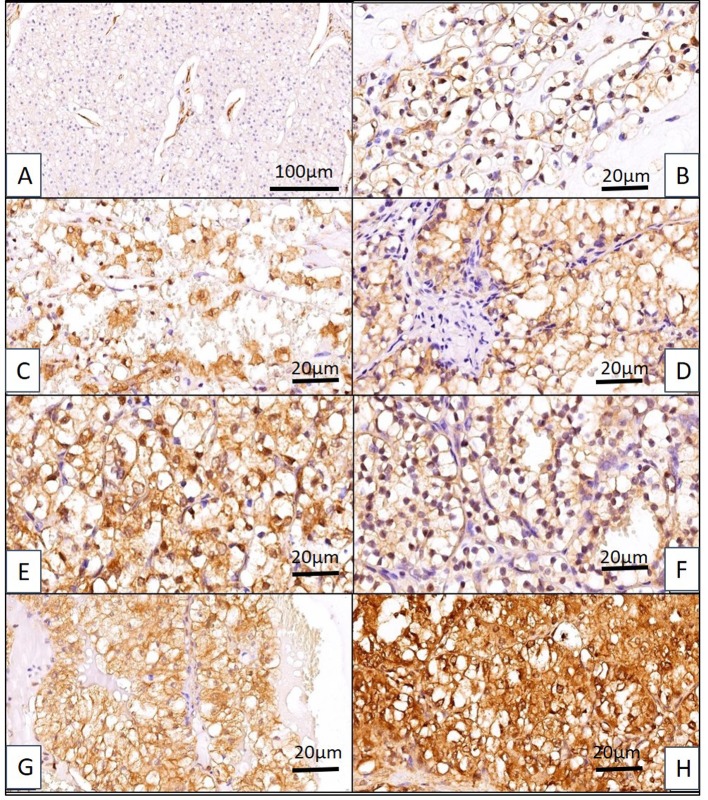
Following an initial evaluation regarding CLIC1 positivity/negativity in cc RCC, we distributed cases depending on CLIC1 presence into i) nucleus (N), ii) cytoplasm (C) and iii) membrane (M) of tumor cells. Few cases had pure N, M, or C location of CLIC1 positive expression, most of them displayed a combined pattern. Thus, we defined 7 subgroups of cc RCC, three with single expression N, M or C and four with a combined expression as follows: iv) nuclear and cytoplasmic (NC), v) nuclear and membrane (NM), vi) cytoplasmic and membrane (CM), and vii) nuclear/membrane/cytoplasmic (NMC) (Figure 4).
Figure 4. Seven expression patterns of CLIC1 have been identified in ccRCC. (A) CLIC1-negative ccRCC (note that tumor blood vessels are CLIC1 positive). (B) Nuclear pattern (N), (C) cytoplasmic pattern (C), (D) membrane pattern (M), (E) nuclear/cytoplamic pattern (NC), (F) nuclear membrane (NM), (G) cytoplasmic/membrane (CM) and (H) nuclear/ cytoplasmic/membrane pattern (NMC).
For G3 tumors, CLIC1 cytoplasmic expression was strongly correlated with CLIC1 high-expressing tumors (p=0.025) and also with tumor heterogeneity (p=0.004). No G4 tumors were identified in the cases evaluated.
