Abstract
Aggressive prolactin pituitary tumors, which exhibit aggressive behaviors and resistance to conventional treatments, are a huge challenge for neurosurgeons. Many studies have investigated the roles of microRNAs (miRNAs) in pituitary tumorigenesis, invasion and metastasis, but few have explored aggressiveness-associated miRNAs in aggressive pituitary tumors. Differentially expressed miRNAs (DEMs) between aggressive and nonaggressive prolactin pituitary tumors were screened using the GSE46294 miRNA expression profile downloaded from the GEO database. The potential target genes of the top three most highly upregulated and downregulated DEMs were predicted by miRTarBase, and potential functional annotation and pathway enrichment analysis were performed using the DAVID database. Protein-protein interaction (PPI) and miRNA-hub gene interaction networks were constructed by Cytoscape software. A total of 43 DEMs were identified, including 19 upregulated and 24 downregulated miRNAs, between aggressive and nonaggressive prolactin pituitary tumors. One hundred and seventy and 680 target genes were predicted for the top three most highly upregulated and downregulated miRNAs, respectively, and these genes were involved in functional enrichment pathways, such as regulation of transcription from RNA polymerase II promoter, DNA-templated transcription, Wnt signaling pathway, protein binding, and transcription factor activity (sequence-specific DNA binding). In the PPI network, the top 10 genes with the highest degree of connectivity of the upregulated and downregulated DEMs were selected as hub genes. By constructing an miRNA-hub gene network, it was found that most hub genes were potentially modulated by hsa-miR-489 and hsa-miR-520b. Targeting hsa-miR-489 and hsa-miR-520b may provide new clues for the diagnosis and treatment of aggressive prolactin pituitary tumors.
Keywords: aggressive pituitary tumor, pituitary carcinoma, prolactinoma, microRNA, bioinformatic analysis
Introduction
Pituitary tumors represent approximately 10–15% of intracranial tumors, of which prolactin-secreting pituitary adenomas (prolactinoma) are the most common subtypes, accounting for 30–40% of pituitary tumors (1,2). Most of these tumors are noninvasive, show slow growth and are easily treated by surgery or medical treatment, including cabergoline and dopamine agonists. However, a small subset, accounting for 2.5–10% of pituitary adenomas, are defined as aggressive pituitary tumors and can exhibit aggressive behaviors, resistance to conventional treatments and/or temozolomide (TMZ), and multiple recurrences despite standard therapies combining surgical, medical and radiotherapy treatment approaches (3,4). Early identification of aggressive pituitary tumors is challenging but is of major clinical importance as these tumors are associated with increased morbidity and mortality (5). Numerous studies have been performed to explore potential predictive and prognostic biomarkers for the molecular pathogenesis underlying the aggressive behavior and malignant transformation of pituitary tumors, yet research results remain fairly unreliable and controversial (4,6,7).
MicroRNAs (miRNAs/miRs) are a large family of short endogenous noncoding RNAs, approximately 21–25 nucleotides in length, that can directly bind to the 3′-untranslated region of messenger RNA (mRNA), thereby leading to suppression of protein translation or mRNA degradation (8,9). Subsequently, miRNAs can negatively regulate the expression of target genes involved in proliferation, apoptosis, cell cycle differentiation, invasion and metabolism (9). Aberrant expression of miRNAs contributes to tumorigenesis, invasion and metastasis by derepressing or silencing key regulatory proteins in various types of tumors, including pituitary adenomas (10–12). Many studies have investigated the roles of miRNAs in pituitary tumorigenesis, dysfunction, neurodegeneration and metastasis by comparing tumoral to normal pituitary tissues (13–16). However, currently, there are few studies that have explored aggressiveness-associated miRNAs in ‘aggressive’ pituitary tumors, especially aggressive prolactinoma, one of the most common subtypes of pituitary adenomas, based on large-scale human tissue datasets.
In recent years, microarray technology and bioinformatic analysis have been widely used to help us discover novel clues to identify reliable and functional miRNAs. In the present study, differentially expressed miRNAs (DEMs, DE-miRNAs) between aggressive and nonaggressive prolactin pituitary tumors were screened using the GSE46294 miRNA expression profile (17). The potential target genes of the top three most highly upregulated and downregulated DE-miRNAs were predicted by miRTarBase. Subsequently, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and protein-protein interaction (PPI) network analyses were performed to help us understand the molecular mechanisms underlying the aggressiveness of pituitary tumors. Finally, 20 hub genes were identified, and an miRNA-hub gene network was constructed by Cytoscape software. In conclusion, our study aimed to explore the aggressiveness-associated miRNAs in aggressive prolactin pituitary tumors and their potential molecular mechanisms based on bioinformatic analysis and to provide candidate biomarkers for early diagnosis and individualized treatment of aggressive prolactin pituitary tumors.
Materials and methods
Microarray data
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) is a public functional genomics data repository of high-throughput gene expression data, chips and microarrays (18). After extensive data screening in the GEO database, only the GSE46294 dataset was selected as it compared the miRNA expression of aggressive and nonaggressive prolactin pituitary tumors (17). GSE46294, based on the GPL13264 platform (Agilent-021827 Human miRNA Microarray), contained four aggressive prolactin pituitary tumor samples and eight nonaggressive prolactin pituitary tumor samples.
Data processing
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is an interactive web tool that can compare different groups of samples from the GEO series to identify DEMs across experimental conditions (19). The DEMs between aggressive and nonaggressive prolactin pituitary tumor samples were screened using GEO2R. Adjusted P-values (adj. P) were applied to correct the false-positive results by using the default Benjamini-Hochberg false discovery rate method. Adj. P<0.01 and |fold change (FC)| >2 were considered the cut-off values for identifying DEMs. A DEM hierarchical clustering heat map was constructed using MeV (Multiple Experiment Viewer, http://mev.tm4.org/), which is a cloud-based application supporting the analysis, visualization, and stratification of large genomic data, particularly RNASeq and microarray data. The potential target genes of the top three most highly upregulated and downregulated DE-miRNAs were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php/), which is a database for experimentally validated miRNA-target interactions (20).
