Abstract
The exact molecular mechanisms underlying cervical tumorigenesis are poorly understood. Polycomb complex protein Bmi1 (Bmi1) is involved in the malignant transformation and biological aggressiveness of several human carcinomas. Therefore, the present study assessed the expression of Bmi1 protein in human cervical cancer tissues and examined the mechanisms involved in cervical carcinogenesis. The expression of Bmi1 protein was examined by immunohistochemistry in cervical carcinoma tissues (n=71), high-grade squamous intraepithelial lesions (n=41) and normal cervical tissues (n=47). Expression of Bmi1 protein gradually increased across samples from the normal cervix (1/47; 2.12%), high-grade squamous intraepithelial lesions (5/42; 16.13%) and cervical carcinomas (31/71; 43.66%; P<0.05). Additionally, Bmi1 protein expression was associated with tumor histopathological grade. The effects of Bmi1 silencing and overexpression on tumor sphere formation and the tumorigenicity of cervical cancer cells were investigated. Overexpression of Bmi1 resulted in significantly attenuated tumor formation and tumor sphere formation. Consistently, Bmi1 silencing significantly inhibited tumor formation and tumor sphere formation. Furthermore, Bmi1 upregulated the expression of Sox2, and the dual-luciferase reporter assay and chromatin immunoprecipitation showed that Bmi1 transactivated Sox2 by binding to the two E-box motifs in the Sox2 promoter. In conclusion, aberrantly elevated Bmi1 promotes cervical cancer tumorigenicity and tumor sphere formation via enhanced transcriptional regulation of Sox2 genes as a potential oncogenic factor that participates in the carcinogenesis of cervical carcinomas.
Keywords: polycomb complex protein Bmi1, Sox2, cervical cancer, cancer stem cell, self-renewal
Introduction
According to cancer stem cell (CSC) theory, only some cells within a tumor will initiate tumorigenic growth (1). It is becoming evident that cancer treatment that fails to eliminate cancer stem cells may allow for regrowth of the tumor (2). CSCs have been identified and isolated from hematopoietic malignancies and solid tumors, including glioblastoma, breast cancer, colon cancer, hepatocellular carcinoma and other types of tumors (3).
Cervical cancer is the fourth most common malignancy affecting women worldwide, and 250,000 deaths have been estimated annually (4). Notably, the high mortality rate of patients is mainly due to poor loco-regional control, including local tissue invasion by the primary tumor and regional lymph node involvement, rather than distant metastasis. Human papillomavirus (HPV) infection is a factor in the development of most cases of cervical cancer (5). At the same time, HPV infection alone is not sufficient to generate a fully malignant phenotype. Thus, the exact molecular and genetic mechanisms of malignant transformation remain to be explored. The prospective discrimination and isolation of cervical cancer stem cells are the most important steps in elucidating cervical carcinogenesis and establishing new therapeutic approaches for this cancer type.
It has been suggested that polycomb complex protein Bmi1 (Bmi1) is associated with cancer initiation and progression in various types of tumor-initiating cells, and plays important roles in the development and progression of carcinomas (6). The Bmi1 gene belongs to the polycomb gene family and is a component of polycomb repressive complex 1, which is implicated in the stable maintenance of gene repression (7). Bmi1 was first isolated as an oncogene that cooperates with c-Myc in the generation of lymphomas in mice (8). Various types of human cancer display a similar pattern in terms of overexpression of Bmi1, such as non-small cell lung cancer, breast cancer, colorectal cancer, prostate cancer and nasopharyngeal carcinoma (9).
Sox2, a major transcription factor belonging to group B of the SOX family, is a key transcription factor in embryonic development and plays a critical role in determining the fate of stem cells. In our previous study, it was demonstrated that Sox2 may participate in the carcinogenesis of cervical carcinomas (10).
Both Bmi1 and Sox2 are specific markers of neural stem cells (11,12). Meanwhile, Bmi1 and Sox2 are both overexpressed in some human cancer types, such as non-small cell lung cancer, breast cancer and cervical cancer (13). Until now, no evidence has supported the interaction between Bmi1 and Sox2 in cervical carcinogenesis, to the best of our knowledge. The present study evaluated Bmi1 expression using immunohistochemical staining of tumor tissues from patients with cervical squamous cell carcinoma, analyzed its role in cervical carcinogenesis, and explored the relationship between Bmi1 and Sox2.
Materials and methods
Clinical specimens
A total of 112 cervical tissue specimens, including high-grade squamous intraepithelial lesions (HSILs; n=41) and cervical cancer (n=71), were examined, and their clinicopathological backgrounds are summarized in Table I. The samples were derived from either surgical resection or biopsy. A total of 47 histologically normal cervical specimens were obtained from biopsy materials. All the specimens were obtained from the Department of Gynecology, First Affiliated Hospital of Xi'an Jiaotong University between January 2016 and December 2017, and were fixed in 10% formalin for 30 min at room temperature and embedded in paraffin. Serial sections, 4 µm in thickness, were cut from each paraffin block. A total of two pathologists, in a blinded fashion, processed several sections for histopathology studies according to the International Federation of Gynecology and Obstetrics classification system (14), while the remaining sections were processed for subsequent immunohistochemistry.
Table I.