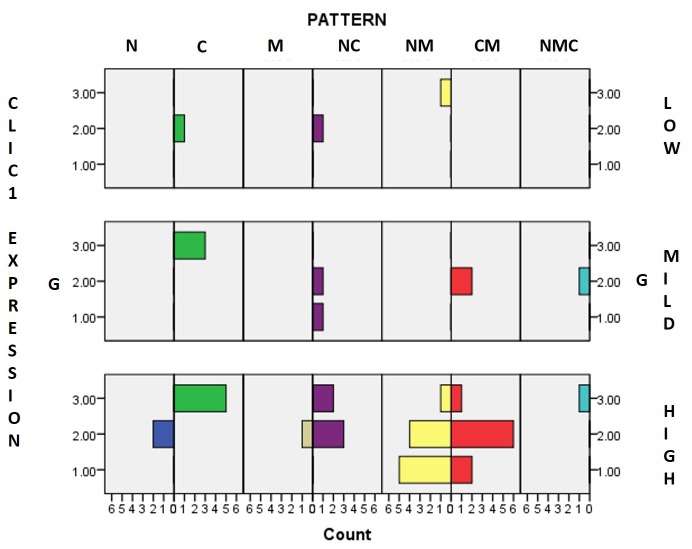
By analyzing our results we observed a high and dynamic heterogeneity of seven patterns related to tumor grade and CLIC1 expressing pattern. We also observed that in tumours with strong expression of CLIC1, the pattern of heterogeneity significantly increased depending on G. Pure N pattern (without any combination) was found only in CLIC1-high expressing group, for G2 tumors. Pure C pattern was persistently expressed for low-, mild- and high-subclasses, but it gradually increased in high-expressing tumors. Another important observation was that in the low-expressing group the C pattern was related to G2, while in the high-expressing group it was related to G3. Pure M pattern was observed only in CLIC1-high expressing tumors related to G2.
Assessement of combined patterns revealed that the NC pattern was present in all three expressing groups, however, it was most prominent in the high-expressing group.
We found particular dynamics in the NM and CM patterns within the group of high CLIC1-expressing tumors. NM pattern gradually decreased from G1 to G3 tumors, while CM pattern increased for G2 tumors compared to G1. Inside G3 subgroup already containing NM and CM patterns it was also observed the expression of NMC pattern which was not observed in G1 and G2 subgroups of tumors with high CLIC1 expression.
Based on these evidences together with significant correlations found between the parameters assessed in the present study we may define four risk groups of cc RCC depending on G, CLIC1 expression and pattern: i) G3/NM/low CLIC1 expression (1), ii) G2/CM/mild CLIC1 expression (2), iii) G1 or G2 /NM or CM /high CLIC1 expression (3) and iv) G2/M /high CLIC1 expression (4) (Figure 5).
Figure 5. Cc RCC subgroups according to tumor grade (G), CLIC1 low, mild or strong expression and pattern as nuclear (N), cytoplasmic (C), membranar (M), nuclear and cytoplasmic (NC), nuclear and membrane (NM), cytoplasmic and membrane (CM), and nuclear/membrane/cytoplasmic (NMC).
Statistically significant correlations between the different parameters used in the present study are summarized in Table I.
Table I. Correlations between CLIC1 parameters and tumor grade. All values highlighted in yellow support our results regarding the influence of CLIC1 expression on tumor grade (G) and pattern heterogeneity.
*Correlation is significant at the 0.05 level (2-tailed). **Correlation is significant at the 0.01 level (2-tailed).
We also assessed the CLIC1 expression in relation to TNM classification parameters. For cc RCC having a combined nuclear and cytoplasmic CLIC1 expression, M parameter was significantly correlated with G2 (p=0.045) while N parameter showed a strong correlation with percentage of CLIC1 positive tumor cells (p=0.008). For concomitant nuclear and membranar CLIC1 expressing tumors, we found that N parameter had three significant correlations with G (p=0.039), the heterogeneity in CLIC1 expression (p=0.031) and the percentage of positive tumor cells (p=0.026). An interesting difference has been found between males and females regarding TNM parameters associated with CLIC1 expression. In males, T parameter seems to significantly depend on the CLIC1 expression pattern (p=0.014) and N parameter on the percentage of positive tumor cells (p=0.043). No significant correlations of TNM with other parameters used in the present study were identified in the women’s group.