Functional and pathway enrichment analyses
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.ncifcrf.gov/) is an online tool for gene functional classification, which is an essential foundation for high-throughput gene analysis to understand the biological significance of genes (21). DAVID was introduced to perform functional annotation and pathway enrichment analysis, including GO (Gene Ontology) enrichment and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, for the predicted target genes of 6 selected DEMs (22,23). A P-value <0.05 was considered statistically significant.
PPI network construction and module analysis
The target genes obtained from the upregulated and downregulated DEMs were first mapped to the STRING database (http://string-db.org) to assess functional associations among these target genes, with a combined score >0.4 defined as significant (24). Then, PPI networks were constructed using Cytoscape, which is a biological graph visualization software for integrated models of biologic molecular interaction networks (25). The Molecular Complex Detection (MCODE) plugin of Cytoscape was used to identify the most significant module in the PPI networks (26). The criteria for selection were as follows: Degree cut-off=2, node score cut-off=0.2, maximum depth=100 and k-core=2. Moreover, GO and KEGG enrichment analyses were performed using DAVID for genes in the modules.
Hub gene analysis and miRNA-hub gene network construction
Hub genes were selected by considering the high degree of connectivity in the PPI networks analyzed by the cytohubba plugin of Cytoscape. The top 10 genes with the highest degree of connectivity were selected as the hub genes of the upregulated and downregulated DEMs, respectively. Subsequently, GO and KEGG enrichment analyses were performed for the selected 20 hub genes. The biological process analysis of hub genes was performed and visualized using the Biological Networks Gene Oncology tool (BiNGO) plugin of Cytoscape (27). The latest information of functional roles of hub genes was downloaded from GeneCards in Nov. 2018 (https://www.genecards.org/). Subsequently, an miRNA-hub gene network was constructed by Cytoscape.
Results
Identification of DEMs and their target genes
Following analysis of the GSE46294 dataset using GEO2R, a total of 43 DEMs were identified, including 19 upregulated and 24 downregulated miRNAs between aggressive and nonaggressive prolactin pituitary tumors. For better visualization, the top 10 most highly upregulated miRNAs and the top 10 most highly downregulated miRNAs are presented in Table I, and the hierarchical clustering heat map of the DEMs is presented in Fig. S1. According to their FC values, hsa-miR-489, hsa-let-7d* and hsa-miR-138-1* were the top 3 most highly upregulated miRNAs, and hsa-miR-520b, hsa-miR-875-5p and hsa-miR-671-3p were the top 3 most highly downregulated miRNAs (Table I). One hundred seventy potential target genes were predicted for the top 3 most highly upregulated miRNAs and 680 potential target genes were predicted for the top 3 most highly downregulated miRNAs by miRTarBase.
Table I.
Top 10 upregulated and downregulated DEMs between aggressive and nonaggressive prolactin pituitary tumors.
| miRNAs (DEMs) | P-value | t | B | logFC |
|---|---|---|---|---|
| Upregulated | ||||
| hsa-miR-489 | 0.00677 | 3.25 | −4.58 | 7.07 |
| hsa-let-7d* | 0.02591 | 2.53 | −4.58 | 6.09 |
| hsa-miR-138-1* | 0.02569 | 2.54 | −4.58 | 5.26 |
| hsa-miR-886-3p | 0.00191 | 3.94 | −4.58 | 4.36 |
| hsa-miR-576-5p | 0.04773 | 2.2 | −4.59 | 3.83 |
| hsa-miR-135b | 0.01671 | 2.77 | −4.58 | 3.72 |
| hsa-miR-137 | 0.03877 | 2.32 | −4.59 | 3.29 |
| hsa-miR-886-3p | 0.00235 | 3.82 | −4.58 | 3.2 |
| hsa-miR-551b | 0.02074 | 2.66 | −4.58 | 3.04 |
| hsa-miR-296-3p | 0.04524 | 2.23 | −4.59 | 3.02 |
| Downregulated | ||||
| hsa-miR-520b | 0.00732 | −3.21 | −4.58 | −6.36 |
| hsa-miR-875-5p | 0.04037 | −2.29 | −4.59 | −5.66 |
| hsa-miR-671-3p | 0.01453 | −2.85 | −4.58 | −5.49 |
| hsa-miR-372 | 0.00348 | −3.61 | −4.58 | −5.49 |
| hsa-miR-586 | 0.02631 | −2.53 | −4.58 | −5.44 |
| hsa-miR-367* | 0.02421 | −2.57 | −4.58 | −4.84 |
| hsa-miR-302b | 0.01052 | −3.02 | −4.58 | −4.49 |
| hsa-miR-187 | 0.0322 | −2.42 | −4.59 | −4.35 |
| hsa-miR-193b* | 0.02207 | −2.62 | −4.58 | −4.31 |
| hsa-miR-452* | 0.00322 | −3.65 | −4.58 | −4.17 |
miRNA names with ‘*’ are also mature miRNAs as annotated in miRBase (http://www.mirbase.org). For example, hsa-let-7d* is hsa-let-7d-3p; hsa-miR-138-1* is hsa-miR-138-1-3p; hsa-miR-367* is hsa-miR-367-5p; hsa-miR-193b* is hsa-miR-193b-5p; hsa-miR-452* is hsa-miR-452-3p. DEMs, differentially expressed miRNAs; hsa, Homo sapiens.