Relationship between Bmi1 or Sox2 expression and the characteristics of the cervical squamous carcinomas.
| Bmi-1 | SOX-2 | ||||||
|---|---|---|---|---|---|---|---|
| Characteristics | n | low | high | P-value | Negative | Positive | P-value |
| Age, years | |||||||
| ≤45 | 33 | 18 | 15 | 0.777 | 8 | 25 | 0.655 |
| >45 | 38 | 22 | 16 | 11 | 27 | ||
| SCC differentiation | |||||||
| Well | 18 | 14 | 4 | 0.034 | 9 | 9 | 0.015 |
| Moderate-poor | 53 | 26 | 27 | 10 | 43 | ||
| Lymph node metastasis | |||||||
| Absence | 57 | 34 | 23 | 0.256 | 13 | 44 | 0.178 |
| Present | 14 | 6 | 8 | 6 | 8 | ||
| Clinical stage | |||||||
| I–II | 60 | 35 | 25 | 0.516 | 15 | 45 | 0.470 |
| III–IV | 11 | 5 | 6 | 4 | 7 | ||
Bmi1, polycomb complex protein Bmi1.
Immunohistochemistry
The slides were deparaffinized in xylene and then were rehydrated through an ethanol gradient. Following a rinse in PBS, the sections were heated at 120°C in 0.01 mol/l sodium citrate buffer (pH 6.0) for 2 min to for antigen retrieval. After blocking nonspecific reactions with the 3% H2O2 for 30 min at room temperature (Sigma-Aldrich; Merck KGaA), the sections were incubated with Bmi1 antibody (1:150; EMD Millipore; cat. no. 05-1322) and Sox2 antibody (1:100; Santa Cruz Biotechnology, Inc.; cat. no. sc-17320) at 4°C overnight. The sections were then stained with horseradish peroxidase (HRP)-labeled goat-mouse and rabbit-goat (1:100; OriGene Technologies, Inc.; cat. nos. SPN-9001 and SPN-9002) at 30°C for 20 min. The expression of proteins was stained with 3,3′-diaminobenzidine (DAB). A similar dilution of the control mouse or control goat IgG (1:100; R&D Systems, Inc.; cat. nos. MAB0031 and AB-108-C) was applied instead of the primary antibody as a negative control. The degree of immunostaining of each formalin-fixed, paraffin-embedded section was reviewed and scored by two independent observers. The immunoreactivity of Bmi1 and Sox2 was semi quantitatively analyzed by light microscope, according methods from previous studies (15). In brief, the percentage of positive cells was divided into five categories of score: <10, 0; 10–25, 1; 25–50, 2; 50–75, 3; and >75%, 4. The intensity of staining was divided into four categories of score: No staining, 0; light brown, 1; brown, 2; and dark brown, 3. Bmi1 and Sox2 staining was assessed by the following formula: Immunohistochemistry score × intensity score. An overall score <3 was defined as negative; >3 and <6 as weak positive; and >6 as strong positive. All scores were evaluated by two different researchers at a magnification of ×400.
Cell lines and cell culture
The human cervical cancer cell lines SiHa, HeLa and C33A were purchased from the American Type Culture Collection. All the cervical cancer cells were grown in DMEM (Sigma Aldrich; Merck KGaA) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/m streptomycin at 37°C in an atmosphere of 5% CO2. Tera-1 cells were cultured in McCoy 5A medium (Sigma-Aldrich; Merck KGaA) with 15% FBS at 37°C in an atmosphere with 5% CO2.
Western blotting
Protein samples from each lysate from fresh cells treated with RIPA lysis buffer (cat. no. sc-24948; Santa Cruz Biotechnology, Inc.) were firstly quantified with a protein bicinchoninic acid kit (Pierce-23225; Thermo Fisher Scientific, Inc.), loaded and separated by 10% SDS-PAGE with 30 ng per lane, and then transferred to PVDF membranes. The PVDF membranes were blocked with fat-free milk at a concentration of 5% for 1 h at room temperature. The membranes were probed with anti-Bmi1 (1:1,000; EMD Millipore; cat. no. 05-1322), anti-Sox2 (1:500; Santa Cruz Biotechnology, Inc.; cat. no. sc-17320), and anti-actin (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-47778) at 4°C overnight. After reacting with HRP-conjugated anti-mouse or anti-goat immunoglobulin (1:10,000; cat. nos. G-21040 and 81-1620; Thermo Fisher Scientific Inc.) for 1 h at room temperature, the proteins were detected using enhanced chemiluminescence (EMD Millipore).
Vector construction and transfection
Full-length Bmi1 cDNA was amplified (16), and then the Bmi1 cDNA fragment was subsequently cloned into the EcoRI and BamHI sites of the internal ribosome entry site vector pIRES2-AcGFP (Clontech Laboratories, Inc.) to generate the pIRES2-AcGFP-Bmi1 recombinant plasmid. The Bmi1 short hairpin (sh)RNA named GenePharma SuperSilencing shRNA™, for which the plasmid was pGPU6/GFP/Neo, was used to specifically silence the expression of Bmi1 (Shanghai GenePharma Co., Ltd.).
All transfection experiments were performed using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. In brief, 105 cells were seeded in 6-well plate and transfected with 2 µg pIRES2-AcGFP-Bmi1 and shBmi1 or empty vector. After culturing in medium containing 1 mg/ml geneticin (cat. no. G418; Genetical; Ameresco, Inc.) for 2–3 weeks, pooled stable clones were isolated. Clones that expressed the Bmi1 cDNA coding region, or Bmi1 silenced cells, were maintained in medium containing 0.8 mg/ml geneticin and used for further investigation.