Discussion
CLIC1 is known as „sensor and effector” of different cellular processes involving the overproduction of reactive oxygen species (ROS) in several benign (28,29) and malignant conditions (10,11). Among malignant tumors, renal cell carcinomas are well known as being highly ROS producing tumors and this is strongly correlated with high tumor grade, advance stage and metastatic behaviour (30). Based on these data of high ROS production by RCC, already stated in the literature and related with our results about CLIC1 expression, we may speculate in this moment that the high expression of CLIC1 certified here by our results may acts as sensor or effector of cellular proliferation, invasion and metastasis. Also, there is some experimental evidence supporting oxidative stress inducing epigenetic changes in malignant renal cell lines (31). Despite this evidence, the role of CLIC1 expression in cc RCC has not been reported in the literature before.
The present study examines for the first time the CLIC1 expression in cc RCC. Surprisingly, a high number of cc RCC cases from our cohort showed CLIC1 positivity not only in the malignant cells of the primary tumor, but also in the neoplastic cells found in the surrounding connective tissue or in blood vessels. It might be that the pattern of CLIC expression is a useful tool for a better assessement of local and vascular invasion in cc RCC, as it is able to detect, not only groups of invasive cells, but also isolated cells spreading around the tumor or invading tumor-associated blood vessels.
The fact that CLIC1 is expressed in the nucleus, cytoplasm and/or membrane of tumor cells suggests different possible biological functions, depending on its location. Usually, CLIC1 has a cytoplasmic localization in normal human tissue and it translocates to the plasma membrane during malignant transformation (32). Setti et al. have proved that CLIC1 integration into the plasma membrane of tumor cells induces a high tumorigenic potential of glioblastoma cells. Also, CLIC1 is constitutively expressed in the plasma membrane of stem/progenitor cells (32). Pure M pattern appeared in our study only for ccRCC tumors from high-CLIC1-expressing group with G2. High-expressing group showed the highest heterogeneity regarding the CLIC1 expression pattern, however, most of them were defined by NM or CM pattern. A special attention should be paid to those subgroups containing the M pattern. According with Peretti et al., the fact that CLIC1 membrane expression is crucial for cancer stem cells proliferation it may represent a potential therapeutic target (14). In accordacne with this, cc RCC subgroups with M pattern may be considered of high risk. Our results support the existence of several cc RCC subgroups, but the data are not yet sufficient to prove whether CLIC1 overexpression may be considered as a marker of aggresiveness. Further research should better define these aspects in cc RCC, and assess whether there is a possible role for CLIC1 as a marker for tumor risk stratification.
Another important aspect of CLIC1 role is its possibile involvement with tumor invasion and metastasis. We observed in our study a strong CLIC1 expression in cc RCC cancer cells invading surrounding tissues as well as in cells detected in the lumen of tumor-associated blood vessels. Moreover, focal areas of the strongest expression levels of CLIC1 were also detected at the periphery of several cc RCC. Gurski et al. have conducted the only study regarding CLIC1 expression in cancer cells derived from patients with metastatic and localized kidney cancer (15). They proved that CLIC1 translocation into the membrane acts as an activator of invadopodia development and sustains metastatic potential of cancer cells derived from kidney cancer. The same team demonstrated that a similar mechanism may act also in endothelial cells and promotes new blood vessel formation. In our study we also observed several intratumoral and peritumoral small blood vessels expressing CLIC1. This aspect may be due to exosome-based exchange between tumor cells and tumor microenvironment cells, such as endothelial cells (33). Such a crosstalk could provide a route responsible for vascular invasion and metastasis. Recently, indeed, Thuringer et al. have reported this very crosstalk between glioblastoma cells and endothelial cells that is mediated by CLIC1 released through extracellular vesicles by tumor cells and captured by endothelial cells, which are induced to sprout and form tubes in 3D matrices (34).
CLIC1 has already been reported as a potential biomarker for some malignant tumors (6,10,16) and a potential therapeutic target for new treatments in cancer. Its expression in both tumor and endothelial cells may pave the way for the development of future anti-CLIC1 therapy, able to target both tumor and its associated vessels.