Functional and pathway enrichment analyses
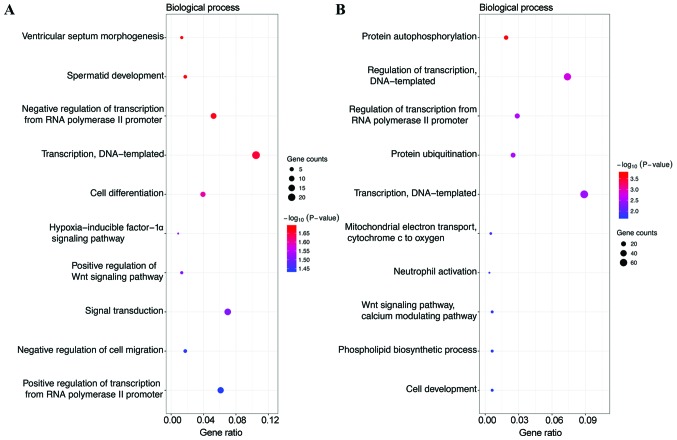
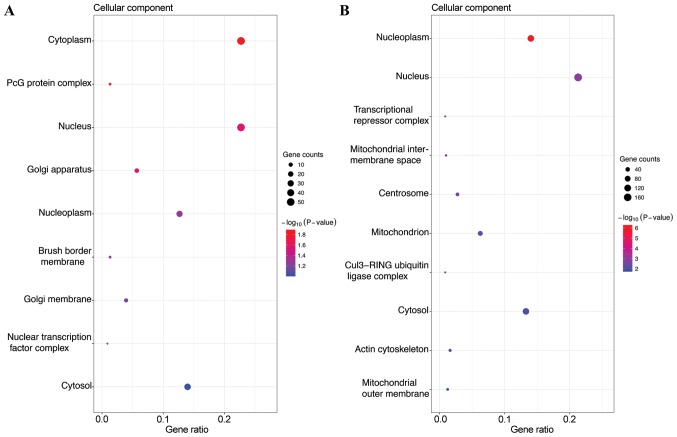
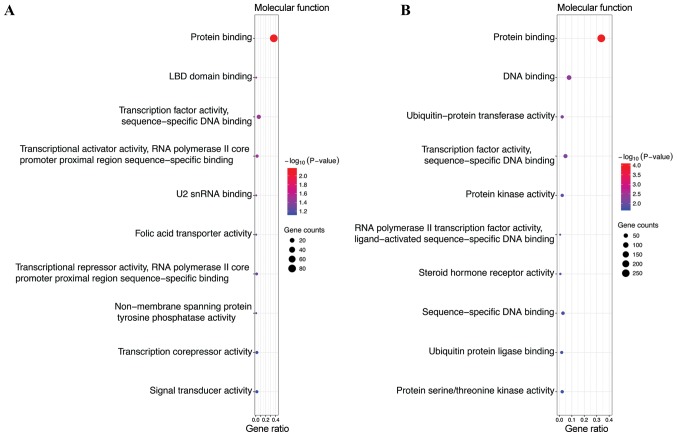
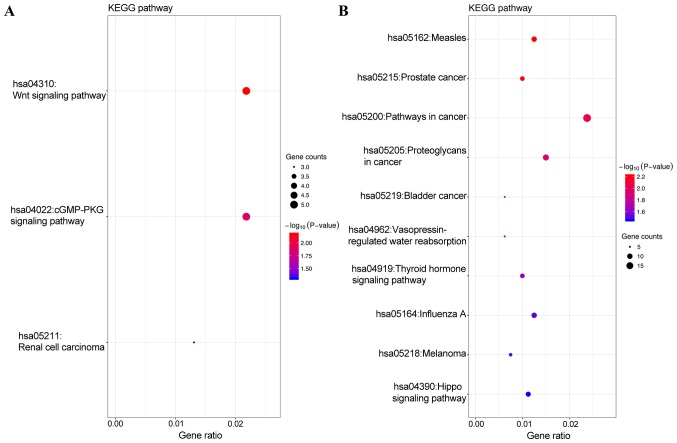
GO analysis, including biological process (BP), cellular component (CC) and molecular function (MF), was performed on the potential target genes of top 3 most highly upregulated miRNAs (Table II) and the top 3 most highly downregulated miRNAs (Table III). GO functional annotation analysis showed that in the BP category, the target genes of the top 3 most highly upregulated miRNAs were significantly enriched in DNA-templated transcription, signal transduction, and positive regulation of transcription from RNA polymerase II promoter (Fig. 1A), while the target genes of the top 3 most highly downregulated miRNAs were enriched in DNA-templated transcription, DNA-templated regulation of transcription, and regulation of transcription from RNA polymerase II promoter (Fig. 1B). In the CC category, the target genes of the top three most highly upregulated miRNAs were significantly enriched in cytoplasm, nucleus and cytosol (Fig. 2A), while the target genes of the top three most highly downregulated miRNAs were enriched in nucleus, nucleoplasm and cytosol (Fig. 2B). In the MF category, the target genes of the top 3 most highly upregulated miRNAs were significantly enriched in protein binding, transcription factor activity, sequence-specific DNA binding, transcriptional activator activity, and RNA polymerase II core promoter proximal region sequence-specific binding (Fig. 3A), while the target genes of the top 3 most highly downregulated miRNAs were enriched in protein binding, DNA binding and transcription factor activity, and sequence-specific DNA binding (Fig. 3B). In addition, KEGG pathway analysis revealed that the target genes of the top 3 most highly upregulated miRNAs were mainly enriched in the Wnt signaling pathway, cGMP-PKG signaling pathway and renal cell carcinoma (Fig. 4A), while the target genes of the top three most highly downregulated miRNAs were mainly enriched in pathways in cancer, proteoglycans in cancer, measles and influenza A (Fig. 4B) (Tables II and III).
Table II.
Functional and pathway enrichment analysis for target genes of the top 3 upregulated miRNAs.