Tumor sphere formation assay
For clinical cervical cancer samples, the tissues were collected immediately after surgical resection and were then washed, minced and dissociated to single cells using collagenase IV (Sigma-Aldrich; Merck KGaA) at 37°C. After 16 h of digestion, the cells were grown in serum-free stem cell medium containing DMEM/F12 (cat. no. 12400-024, Gibco; Thermo Fisher Scientific, Inc.) supplemented with basic fibroblast growth factor, epidermal growth factor, N2 supplement and B27 supplement. For in vivo propagation, the tumor spheres were dissociated to single cells after 7 days and quantified. Next, 1,000 cells were injected into the subcutaneous tissue in the dorsum of 4- to 6-week-old female BALB/c nude mice (Shanghai SLAC Laboratory Animal Co., Ltd.). A total of 30 mice weighing 12–14 g were housed in SPF conditions with a temperature of 24±2°C and a humidity of 40–60%. Mice had free access to regular chow which was replaced every three days, and were housed on a 12 h light/12 h dark cycle. The xenotransplanted tumor passage in vivo were performed in 3 number of mice (30 number mice for 10-serial-passage). After 60 days, the mice were sacrificed, and the formed tumors were dissected and handled as described previously (17).
For cervical cancer cell lines, the cells were plated at a density of 200 cells/well in 24-well ultralow attachment plates, or at a density of 1 cell/well in 96-well plates, and maintained in stem cell medium at 37°C in an atmosphere of 5% CO2. Tumor spheres that arose within 2 weeks were recorded. For serial tumor sphere formation assays, the spheres were harvested, disaggregated with 0.25% trypsin/EDTA, filtered through a 40-µm mesh and re-plated according to the aforementioned method. For each cell type, triplicate samples were used, and two individuals counted the number of spheres in a blind fashion.
Tumor formation assays
Female 6-week-old NOD/SCID mice (Shanghai SLAC Laboratory Animal Co., Ltd.) weighing 12–14 g were used to assess the tumor formation properties in vivo; A total number of 9 mice (3 mice/group: SiHa-GFP cell and SiHa-Bmi1 cell group; HeLa-GFP cell and HeLa-Bmi1 cell group; and C33A-shControl cell and C33A-shBmi1 cell group) were housed in SPF conditions with a temperature of 24±2°C and a humidity of 40–60%. Mice had free access to regular chow which was replaced every three days, and were housed on a 12 h light/12 h dark cycle. A total of 106 cells were injected into the subcutis on the dorsum. Engrafted mice were inspected twice per week, until the observation was terminated at 8–12 weeks, by visual observation and palpation for the appearance of tumors. The tumor volume (V) was determined by the length (a) and width (b) as follows: V=ab2/2. The experimental protocols were evaluated and approved by the Animal Care and Use Committee of the Medical School of Xi'an Jiaotong University. A portion of each tumor was fixed for 24 h in 10% formaldehyde at room temperature and embedded in paraffin for immunohistochemistry analysis according to the aforementioned protocol.
Dual luciferase reporter assay
Sox2 promoter regions containing 2 E-Box motifs that can bind to the Bmi1 protein were predicted using Promoter 2.0 Prediction Server (http://www-bimas.cit.nih.gov/molbio/proscan/). Promoter luciferase reporters containing the Sox2 promoter regions of −1,000 bp-+1 bp, −900 bp-+1 bp, −400 bp-+1 bp and −100 bp-+1 bp, and pTK-RL plasmids (Promega Corporation), were transiently co-transfected with the Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) into tumor cells (5×104) plated in 24-well plate dishes, while the activities of both firefly and Renilla luciferase reporters were determined 48 h post-transfection using the Dual Luciferase Assay kit (Promega Corporation), according to the manufacturer's instructions. The promoter luciferase reporter activity is presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. The specific activity was displayed as a fold change of the experimental group compared with the control group. All experiments were performed in triplicate.
Quantitative chromatin immunoprecipitation (ChIP)
Quantitative ChIP assays were performed according to the manufacturer's protocol for the EZ-ChIP™ assay kit (cat. no. 17-371; EMD Millipore). Each sample was assayed in triplicate, and the amount of precipitated DNA was calculated as 10% of the input sample. A total of 20 µg Bmi1 antibody was validated to immunoprecipitate the chromatin DNA cross-linked complex with 1% formaldehyde. Normal mouse IgG and histone H3 rabbit monoclonal antibodies were used as the negative and positive controls, respectively. Regions of interest were amplified from precipitated samples by reverse transcription-quantitative PCR (RT-qPCR). Each sample was assayed in triplicate, and the amount of precipitated DNA was calculated as the percentage of the input sample.
RT-qPCR
Total RNA was extracted using TRIzol reagent, according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2 µg total RNA was reverse transcribed using Takara reverse transcriptase (Takara Biotechnology Co., Ltd.). A volume of 2.0 µl of each diluted cDNA (1:20) was subjected to RT-qPCR in a final volume of 20 µl containing 100 nM of each specific primer and SYBR Green Mix (Takara Biotechnology, Co., Ltd.). The sequences of the primers are supplied in Table SI.