There are some data available concerning the inhibitory effects of metformin (a well-known oral antidiabetic drug, recently and intensely spotlighted as having anti-cancer effects by suppressing oncogenic signaling pathways, including receptor tyrosine kinase, PI3K/Akt, and mTOR pathways) on CLIC1 expression in malignant tumors, especially against cancer stem cells (35). Among all the different mechanisms proposed for metformin-mediated antiproliferative activity there is also the inhibition of CLIC1 activity. Gastric cancer cell lines SGC-7901 and MGC-803 express CLIC1 (36). The specific inhibitor indanyloxyacetic acid-94 (IAA94) prevents CLIC1 from translocating in the cell membrane and through this action it prevents cell aggregation and proliferation (37). Silencing of the CLIC1 promotes apoptosis and decreases gallbladder cancer cells proliferation and migration (38).
In this study we report, for the first time, the expression and histopathological significance of CLIC1 in human cc RCC specimens, as detected by immunohistochemistry. CLIC1 expression stratified cc RCC in four subgroups depending on tumor grade, CLIC1 expression pattern and percent of tumor-positive cells. Further investigations are needed to elucidate the prognostic and therapeutic impact of CLIC1in cc RCC.
Conflicts of Interest
The Authors have no conflict of interests to declare.
Authors’ Contributions
AN wrote the paper and participated into the study. MR designed the study. MR and AMC evaluated histopathology and immunohisto-chemistry of the specimens, revised the first draft of the manuscript. AA and II are surgeons participating to tumor removal. TCC, CP and MM provided the antibody and participated to the manuscript writing.
Acknowledgements
The Authors are grateful for the financial support provided by Victor Babes University of Medicine and Pharmacy Timisoara, from its internal research funds during implementation of the project RCC AMIRA, pIII-C4-PCFI-2016/2017-01. They would also like to thank the histotechnologists Ciprian Onica and Patricia Berzava for their excellent technical support.
Figure 2. Homogeneous (A) versus heterogeneous (B) distribution of CLIC1 immunopositive reaction inside cc RCC (CLIC1 immunohistochemistry, ×40 magnification).
References
- 1.Wang W, Xu X, Wang W, Shao W, Li L, Yin W, Xiu L, Mo M, Zhao J, He Q, He J. The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumour Biol. 2011;32:1199–1208. doi: 10.1007/s13277-011-0223-0. PMID: 21858536. DOI: 10.1007/s13277-011-0223-0. [DOI] [PubMed] [Google Scholar]
- 2.Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN, Curmi PM. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279:9298–9305. doi: 10.1074/jbc.M308444200. PMID: 14613939. DOI: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- 3.Goodchild SC, Howell MW, Cordina NM, Littler DR, Breit SN, Curmi PM, Brown LJ. Oxidation promotes insertion of the CLIC1 chloride intracellular channel into the membrane. Eur Biophys J. 2009;39(1):129–138. doi: 10.1007/s00249-009-0450-0. PMID: 19387633. DOI: 10.1007/s00249-009-0450-0. [DOI] [PubMed] [Google Scholar]