| Category | Term | Pathway description | Count | P-value |
|---|---|---|---|---|
| Upregulated miRNAs | ||||
| GO BP | GO:0060412 | Ventricular septum morphogenesis | 3 | 0.020464503 |
| GO BP | GO:0007286 | Spermatid development | 4 | 0.021020749 |
| GO BP | GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 12 | 0.021742388 |
| GO BP | GO:0006351 | Transcription, DNA-templated | 24 | 0.022393279 |
| GO BP | GO:0030154 | Cell differentiation | 9 | 0.025194909 |
| GO BP | GO:0097411 | Hypoxia-inducible factor-1α signaling pathway | 2 | 0.030146509 |
| GO BP | GO:0030177 | Positive regulation of Wnt signaling pathway | 3 | 0.030678983 |
| GO BP | GO:0007165 | Signal transduction | 16 | 0.030948235 |
| GO BP | GO:0030336 | Negative regulation of cell migration | 4 | 0.036066871 |
| GO BP | GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 14 | 0.03646379 |
| GO CC | GO:0005737 | Cytoplasm | 52 | 0.0134897 |
| GO CC | GO:0031519 | PcG protein complex | 3 | 0.016939042 |
| GO CC | GO:0005634 | Nucleus | 52 | 0.026624876 |
| GO CC | GO:0005794 | Golgi apparatus | 13 | 0.026655792 |
| GO CC | GO:0005654 | Nucleoplasm | 29 | 0.053523267 |
| GO CC | GO:0031526 | Brush border membrane | 3 | 0.054869988 |
| GO CC | GO:0000139 | Golgi membrane | 9 | 0.072820488 |
| GO CC | GO:0044798 | Nuclear transcription factor complex | 2 | 0.078554642 |
| GO CC | GO:0005829 | Cytosol | 32 | 0.094144731 |
| GO MF | GO:0005515 | Protein binding | 83 | 0.007060503 |
| GO MF | GO:0050693 | LBD domain binding | 2 | 0.030452531 |
| GO MF | GO:0003700 | Transcription factor activity, sequence-specific DNA binding | 14 | 0.034027538 |
| GO MF | GO:0001077 | Transcriptional activator activity, RNA polymerase II core sequence-specific binding | 6 | 0.035934263 |
| GO MF | GO:0030620 | U2 snRNA binding | 2 | 0.045331887 |
| GO MF | GO:0008517 | Folic acid transporter activity | 2 | 0.052686367 |
| GO MF | GO:0001078 | Transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding | 4 | 0.054345955 |
| GO MF | GO:0004726 | Non-membrane spanning protein tyrosine phosphatase activity | 2 | 0.059984623 |
| GO MF | GO:0003714 | Transcription corepressor activity | 5 | 0.071931973 |
| GO MF | GO:0004871 | Signal transducer activity | 5 | 0.07295342 |
| KEGG | hsa04310 | Wnt signaling pathway | 5 | 0.006641183 |
| KEGG | hsa04022 | cGMP-PKG signaling pathway | 5 | 0.01255563 |
| KEGG | hsa05211 | Renal cell carcinoma | 3 | 0.049309583 |
In the event there were more than five terms enriched in this category, the top 5 terms were selected per P-value. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; Count, numbers of enriched genes in each term; hsa, Homo sapiens.
Table III.
Functional and pathway enrichment analysis for target genes of the top 3 downregulated miRNAs.
| Category | Term | Description | Count | P-value |
|---|---|---|---|---|
| Downregulated miRNAs | ||||
| GO BP | GO:0046777 | Protein autophosphorylation | 15 | 0.000170888 |
| GO BP | GO:0006355 | Regulation of transcription, DNA-templated | 59 | 0.001639464 |
| GO BP | GO:0006357 | Regulation of transcription from RNA polymerase II promoter | 23 | 0.002882721 |
| GO BP | GO:0016567 | Protein ubiquitination | 20 | 0.002898481 |
| GO BP | GO:0006351 | Transcription, DNA-templated | 71 | 0.003602574 |
| GO BP | GO:0006123 | Mitochondrial electron transport, cytochrome c to oxygen | 4 | 0.014446477 |
| GO BP | GO:0042119 | Neutrophil activation | 3 | 0.017126856 |
| GO BP | GO:0007223 | Wnt signaling pathway, calcium modulating pathway | 5 | 0.018278676 |
| GO BP | GO:0008654 | Phospholipid biosynthetic process | 5 | 0.019902891 |
| GO BP | GO:0048468 | Cell development | 5 | 0.019902891 |
| GO CC | GO:0005654 | Nucleoplasm | 112 | 6.68468E-07 |
| GO CC | GO:0005634 | Nucleus | 170 | 0.001202665 |
| GO CC | GO:0017053 | Transcriptional repressor complex | 7 | 0.002719148 |
| GO CC | GO:0005758 | Mitochondrial intermembrane space | 8 | 0.002811554 |
| GO CC | GO:0005813 | Centrosome | 22 | 0.003323275 |
| GO CC | GO:0005739 | Mitochondrion | 50 | 0.006368195 |
| GO CC | GO:0031463 | Cul3-RING ubiquitin ligase complex | 7 | 0.007248149 |
| GO CC | GO:0005829 | Cytosol | 106 | 0.009417134 |
| GO CC | GO:0015629 | Actin cytoskeleton | 13 | 0.010602362 |
| GO CC | GO:0005741 | Mitochondrial outer membrane | 10 | 0.014535489 |
| GO MF | GO:0005515 | Protein binding | 269 | 9.14069E-05 |
| GO MF | GO:0003677 | DNA binding | 62 | 0.004456722 |
| GO MF | GO:0004842 | Ubiquitin-protein transferase activity | 18 | 0.005935513 |
| GO MF | GO:0003700 | Transcription factor activity, sequence-specific DNA binding | 39 | 0.006954496 |
| GO MF | GO:0004672 | Protein kinase activity | 18 | 0.013435998 |
| GO MF | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding | 5 | 0.013877869 |
| GO MF | GO:0003707 | Steroid hormone receptor activity | 6 | 0.015112205 |
| GO MF | GO:0043565 | Sequence-specific DNA binding | 23 | 0.017535243 |
| GO MF | GO:0031625 | Ubiquitin protein ligase binding | 15 | 0.018757483 |
| GO MF | GO:0004674 | Protein serine/threonine kinase activity | 18 | 0.020173241 |
| KEGG | hsa05162 | Measles | 10 | 0.005987506 |
| KEGG | hsa05215 | Prostate cancer | 8 | 0.006312095 |
| KEGG | hsa05200 | Pathways in cancer | 19 | 0.009215433 |
| KEGG | hsa05205 | Proteoglycans in cancer | 12 | 0.011467731 |
| KEGG | hsa05219 | Bladder cancer | 5 | 0.018691786 |
| KEGG | hsa04962 | Vasopressin-regulated water reabsorption | 5 | 0.023655555 |
| KEGG | hsa04919 | Thyroid hormone signaling pathway | 8 | 0.023895596 |
| KEGG | hsa05164 | Iinfluenza A | 10 | 0.030429321 |
| KEGG | hsa05218 | Melanoma | 6 | 0.032052212 |
| KEGG | hsa04390 | Hippo signaling pathway | 9 | 0.035379929 |
If there were more than five terms enriched in this category, the top 5 terms were selected per the P-value. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; Count, numbers of enriched genes in each term; hsa, Homo sapiens.