The amplification was carried out as follows: Initial enzyme activation at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and then a dissociation stage using an iQ5 multicolor Real-time PCR Detection System (Bio-Rad, Laboratories, Inc.). The quantification cycle (Cq) value was determined as the point at which the fluorescence exceeded a threshold value preset by the instrument software. Relative expression in each experiment set (fold-change vs. control) was calculated according to comparative Cq method using the formula: RQ=2−ΔΔCq (18).
Database
Expression of Bmi1 and SOX2 in patients with cervical cancer compared to normal cervix tissue was assessed using data from the Gene Expression Profiling Interactive Analysis website (GEPIA; http://gepia.cancer-pku.cn/index.html) based on The Cancer Genome Atlas database (normal cervix, n=13; cervical cancer, n=306).
Statistical analysis
All statistical analyses were carried out using the SPSS 16.0 statistical software package (SPSS, Inc.). A Tukey HSD post hoc test after two-way ANOVA was used to analyze the significance of protein expression in the normal cervix, HSIL and cervical carcinoma samples. The relationship between the expression of Bmi1 and clinicopathological characteristics was assessed by χ2 test. Spearman correlation analysis was used to demonstrate the relationship between Bmi1 and Sox2. All the data are presented as mean ± SD and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of Bmi1 protein in paraffin-embedded cervical tissue specimens
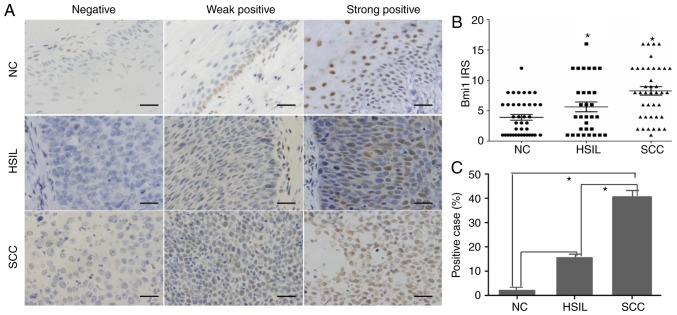
The expression and subcellular localization of Bmi1 protein were determined by immunohistochemistry in 47 paraffin-embedded normal cervix (NC) samples, 41 HSILs, and 71 cervical squamous cell carcinoma (SCC) samples. As shown in Fig. 1, Bmi1 was localized to the nucleus. Among the 47 NC samples, 46 cases exhibited low Bmi1 expression (weakly positive or no staining), and high Bmi1 expression (strongly positive) was found only in 1 case (2.12%; Fig. 1A). Of the low Bmi1 expression cases, 14 cases showed no staining and 28 cases showed weak staining. In our study, basal cells in the cervical epithelium were Bmi1-positive with 32 weakly staining samples and 1 strongly positive staining sample. A significantly higher frequency of Bmi1 staining was observed in the HSIL (7/41; 16.13%) and SCC samples (31/71; 43.66%; P<0.05; Fig. 1B and C).
Figure 1.
Expression of Bmi1 protein in paraffin-embedded cervical tissue specimens. (A) Immunohistochemical staining for Bmi1 protein in 47 NC, 41 HSIL and 71 SCC specimens was performed and then the expression was divided into three staining levels: Negative, weakly positive and strongly positive. (B) Semiquantitative analysis and (C) the rate of positive Bmi1 expression are illustrated. Bars indicate SD. *P<0.05. NC, normal cervical; HSIL, high-grade squamous intraepithelial lesion; SCC, squamous cervical carcinoma; Bmi1, polycomb complex protein Bmi1; IRS, immunoreactivity score.
Statistical evaluation of the immunohistochemistry was compared with clinical reports and pathological findings (Table I). The expression level of Bmi1 was significantly associated with histological grade (P<0.05) but was not associated with age, tumor size, lymph node metastasis or clinical stage.