- 4.Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta. 2015;1848:2523–2531. doi: 10.1016/j.bbamem.2014.12.012. PMID: 25546839. DOI: 10.1016/j.bbamem.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Li BP, Mao YT, Wang Z, Chen YY, Wang Y, Zhai CY, Shi B, Liu SY, Liu JL, Chen JQ. CLIC1 Promotes the Progression of Gastric Cancer by Regulating the MAPK/AKT Pathways. Cell Physiol Biochem. 2018;46:907–924. doi: 10.1159/000488822. PMID: 29669336. DOI: 10.1159/000488822. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Xu J, Feng J, Li J, Jiang C, Li X, Zou S, Wang Q, Li Y. Expression of CLIC1 as a potential biomarker for oral squamous cell carcinoma: a preliminary study. Onco Targets Ther. 2018;11:8073–8081. doi: 10.2147/OTT.S181936. PMID: 30519049. DOI: 10.2147/ OTT.S181936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Setti M, Osti D, Richichi C. Extracellular vesicle-mediated transfer of CLIC1 protein is a novel mechanism for the regulation of glioblastoma growth. Oncotarget. 2015;6:31413–31427. doi: 10.18632/oncotarget.5105. PMID: 26429879. DOI: 10.18632/oncotarget.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbieri F, Würth R, Pattarozzi A, Verduci I, Mazzola C, Cattaneo M G, Tonelli M, Solari A, Bajetto A, Daga A, Vicentini L M, Mazzanti M, Florio T. Inhibition of chloride intracellular channel 1 (CLIC1) as biguanide class-effect to impair human glioblastoma stem cell viability. Front Pharmacol. 2018;9:899. doi: 10.3389/fphar.2018.00899. PMID: 30186163. DOI: 10.3389/fphar.2018. 00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Dong Q, Zhang B, Wang X, Ye B, Zhang F, Song X, Gao G, Mu J, Wang Z, Ma F, Gu J. Chloride intracellular channel 1 (CLIC1) is activated and functions as an oncogene in pancreatic cancer. Med Oncol. 2015;32:616. doi: 10.1007/s12032-015-0616-9. PMID: 25920608. DOI: 10.1007/s12032-015-0616-9. [DOI] [PubMed] [Google Scholar]
- 10.Singha B, Harper SL, Goldman AR, Bitler BG, Aird KM, Borowsky ME, Cadungog MG, Liu Q, Zhang R, Jean S, Drapkin R, Speicher DW. CLIC1 and CLIC4 complement CA125 as a diagnostic biomarker panel for all subtypes of epithelial ovarian cancer. Sci Rep. 2018;8:14725. doi: 10.1038/s41598-018-32885-2. PMID: 30282979. DOI: 10.1038/s41598-018-32885-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi T, Shiozaki A, Nako Y, Ichikawa D, Kosuga T, Shoda K, Arita T, Konishi Η, Komatsu S, Kubota T, Fujiwara H, Okamoto K, Kishimoto M, Konishi E, Marunaka Y, Otsuji E. Chloride intracellular channel 1 as a switch among tumor behaviors in human esophageal squamous cell carcinoma. Oncotarget. 2018;9:23237–23252. doi: 10.18632/oncotarget.25296. DOI: 10.18632/oncotarget.25296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao PC, Chang YH. CLIC1 (chloride intracellular channel 1) Atlas of genetics and cytogenetics in oncology and haematology. 2011;16:266–269. [Google Scholar]
- 13.Hossain KR, Holt SA, Le Brun AP, Al Khamici H, Valenzuela SM. X-ray and neutron reflectivity study shows that CLIC1 undergoes cholesterol-dependent structural reorganization in lipid monolayers. Langmuir. 2017;33:12497–12509. doi: 10.1021/acs.langmuir.7b02872. PMID: 29016141. DOI: 10.1021/acs.langmuir.7b02872. [DOI] [PubMed] [Google Scholar]
- 14.Peretti M, Raciti FM, Carlini V, Verduci I, Sertic S, Barozzi S, Garré M, Pattarozzi A, Daga A, Barbieri F, Costa A, Florio T, Mazzanti M. Mutual Influence of ROS, pH, and CLIC1 membrane protein in the regulation of G1-S phase progression in human glioblastoma stem cells. Mol Cancer Ther. 2018;17:2451–2461. doi: 10.1158/1535-7163.MCT-17-1223. PMID: 30135216. DOI: 10.1158/1535-7163.MCT-17-1223. [DOI] [PubMed] [Google Scholar]