Figure 1.
Gene Ontology (GO) functions for the target genes of the top 3 most highly upregulated miRNAs and the top 3 most highly downregulated miRNAs. (A) Enriched biological processes of the upregulated miRNAs; (B) enriched biological processes of the downregulated miRNAs.
Figure 2.
Gene Ontology (GO) functions for the target genes of the top 3 most highly upregulated miRNAs and the top 3 most highly downregulated miRNAs. (A) Enriched cellular components of the upregulated miRNAs; (B) enriched cellular components of the downregulated miRNAs.
Figure 3.
Gene Ontology (GO) functions for the target genes of the top 3 most highly upregulated miRNAs and the top 3 most highly downregulated miRNAs. (A) Enriched molecular functions of the upregulated miRNAs; (B) enriched molecular functions of the downregulated miRNAs.
Figure 4.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the target genes of the top 3 most highly upregulated miRNAs and the top 3 most highly downregulated miRNAs. (A) Enriched KEGG pathways of the upregulated miRNAs; (B) enriched KEGG pathways of the downregulated miRNAs.
PPI network construction and module analysis
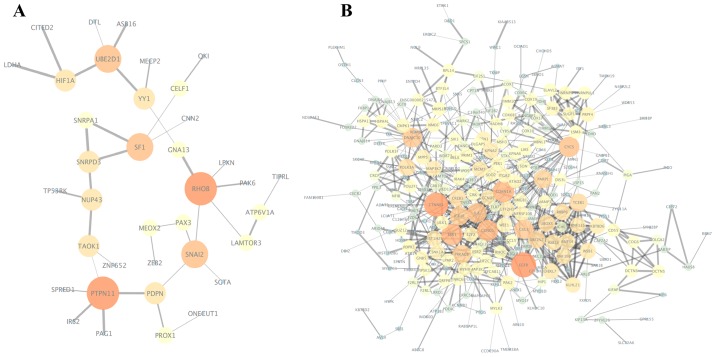
The PPI networks of the target genes of the top 3 most highly upregulated and downregulated DEMs were constructed (Fig. 5), and the most significant module was obtained using the MCODE plugin of Cytoscape. The genes in the most significant module of the upregulated DEMs were SF1, SNRPD3 and SNRPA1, while the genes in the most significant module of the downregulated DEMs were RNF34, RNF19B, ASB16, FBXL7, UBE2V2, RBBP6, KBTBD6, WSB1, KLHL21, CUL3, TCEB1, UBOX5 and RNF115. Functional analyses of the genes involved in the module of the downregulated DEMs were performed using DAVID, showing that genes in this module were mainly enriched in protein K48-linked ubiquitination (BP), polar microtubule (CC), ubiquitin-protein transferase activity (MF), and ubiquitin-mediated proteolysis(KEGG).
Figure 5.
(A) Protein-protein interaction (PPI) network of the target genes of the top 3 most highly upregulated differentially expressed miRNAs (DEMs). (B) PPI network of the target genes of the top 3 most highly downregulated DEMs. Node size indicates the connectivity degree, and larger circles indicate a higher degree. Edge size indicates the combined scores between genes, which represent the confidence of protein interactions. The color gradually increases from dark (blue) to bright (red), representing the gradually increase in the number of interacting genes.
Hub gene analysis and miRNA-hub gene network construction
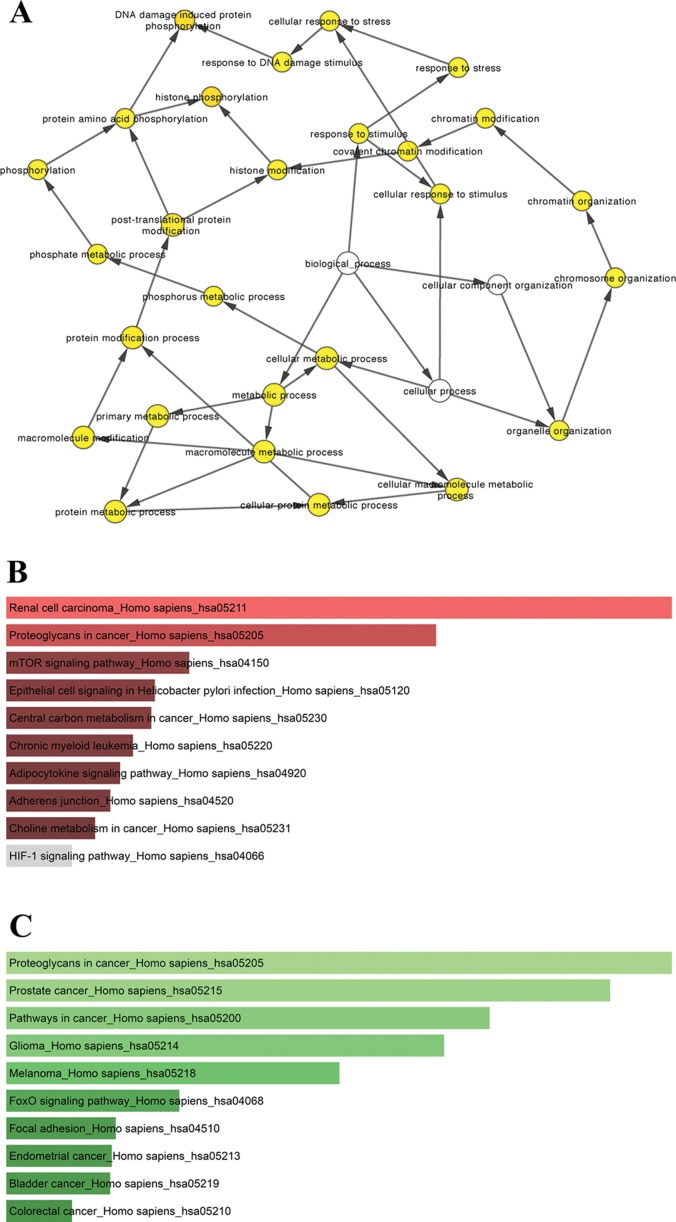
For the upregulated miRNAs, the hub genes included RHOB, PTPN11, SNAI2, UBE2D1, SF1, PDPN, NUP43, YY1, HIF1A and SNRPD3. For the downregulated miRNAs, the hub genes were EGFR, CTNNB1, ESR1, CDKN1A, CCND1, CYCS, DNAJC10, IL8, CUL3 and IGF1R. The abbreviations, full names and functions of these 20 hub genes are shown in Table IV. Among these genes, EGFR (epidermal growth factor receptor) demonstrated the highest node degrees, which suggested that EGFR may be a key target associated with prolactin pituitary tumor aggressiveness. Biological process analysis of the hub genes is shown in Fig. 6A. Functional and pathway enrichment analyses for the hub genes of the top 3 upregulated and downregulated miRNAs are presented in Tables V and VI. As shown in Fig. 6, KEGG analysis showed that the hub genes of the upregulated miRNAs were mainly enriched in renal cell carcinoma and proteoglycans in cancer (Fig. 6B, Table V), while the hub genes of the downregulated miRNAs were mainly enriched in proteoglycans in cancer, prostate cancer and pathways in cancer (Fig. 6C, Table VI).