Bmi1 promotes tumorigenicity in cervical cancer
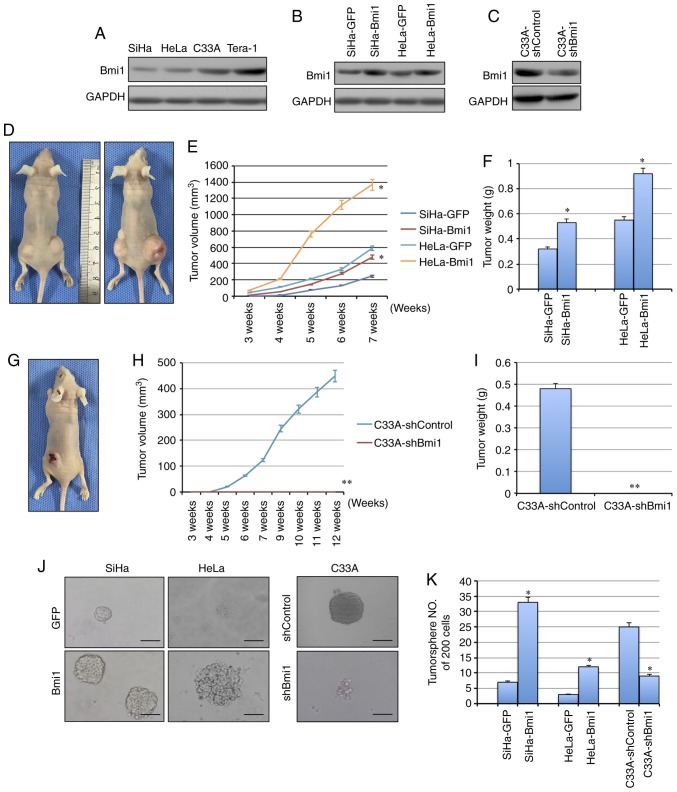
Bmi1 is associated with the initiation and progression of various types of tumor-initiating cells and plays important roles in the development and progression of carcinomas. Here, to determine whether Bmi1 affects the tumorigenicity of cervical cancer cell lines, the expression of Bmi1 was evaluated in cervical cancer cell lines by western blotting (Fig. 2A). Next, exogenous Bmi1 in SiHa and HeLa cells was overexpressed by stable gene transfection, and the expression of Bmi1 was knocked down in C33A cells by shRNA (Fig. 2B and C). To test the tumor formation ability, cells were subcutaneously injected into nude mice. When 105 cells were inoculated into nude mice, the tumors formed by Bmi1-overexpressed SiHa cells grew more quickly and to a larger size, with an average net weight of 532±6.23 mg, than those formed by control cells, with an average net weight of 324±4.76 mg, in three mice/group (P<0.05; Fig. 2D-F). Similarly, HeLa-Bmi1 cells formed tumors of 925±12.32 mg, which were twice as large as those with HeLa-GFP cells (P<0.05; Fig. 2D-F). Additionally, Bmi1-silenced C33A cells formed tumors that grew more slowly and to a smaller size than those formed by C33A-shControl cells (482±6.02 mg vs. 0; P<0.001; Fig. 2G-I), suggesting that Bmi1 contributes to the tumorigenesis of cervical cancer.
Figure 2.
Bmi1 promotes carcinogenicity and tumor sphere formation ability in cervical cancer. (A) The expression of Bmi1 protein in three cervical cancer cell lines was detected by western blotting; the teratoma cell line Tera-1 was used as the positive control. (B) Bmi1 protein expression was evaluated in Bmi1-overexpressing SiHa and HeLa cells by western blotting. (C) Bmi1 protein expression was evaluated in Bmi1-silenced C33A cells by western blotting. (D) Tumor growth was assessed over (E) 7 weeks, as well as the (F) the tumor net weight of nude mice inoculated with SiHa-Bmi1, SiHa-GFP, HeLa-Bmi1 and HeLa-GFP cells with three replicates (1×105 cells). Additionally, (G) tumor growth was measured over (H) 12 weeks, as well as the (I) tumor weights of nude mice inoculated with C33A-shBmi1 and C33A-shControl cells with three replicates (1×106 cells). (J) The tumor sphere formation assay was performed in FBS-free medium in low-density cultures per 200 cells with six replicates, and then (K) the number of tumor spheres formed by Bmi1-overexpressing SiHa and HeLa cells and Bmi1-silenced C33A cells was calculated. Bars=SE. *P<0.05, **P<0.01 vs. respective control. Bmi1, polycomb complex protein Bmi1; GFP, green fluorescent protein; sh, short hairpin.
Bmi1 promotes tumor sphere formation ability in cervical cancer
Besides the tumor formation ability, CSCs share critical properties with stem cells: Self-renewal and differentiation. The tumor sphere formation assay is recognized as a classical assay for self-renewal. Cells were cultured in serum-free medium under conditions that were optimal for growing tumor spheres. As shown in Fig. 2J, Bmi1-overexpressing SiHa and HeLa cells were generated by consecutive passages of tumor sphere cultures, and these cells formed more tumor spheres (33±4.12/200 cells in SiHa-Bmi1 and 12±1.29/200 cells in HeLa-Bmi1) than the control cells (7±0.87/200 cells in SiHa-GFP and 3±0.41/200 cells in HeLa-GFP) in each culture passage (Fig. 2K; P<0.05). Furthermore, the Bmi1-silenced C33A cells showed significantly decreased tumor sphere formation capacity compared with the control cells (9±1.23/200 cells in C33A-shBmi1 vs 25±3.14/200 cells in C33A-shControl; P<0.05; Fig. 2J and K). Taken together, these findings indicated that Bmi1-expressing cells possibly contain more cells that have self-renewal ability.