- 15.Gurski LA, Knowles LM, Basse PH, Maranchie JK, Watkins SC, Pilch J. Relocation of CLIC1 promotes tumor cell invasion and colonization of fibrin. Mol Cancer Res. 2014;13:273–280. doi: 10.1158/1541-7786.MCR-14-0249. PMID: 25205595. DOI: 10.1158/1541-7786.MCR-14-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Yin M, Huang B, Wang Y, Li X, Lou G. CLIC1 a novel biomarker of intraperitoneal metastasis in serous epithelial ovarian cancer. Tumour Biol. 2015;36:4175–4179. doi: 10.1007/s13277-015-3052-8. PMID: 25582317. DOI: 10.1007/s13277-015-3052-8. [DOI] [PubMed] [Google Scholar]
- 17.Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2018;47:900–908. doi: 10.1093/nar/gky939. DOI: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel RL, Mille KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. PMID: 30620402. DOI: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 19.Massari F, Di Nunno V, Mollica V, Graham J, Gatto L, Heng D. Adjuvant tyrosine kinase inhibitors in treatment of renal cell carcinoma: A meta-analysis of available clinical trials. Clin Genitourin Cancer. 2019;1558-7673:30005–30009. doi: 10.1016/j.clgc.2018.12.011. PMID: 30704796. DOI: 10.1016/j.clgc.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Volpe A, Bollito E, Bozzola C, Di Domenico A, Bertolo R, Zegna L, Duregon E, Boldorini R, Porpiglia F, Terrone C. Classification of histologic patterns of pseudocapsular invasion in organ-confined renal cell carcinoma. Clin Genitourin Cancer. 2016;14:69–75. doi: 10.1016/j.clgc.2015.07.020. PMID: 26337654. DOI: 10.1016/j.clgc.2015. 07.020. [DOI] [PubMed] [Google Scholar]
- 21.Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG, Kolenko VM. Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Mol Cancer Ther. 2018;17:1355–1364. doi: 10.1158/1535-7163.MCT-17-1299. PMID: 29967214. DOI: 10.1158/1535-7163.MCT-17-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duran I, Lambea J, Maroto P, González-Larriba JL, Flores L, Granados-Principal S, Graupera M, Sáez B, Vivancos A, Casanovas O. Resistance to targeted therapies in renal cancer: The importance of changing the mechanism of action. Target Oncol. 2017;12:19–35. doi: 10.1007/s11523-016-0463-4. PMID: 27844272. DOI: 10.1007/s11523-016-0463-4. [DOI] [PubMed] [Google Scholar]
- 23.Lichner Z, Saleeb R, Butz H, Ding Q, Nofech-Mozes R, Riad S, Farag M, Varkouhi AK, Dos Santos CC, Kapus A, Yousef GM. Sunitinib induces early histomolecular changes in a subset of renal cancer cells that contribute to resistance. FASEB J. 2019;33:1347–1359. doi: 10.1096/fj.201800596R. PMID: 30148679. DOI: 10.1096/fj.2018 00596R. [DOI] [PubMed] [Google Scholar]
- 24.Motzer RJ, Mazumdar M, Bacik J, Berg W, Amsterdam A, Ferrara J. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J Clin Oncol. 1999;17:2530–2540. doi: 10.1200/JCO.1999.17.8.2530. PMID: 10561319. DOI: 10.1200/JCO.1999.17.8.2530. [DOI] [PubMed] [Google Scholar]
- 25.Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, Shi T, Belldegrun AS, Horvath S, Figlin RA. Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. J Urol. 2005;173:1496–1501. doi: 10.1097/01.ju.0000154351.37249.f0. PMID: 15821467. DOI: 10.1097/01.ju.0000154351.37249.f0. [DOI] [PubMed] [Google Scholar]
- 26.Tulk BM, Edwards JC. NCC27, a homolog of intracellular Cl- channel p64, is expressed in brush border of renal proximal tubule. Am J Physiol. 1998;274:1140–1149. doi: 10.1152/ajprenal.1998.274.6.F1140. PMID: 9841507. DOI: 10.1152/ajprenal.1998.274.6.F1140. [DOI] [PubMed] [Google Scholar]
- 27.Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. PMID: 24025520. DOI: 10.1097/ PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 28.Averaimo S, Milton RH, Duchen MR, Mazzanti M. Chloride intracellular channel 1 (CLIC1): Sensor and effector during oxidative stress. FEBS Lett. 2010;584:2076–2084. doi: 10.1016/j.febslet.2010.02.073. PMID: 20385134. DOI: 10.1016/j.febslet.2010.02.073. [DOI] [PubMed] [Google Scholar]