Table IV.
Functional roles of the hub genes of the top 3 upregulated/downregulated miRNAs identified in the PPI interaction.
| Gene symbol | Degree | Full name | Function |
|---|---|---|---|
| Upregulated miRNAs | |||
| RHOB | 16 | Ras homolog family member B | Protein coding gene. Among its related pathways are ERK signaling and focal adhesion. GO annotations related to this gene include GTP binding and GDP binding. |
| PTPN11 | 15 | Protein tyrosine phosphatase, non-receptor type 11 | Protein coding gene. Among its related pathways are immune response Fcε RI pathway and EGF/EGFR signaling pathway. GO annotations related to this gene include protein domain-specific binding and protein tyrosine phosphatase activity. |
| SNAI2 | 15 | Snail family transcriptional repressor 2 | Protein coding gene. Among its related pathways are ERK signaling and adherens junction. GO annotations related to this gene include sequence-specific DNA binding and tran scriptional repressor activity, RNA polymerase II proximal promoter sequence-specific DNA binding. |
| UBE2D1 | 14 | Ubiquitin conjugating enzyme E2 D1 | Protein coding gene. Among its related pathways are gene expression and cell cycle, mitotic. GO annotations related to this gene include ligase activity and acid-amino acid ligase activity. |
| SF1 | 14 | Splicing factor 1 | Protein Coding gene. Among its related pathways are Oct4 in mammalian ESC pluripotency and mRNA splicing-major pathway. GO annotations related to this gene include nucleic acid binding and RNA binding. |
| PDPN | 14 | Podoplanin | Protein coding gene. Among its related pathways are cytoskel etal signaling and response to elevated platelet cytosolic Ca2+. GO annotations related to this gene include amino acid trans membrane transporter activity and folic acid transmembrane transporter activity. |
| NUP43 | 13 | Nucleoporin 43 | Protein coding gene. Among its related pathways are cell cycle, mitotic and transport of the SLBP independent mature mRNA. |
| YY1 | 13 | YY1 transcription factor | Protein coding gene. Among its related pathways are gene expression and translational control. GO annotations related to this gene include DNA binding transcription factor activity and transcription coactivator activity. |
| HIF1A | 11 | Hypoxia inducible factor 1 subunit α | Protein coding gene. Among its related pathways are ERK signaling and central carbon metabolism in cancer. GO anno tations related to this gene include DNA binding transcription factor activity and protein heterodimerization activity. |
| SNRPD3 | 11 | Small nuclear ribonu cleoprotein D3 polypeptide | Protein coding gene. Among its related pathways are mRNA splicing-major pathway and processing of capped intronless pre-mRNA. GO annotations related to this gene include histone pre-mRNA DCP binding. |
| Downregulated miRNAs | |||
| EGFR | 33 | Epidermal growth factor receptor | Protein coding gene. Among its related pathways are ERK signaling and gene expression. GO annotations related to this gene include identical protein binding and protein kinase activity. |
| CTNNB1 | 31 | Catenin β1 | Protein coding gene. Among its related pathways are ERK signaling and focal adhesion. GO annotations related to this gene include DNA binding transcription factor activity and binding. |
| ESR1 | 25 | Estrogen receptor 1 | Estrogen resistance and myocardial infarction. Among its related pathways are gene expression and integrated breast cancer pathway. GO annotations related to this gene include DNA binding transcription factor activity and identical protein binding. |
| CDKN1A | 25 | Cyclin dependent kinase inhibitor 1A | Protein coding gene. Among its related pathways are gene expression and Akt signaling. GO annotations related to this gene include ubiquitin protein ligase binding and cyclin binding. |
| CCND1 | 24 | Cyclin D1 | Protein coding gene. Diseases associated with CCND1 include myeloma, multiple and Von Hippel-Lindau syndrome. Among its related pathways are ERK signaling and focal adhesion. GO annotations related to this gene include protein kinase activity and enzyme binding. |
| CYCS | 23 | Cytochrome c, somatic | Protein coding gene. Diseases associated with CYCS include thrombocytopenia 4 and autosomal thrombocytopenia with normal platelets. Among its related pathways are gene expression and activation of caspases through apoptosome-mediated cleavage. GO annotations related to this gene include iron ion binding and electron transfer activity. |
| DNAJC10 | 21 | DNAJ heat shock protein family (Hsp40) member C10 | Protein coding gene. Among its related pathways are protein processing in endoplasmic reticulum. GO annotations related to this gene include chaperone binding and protein disulfide oxidoreductase activity. |
| IL8 | 21 | C-X-C motif chemokine ligand 8 | Protein coding gene. Among its related pathways are Akt signaling and rheumatoid arthritis. GO annotations related to this gene include chemokine activity and interleukin-8 receptor binding. |
| CUL3 | 20 | Cullin 3 | Protein Coding gene. Among its related pathways are RET signaling and Class I MHC mediated antigen processing and presentation. GO annotations related to this gene include protein homodimerization activity and ubiquitin-protein trans ferase activity. |
| IGF1R | 19 | Insulin like growth factor 1 receptor | Protein coding gene. Among its related pathways are ERK signaling and mTOR pathway. GO annotations related to this gene include identical protein binding and protein kinase activity. |
PPI, protein-protein interaction; GO, Gene Ontology. Online database GeneCards (https://www.genecards.org).