Bmi1-positive cervical cancer cells express more stem cell-related proteins
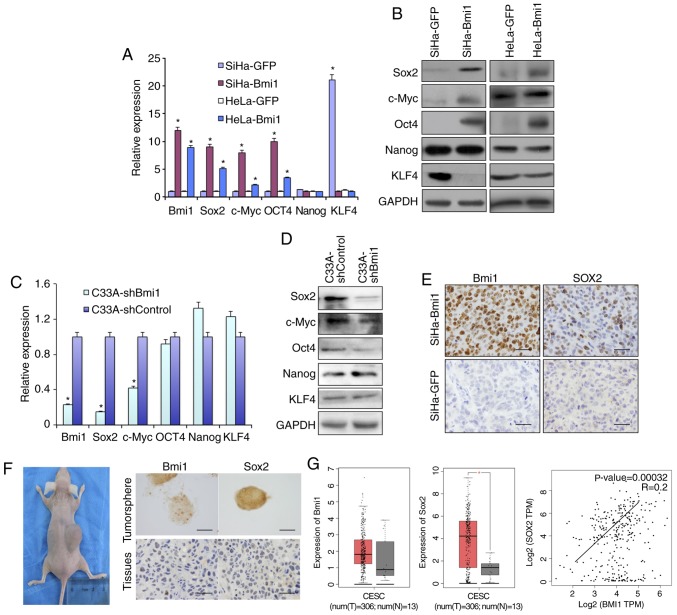
Sox2, Oct4, c-Myc and Nanog are recognized as stem cell self-renewal-related nuclear transcription factors. The present study compared the expression of these transcription factors in Bmi1-overexpressing SiHa and HeLa cells with their respective controls. As shown in Fig. 3, qPCR and western blot analysis (Fig. 3A and B) revealed that Bmi1-overexpressing SiHa and HeLa cells expressed higher levels of Sox2, Oct4 and c-Myc than control cells at both the transcriptional and translational levels. Kruppel like factor 4 (KLF4), a tumor suppressor in Bmi1-overexpressing cells, was downregulated. However, Nanog protein expression was unchanged. Additionally, in Bmi1-silenced C33A cells, the factors Sox2, Oct4 and c-Myc showed lower expression than in C33A-shControl cells by both qPCR and western blotting (Fig. 3C and D).
Figure 3.
Bmi1-positive cervical cancer cells express more stem cell-related genes, including Sox2. The differential expression of several stem cell-related genes, including Sox2, c-Myc, Oct4, Nanog and KLF4, in Bmi1-overexpressing SiHa and HeLa cells and controls was validated by (A) qPCR and (B) western blotting. Additionally, the stem cell-related factors were detected in C33A-shBmi1 and C33A-shControl cells by (C) qPCR and (D) western blotting. (E) Expression of the stem cell-related factor Sox2 in tumor xenografts formed by SiHa-Bmi1 and SiHa-GFP cells was detected by immunohistochemistry. (F) Bmi1 and Sox2 immunoreactivity was predominantly detected in primary cervical tumor spheres and xenografts formed by tumor sphere propagation in vivo. (G) The dataset from the GEPIA repository showed the expression and correlation of Bmi1 and Sox2 in cervical cancer tissues and normal cervical tissues. Error bars represent SD (with three replicates). *P<0.05 vs. respective control. qPCR, quantitative PCR; KLF4, Kruppel like factor 4; Bmi1, polycomb complex protein 1; GFP, green fluorescent protein; sh, short hairpin.
In a previous study, immunofluorescence analysis showed that Bmi1 colocalizes to the nucleus with Sox2 in both normal cervical and cervical carcinoma tissues (16). However, their mechanism of action should be further explored. Here, the Sox2 level was increased in Bmi1-overexpressing cells and was decreased in Bmi1-silenced cells, as revealed by both qPCR and western blotting (Fig. 3A-D). The expression levels of Sox2 were also positively correlated with those of Bmi1 in the Sox2-overexpressing xenograft tumor tissues (Fig. 3E). The present study also detected Sox2 and Bmi1 protein in fresh tissue-derived cervical cancer tumor sphere cells and 10-serial-passage xenograft tissue formed by primary cervical cancer cells. Both Sox2 and Bmi1 staining was found in the nuclei of cervical cancer tumor sphere cells and in 10th xenograft tissue (Fig. 3F). Additionally, as shown in Fig. 3G, by analyzing data from GEPIA, the expression levels of Bmi1 and Sox2 were both upregulated and positively correlated in human cervical tissues (r=+0.2; P<0.05). These results suggest that Bmi1 maintains cervical cancer ‘stemness’, possibly by upregulating the expression of Sox2.
Bmi1 upregulates Sox2 expression by binding to the E-Box region of the Sox2 promoter
It was found that the Sox2 promoter contains 2 E-Box motifs using the prediction software Promoter Scan (Fig. 4A) that could be recognized by Bmi1. To identify the relationship between Bmi1 and Sox2 proteins in cervical carcinoma, the Sox2 promoter-luciferase constructs were transfected into SiHa-Bmi1 and SiHa-GFP cells. The construct with either one or both E-Box motifs in the Sox2 promoter showed the strongest luciferase signals in SiHa-Bmi1 cells compared with those in SiHa-GFP cells (Fig. 4B; P<0.05), suggesting that Bmi1 transactivates Sox2 expression through the E-Box motifs in the Sox2 promoter in cervical carcinoma.
Figure 4.
Bmi1 upregulates Sox2 expression by binding to the E-Box region of the Sox2 promoter. (A) The schematic structure of the Sox2 promoter is shown, including the locations and sequences of putative Bmi1-binding sites and quantitative ChIP primers flanking the regulatory region. (B) The activity of a series of Sox2 promoter deletion mutants was measured using the dual luciferase assay and is presented as a fold change of SiHa-Bmi1 vs. SiHa-GFP cells. (C) Quantitative ChIP assay of the Sox2 promoter region in SiHa cells overexpressing Bmi1 or transfected with empty vector. IgG was used as the negative control. The binding activity is presented as a percentage of the total input. Bars indicate SE. *P<0.05 vs. control. NS, P>0.05. ChIP, chromatin immunoprecipitation; Bmi1, polycomb complex protein Bmi1; GFP, green fluorescent protein; Luc, luciferase; UTR, untranslated region; NS, not significant.