- 29.Liu B, Billington C K, Henry A P, Bhaker S K, Kheirallah A K, Swan C, Hall IP. Chloride intracellular channel 1 (CLIC1) contributes to modulation of cyclic AMP-activated whole-cell chloride currents in human bronchial epithelial cells. Physiol Rep. 2018;6:13508. doi: 10.14814/phy2.13508. PMID: 29368798. DOI: 10.14814/ phy2.13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesamoni R, Bhattacharyya S, Kumar S, Chauhan A, Mete UK, Agarwal MM, Mavuduru R, Kaushik G, Mandal AK, Singh SK. Status of oxidative stress in patients with renal cell carcinoma. J Urol. 2012;187:1172–1176. doi: 10.1016/j.juro.2011.11.105. PMID: 22335872. DOI: 10.1016/j.juro.2011.11.105. [DOI] [PubMed] [Google Scholar]
- 31.Mahalingaiah P K, Ponnusamy L, Singh K P. Oxidative stress-induced epigenetic changes associated with malignant transformation of human kidney epithelial cells. Oncotarget. 2016;8:11127–11143. doi: 10.18632/oncotarget.12091. PMID: 27655674. DOI: 10.18632/ oncotarget.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setti M, Savalli N, Osti D, Richichi C, Angelini M, Brescia P, Fornasari L, Carro M S, Mazzanti M, Pelicci G. Functional role of CLIC1 ion channel in glioblastoma-derived stem/ progenitor cells. J Natl Cancer Inst. 2013;105:1644–1655. doi: 10.1093/jnci/djt278. PMID: 24115360. DOI: 10.1093/jnci/djt278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maia J, Caja S, Strano Moraes ΜC, Couto N, Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev Biol. 2018;6:18. doi: 10.3389/fcell.2018.00018. PMID: 29515996. DOI: 10.3389/fcell.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thuringer D, Chanteloup G, Winckler P, Garrido C. The vesicular transfer of CLIC1 from glioblastoma to microvascular endothelial cells requires TRPM7. Oncotarget. 2018;9:33302–33311. doi: 10.18632/oncotarget.26048. PMID: 30279961. DOI: 10.18632/oncotarget.26048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gritti M, Würth R, Angelini M, Barbieri F, Peretti M, Pizzi E, Pattarozzi A, Carra E, Sirito R, Daga A, Curmi P M, Mazzanti M, Florio T. Metformin repositioning as antitumoral agent: selective antiproliferative effects in human glioblastoma stem cells, via inhibition of CLIC1-mediated ion current. Oncotarget. 2014;5:11252–11268. doi: 10.18632/oncotarget.2617. PMID: 25361004. DOI: 10.18632/ oncotarget.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma P F, Chen J Q, Wang Z, Liu J L, Li B P. Function of chloride intracellular channel 1 in gastric cancer cells. World J Gastroenterol. 2012;18:3070–3080. doi: 10.3748/wjg.v18.i24.3070. PMID: 22791942. DOI: 10.3748/wjg.v18.i24.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Zhu J, Hu X, Wang C, Lu D, Gong C, Yang J, Zong L. CLIC1 inhibition attenuates vascular inflammation, oxidative stress, and endothelial injury. PLoS One. 2016;11:0166790. doi: 10.1371/journal.pone.0166790. PMID: 27861612. DOI: 10.1371/journal.pone.0166790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y M, Zhang Z L, Liu Q Y, Xiao Y S, Wei L, Xi C, Nan X. Effect of CLIC1 gene silencing on proliferation, migration, invasion and apoptosis of human gallbladder cancer cells. JCMM. 2018;22:2569–2579. doi: 10.1111/jcmm.13499. PMID: 29516682. DOI: 10.1111/ jcmm.13499. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]