Figure 6.
(A) The biological process analysis of hub genes. Node color depth refers to the corrected ontology P-values. Node size indicates the number of genes involved in the ontologies. P<0.01 was considered statistically significant. (B) Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the hub genes of the top 3 most highly upregulated miRNAs. (C) Enriched KEGG pathways for the hub genes of the top 3 most highly downregulated miRNAs.
Table V.
Functional and pathway enrichment analysis for the hub genes of the top 3 upregulated miRNAs.
| Category | Term | Pathway description | Genes |
|---|---|---|---|
| Upregulated miRNAs | |||
| GO BP | GO:0032364 | Oxygen homeostasis | HIF1A |
| GO BP | GO:0032909 | Regulation of transforming growth factor β2 production | HIF1A |
| GO BP | GO:0033483 | Gas homeostasis | HIF1A |
| GO BP | GO:0032642 | Regulation of chemokine production | SNAI2, HIF1A |
| GO BP | GO:0046885 | Regulation of hormone biosynthetic process | HIF1A |
| GO BP | GO:0043619 | Regulation of transcription from RNA polymerase II promoter in response to oxidative stress | HIF1A |
| GO BP | GO:0070099 | Regulation of chemokine-mediated signaling pathway | HIF1A |
| GO BP | GO:0032352 | Positive regulation of hormone metabolic process | HIF1A |
| GO BP | GO:0010839 | Negative regulation of keratinocyte proliferation | SNAI2 |
| GO BP | GO:0071364 | Cellular response to epidermal growth factor stimulus | SNAI2, PTPN11 |
| GO CC | GO:0031528 | Microvillus membrane | PDPN |
| GO CC | GO:0000243 | Commitment complex | SNRPD3 |
| GO CC | GO:0005683 | U7 snRNP | SNRPD3 |
| GO CC | GO:0005687 | U4 snRNP | SNRPD3 |
| GO CC | GO:0034709 | Methylosome | SNRPD3 |
| GO CC | GO:0031527 | Filopodium membrane | PDPN |
| GO CC | GO:0071437 | Invadopodium | PDPN |
| GO CC | GO:0031011 | Ino80 complex | YY1 |
| GO CC | GO:0005685 | U1 snRNP | SNRPD3 |
| GO CC | GO:0031258 | Lamellipodium membrane | PDPN |
| GO MF | GO:0000400 | Four-way junction DNA binding | YY1 |
| GO MF | GO:0001227 | Transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding | YY1, SNAI2 |
| GO MF | GO:0019956 | Chemokine binding | PDPN |
| GO MF | GO:0043565 | Sequence-specific DNA binding | YY1, SNAI2, HIF1A |
| GO MF | GO:0061631 | Ubiquitin conjugating enzyme activity | UBE2D1 |
| GO MF | GO:0000217 | DNA secondary structure binding | YY1 |
| GO MF | GO:0061650 | Ubiquitin-like protein conjugating enzyme activity | UBE2D1 |
| GO MF | GO:0005158 | Insulin receptor binding | PTPN11 |
| GO MF | GO:0035326 | Enhancer binding | YY1 |
| GO MF | GO:0001078 | Transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding | YY1, SNAI2 |
| KEGG | hsa05211 | Renal cell carcinoma | PTPN11, HIF1A |
| KEGG | hsa05205 | Proteoglycans in cancer | PTPN11, HIF1A |
| KEGG | hsa04150 | mTOR signaling pathway | HIF1A |
| KEGG | hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | PTPN11 |
| KEGG | hsa05230 | Central carbon metabolism in cancer | HIF1A |
| KEGG | hsa05220 | Chronic myeloid leukemia | PTPN11 |
| KEGG | hsa04920 | Adipocytokine signaling pathway | PTPN11 |
| KEGG | hsa04520 | Adherens junction | SNAI2 |
| KEGG | hsa05231 | Choline metabolism in cancer | HIF1A |
| KEGG | hsa04066 | HIF-1 signaling pathway | HIF1A |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
Table VI.
Functional and pathway enrichment analysis for the hub genes of top 3 downregulated miRNAs.