Furthermore, quantitative ChIP assays were used to identify the E-Box of the Sox2 promoter to which Bmi1 binds. Primers were designed to amplify the two specific regions, and a primer pair was used to amplify a 150-bp fragment in the Sox2 3′ untranslated region as a control (Fig. 4A). Following immunoprecipitation using the Bmi1 antibody, the E-Box primer was amplified 8–10-fold more from SiHa-Bmi1 cells than from SiHa-GFP cells (Fig. 4C). However, no significant differences were observed in the control IgG immunoprecipitate for all primers, demonstrating the specificity of Bmi1 binding to the Sox2 promoter in cervical carcinoma cells.
Discussion
Presently, the molecular mechanisms of the initiation and progression of cervical cancer remain unclear, although many genetic factors have been found to be associated with cervical cancer.
Previously, it was shown that deregulation of Bmi1 is associated with the pathogenesis of different human cancer types, including breast cancer (19,20), Ewing sarcoma (21) and leukemia (22,23). Furthermore, Bmi1 was shown to be a useful molecular marker to predict the prognosis of bladder cancer (24) and nasopharyngeal carcinoma (25,26). The significantly high frequency of Bmi1 expression in invasive SCC compared with that in HSIL and NC samples is a finding of great interest. First, Bmi1 staining in the normal cervical epithelium was found only in basal cells where epithelial ‘reserve’ cells are located. It has been suggested that reserve cells appear to be the candidate for cervical stem cells (27), and they play a central role in the pathogenesis of cervical intraepithelial neoplasia. Because Bmi1 is necessary to maintain normal stem cells and cancer stem cells (28), the present experiments indicated that Bmi1 expression is more upregulated in HSILs and invasive cervical cancer than in normal cancer. At the same time, squamous carcinoma stage II and III showed a relatively higher intensity of Bmi1 staining than squamous carcinoma stage I. These results were consistent with those of the previous study. Furthermore, exogenously expressed Bmi1 enhanced tumorigenicity, and knockdown of Bmi1 suppressed tumorigenicity, indicating that KLF4 works as a promoter for cervical cancer. These results support the notion that highly expressed Bmi1 promotes the pathogenesis of cervical carcinoma.
Furthermore, it was found that Bmi1 upregulated the expression of stem cell-related factors such as Sox2, Oct4 and c-Myc. Previous studies have reported that Bmi1 showed colocalization with Sox2 in cervical tissues (16,29). Interestingly, Bmi1 positively regulates factors through binding to the E-Box motif in the proximal region. In this study, promoter analysis revealed that Bmi1 activated Sox2 expression by directly binding to the proximal E-Box sequence of the Sox2 promoter in cervical carcinoma cells. Therefore, the results from our study revealed that the stemness characteristics induced by Bmi1 in cervical carcinoma likely occurs through activating the expression of Sox2.
Additionally, Sox2 belongs to a highly conserved family of high mobility group transcription factors with the sex-determining gene Sry as the first-identified member. Sox proteins are important factors in regulating stem cell biology, and the determination of cell fate and maintenance. Sox2 has also been reported to play a critical role in developmental processes (30), including neural stem cell specification and maintenance (31,32) and lung morphogenesis (33,34). A previous study showed that Sox2 may participate in the carcinogenesis of cervical carcinomas (28). The present study investigated the expression of Bmi1 in human cervical squamous cell carcinoma and its relationship to the expression of Sox2. Interestingly, a high frequency of expression of Bmi1 and Sox2 was observed in invasive cervical carcinomas compared with that in the normal cervix or HSIL. The present study characterized the role of Sox2 and Bmi1 in the maintenance cervical cancer stem cells using tumor sphere formation assays and propagation in vivo. These factors are necessary for stem-cell-like cancer cells in cervical cancer because they are clearly and strongly expressed in stem-cell-like cells of cervical cancer formed under in vitro culture conditions and in vivo, facilitating the proliferation of only CSCs and progenitor cells. The results suggested that the co-deregulation of Sox2 and Bmi1 is a general phenomenon in cervical cancer.
In summary, the present study suggested a general role for Bmi1 and Sox2 in cervical cancer development. The finding that cervical cancer cells require Bmi1 and Sox2 for their tumorigenic activity suggests that interference with Bmi1 and Sox2 activity could be a therapeutic strategy for cervical cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by a grant to Dr Rui Xu from the National Natural Science Foundation of Shaanxi province (grant no. 2012JM4006) in the design of the study, collection, analysis and interpretation of data, and manuscript writing.
Availability of data and materials
The datasets used and analyzed during the current study are available in the GEPIA repository (http://gepia.cancer-pku.cn/detail.php?gene=BMI1; http://gepia.cancer-pku.cn/detail.php?gene=BMI1 and http://gepia.cancer-pku.cn/detail.php?gene=BMI1).
Authors' contributions
WTY analyzed and interpreted the patient data regarding the cervical dissection. RX and LC performed histological examination of the cervix, cell experiments and animal assays, and were major contributors in the writing of the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The institutional review board, named as ‘Ethics Committee of Medical School of Xi'an Jiaotong University’, approved the population study. The Ethics Committee of Medical School of Xi'an Jiaotong University approved the design of the cervical cancer study, including tissue sample collection. The experimental protocols involving animals were evaluated and approved by the Animal Care and Use Committee of the Medical School of Xi'an Jiaotong University.
Patient consent for publication
The patients provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dick JE. Breast cancer stem cells revealed. Proc Natl Acad Sci USA. 2003;100:3547–3549. doi: 10.1073/pnas.0830967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham SM, Jørgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, Holyoake TL. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.V99.1.319. [DOI] [PubMed] [Google Scholar]
- 3.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, Wicinski J, Cabaud O, Charafe-Jauffret E, Birnbaum D, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120:485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 5.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Rountree CB, Ding W, He L, Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cells. 2009;27:290–299. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paavonen J. Human papillomavirus infection and the development of cervical cancer and related genital neoplasias. Int J Infect Dis. 2007;11(Suppl 2):S3–S9. doi: 10.1016/S1201-9712(07)60015-0. [DOI] [PubMed] [Google Scholar]
- 10.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertolini JA, Favaro R, Zhu Y, Pagin M, Ngan CY, Wong CH, Tjong H, Vermunt MW, Martynoga B, Barone C, et al. Mapping the global chromatin connectivity network for Sox2 function in neural stem cell maintenance. Cell Stem Cell. 2019;24:462–476.e6. doi: 10.1016/j.stem.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas D, Hsu JH, Hung L, Cooper A, Abdueva D, van Doorninck J, Peng G, Shimada H, Triche TJ, Lawlor ER. BMI-1 promotes ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res. 2008;68:6507–6515. doi: 10.1158/0008-5472.CAN-07-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spremović-Rađenović S, Stefanović A, Kadija S, Jeremić K, Sparić R. Classification and the diagnostics of abnormal uterine bleeding in nongravid women of reproductive age: The PALM-COEIN classification system adopted by the international federation of gynecology and obstetrics. Vojnosanit Pregl. 2016;73:1154–1159. doi: 10.2298/VSP160709289S. [DOI] [PubMed] [Google Scholar]
- 15.Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH, Band V, Dimri GP. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 2007;67:5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, Yang WT, Zheng PS. Coexpression of B-lymphoma Moloney murine leukemia virus insertion region-1 and sex-determining region of Y chromosome-related high mobility group box-2 in cervical carcinogenesis. Hum Pathol. 2013;44:208–217. doi: 10.1016/j.humpath.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Liu XF, Yang WT, Xu R, Liu JT, Zheng PS. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9:e87092. doi: 10.1371/journal.pone.0087092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Wiederschain D, Chen L, Johnson B, Bettano K, Jackson D, Taraszka J, Wang YK, Jones MD, Morrissey M, Deeds J, et al. Contribution of polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol Cell Biol. 2007;27:4968–4979. doi: 10.1128/MCB.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JH, Yoon SY, Jeong SH, Kim SY, Moon SK, Joo JH, Lee Y, Choe IS, Kim JW. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast. 2004;13:383–388. doi: 10.1016/j.breast.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Chai L, Liu F, Fink LM, Lin P, Silberstein LE, Amin HM, Ward DC, Ma Y. Bi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci USA. 2007;104:10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 24.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, He SS, Cai XY, Chen HY, Yang XL, Lu LX, Chen Y. The novel prognostic score combining red blood cell distribution width and body mass index (COR-BMI) has prognostic impact for survival outcomes in nasopharyngeal carcinoma. J Cancer. 2018;9:2295–2301. doi: 10.7150/jca.24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Hu D, Yang G, Zhou J, Yang C, Gao Y, Zhu Z. Down-regulation of BMI-1 cooperates with artemisinin on growth inhibition of nasopharyngeal carcinoma cells. J Cell Biochem. 2011;112:1938–1948. doi: 10.1002/jcb.23114. [DOI] [PubMed] [Google Scholar]
- 27.Peters WM. Nature of ‘basal’ and ‘reserve’ cells in oviductal and cervical epithelium in man. J Clin Pathol. 1986;39:306–312. doi: 10.1136/jcp.39.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji J, Zheng PS. Expression of Sox2 in human cervical carcinogenesis. Hum Pathol. 2010;41:1438–1447. doi: 10.1016/j.humpath.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Maurizi G, Verma N, Gadi A, Mansukhani A, Basilico C. Sox2 is required for tumor development and cancer cell proliferation in osteosarcoma. Oncogene. 2018;37:4626–4632. doi: 10.1038/s41388-018-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreu-Agullo C, Maurin T, Thompson CB, Lai EC. Ars2 maintains neural stem-cell identity through direct transcriptional activation of Sox2. Nature. 2011;481:195–198. doi: 10.1038/nature10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyagi S, Nishimoto M, Saito T, Ninomiya M, Sawamoto K, Okano H, Muramatsu M, Oguro H, Iwama A, Okuda A. The Sox2 regulatory region 2 functions as a neural stem cell-specific enhancer in the telencephalon. J Biol Chem. 2006;281:13374–13381. doi: 10.1074/jbc.M512669200. [DOI] [PubMed] [Google Scholar]
- 33.Danopoulos S, Alonso I, Thornton ME, Grubbs BH, Bellusci S, Warburton D, Al Alam D. Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am J Physiol Lung Cell Mol Physiol. 2018;314:L144–L149. doi: 10.1152/ajplung.00379.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: Branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available in the GEPIA repository (http://gepia.cancer-pku.cn/detail.php?gene=BMI1; http://gepia.cancer-pku.cn/detail.php?gene=BMI1 and http://gepia.cancer-pku.cn/detail.php?gene=BMI1).