| Category | Term | Pathway description | Genes |
|---|---|---|---|
| Downregulated miRNAs | |||
| GO BP | GO:0070141 | Response to UV-A | CCND1, EGFR |
| GO BP | GO:0097193 | Intrinsic apoptotic signaling pathway | CDKN1A, CUL3, DNAJC10, CYCS |
| GO BP | GO:0032355 | Response to estradiol | CTNNB1, ESR1, EGFR |
| GO BP | GO:1903798 | Regulation of production of miRNAs involved in gene silencing by miRNA | ESR1, EGFR |
| GO BP | GO:0033674 | Positive regulation of kinase activity | CDKN1A, EGFR, IGF1R |
| GO BP | GO:0001934 | Positive regulation of protein phosphorylation | CDKN1A, CCND1, EGFR, IGF1R |
| GO BP | GO:0045737 | Positive regulation of cyclin-dependent protein serine/threonine kinase activity | CCND1, EGFR |
| GO BP | GO:0045740 | Positive regulation of DNA replication | EGFR, IGF1R |
| GO BP | GO:0006367 | Transcription initiation from RNA polymerase II promoter | CDKN1A, CCND1, ESR1 |
| GO BP | GO:0034333 | Adherens junction assembly | CTNNB1 |
| GO CC | GO:0030128 | Clathrin coat of endocytic vesicle | EGFR |
| GO CC | GO:0030122 | AP-2 adaptor complex | EGFR |
| GO CC | GO:0030131 | Clathrin adaptor complex | EGFR |
| GO CC | GO:1990907 | β-catenin-TCF complex | CTNNB1 |
| GO CC | GO:0005719 | Nuclear euchromatin | CTNNB1 |
| GO CC | GO:0000791 | Euchromatin | CTNNB1 |
| GO CC | GO:0035327 | Transcriptionally active chromatin | ESR1 |
| GO CC | GO:0000790 | Nuclear chromatin | CTNNB1, ESR1 |
| GO CC | GO:0005758 | Mitochondrial intermembrane space | CYCS |
| GO CC | GO:0016342 | Catenin complex | CTNNB1 |
| GO MF | GO:0097472 | Cyclin-dependent protein kinase activity | CDKN1A, CCND1 |
| GO MF | GO:0019900 | Kinase binding | CDKN1A, CCND1, CTNNB1, ESR1 |
| GO MF | GO:0004693 | Cyclin-dependent protein serine/threonine kinase activity | CDKN1A, CCND1 |
| GO MF | GO:0004709 | MAP kinase kinase kinase activity | EGFR, IGF1R |
| GO MF | GO:0001223 | Transcription coactivator binding | ESR1 |
| GO MF | GO:0044389 | Ubiquitin-like protein ligase binding | CDKN1A, CUL3, EGFR |
| GO MF | GO:0019901 | Protein kinase binding | CDKN1A, CCND1, ESR1, EGFR, IGF1R |
| GO MF | GO:0030331 | Estrogen receptor binding | CTNNB1, ESR1 |
| GO MF | GO:0016671 | Oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | DNAJC10 |
| GO MF | GO:0046934 | Phosphatidylinositol-4,5-bisphosphate 3-kinase activity | ESR1, EGFR |
| KEGG | hsa05205 | Proteoglycans in cancer | CDKN1A, CCND1, ESR1, CTNNB1, EGFR, IGF1R |
| KEGG | hsa05215 | Prostate cancer | CDKN1A, CCND1, CTNNB1, EGFR, IGF1R |
| KEGG | hsa05200 | Pathways in cancer | CDKN1A, CCND1, CTNNB1, CYCS, EGFR, IGF1R |
| KEGG | hsa05214 | Glioma | CDKN1A, CCND1, EGFR, IGF1R |
| KEGG | hsa05218 | Melanoma | CDKN1A, CCND1, EGFR, IGF1R |
| KEGG | hsa04068 | FoxO signaling pathway | CDKN1A, CCND1, EGFR, IGF1R |
| KEGG | hsa04510 | Focal adhesion | CCND1, CTNNB1, EGFR, IGF1R |
| KEGG | hsa05213 | Endometrial cancer | CCND1, CTNNB1, EGFR |
| KEGG | hsa05219 | Bladder cancer | CDKN1A, CCND1, EGFR |
| KEGG | hsa05210 | Colorectal cancer | CCND1, CYCS, CTNNB1 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; hsa, Homo sapiens.
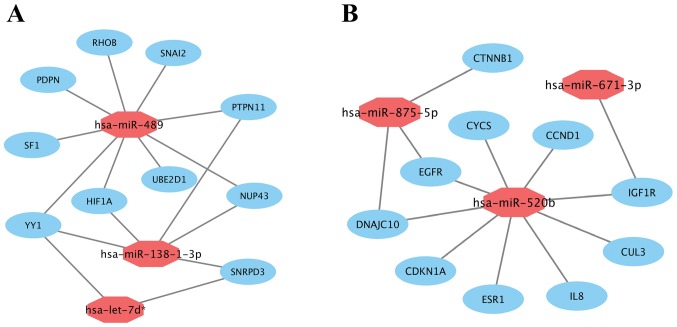
Subsequently, miRNA-hub gene networks were constructed by Cytoscape (Fig. 7). As shown in Fig. 7A, hsa-miR-489, the most highly upregulated DEM, potentially could target 9 (RHOB, PTPN11, SNAI2, UBE2D1, SF1, PDPN, NUP43, YY1 and HIF1A) of 10 hub genes. Five hub genes and 2 hub genes potentially were regulated by upregulated hsa-miR-138-1-3p and hsa-let-7d*, respectively. Additionally, according to Fig. 7B, hsa-miR-520b, the most highly downregulated DEM, potentially could also target 9 (EGFR, ESR1, CDKN1A, CCND1, CYCS, DNAJC10, IL8, CUL3 and IGF1R) of 10 hub genes. Three hub genes and 1 hub gene potentially were regulated by downregulated hsa-miR-875-5p and hsa-miR-671-3p, respectively. The results suggested that hsa-miR-489 and hsa-miR-520b may be the most important regulators of prolactin pituitary tumor aggressiveness.
Figure 7.
miRNA-hub gene network (A) for the top 3 most highly upregulated miRNAs and their hub genes; (B) for the top 3 most highly downregulated miRNAs and their hub genes.
Discussion
Prolactin-secreting pituitary adenoma is the most common (30–40%) subtype of pituitary tumors and commonly presents with headache, visual disturbances, amenorrhea, galactorrhea, infertility and hyposexuality (1,2). Most prolactinomas are noninvasive and easily treated by surgery, radiotherapy or medical treatment, including cabergoline and dopamine agonists, which are highly effective drugs for prolactinoma. However, aggressive prolactin pituitary tumors, with unknown incidence, are entities whose pathological behaviors lie between those of benign pituitary adenomas and malignant pituitary carcinomas. They display a rather distinct aggressive behavior with marked invasion of nearby anatomical structures, a tendency for resistance to conventional treatments and/or TMZ, and early postoperative recurrences (3,4). Extensive research has been performed to explore potential biomarkers for early diagnosis and treatment of aggressive pituitary tumors. The Raf/MEK/ERK, PI3K/Akt/mTOR, and VEGFR pathways were found to be upregulated in pituitary tumors, suggesting that these pathways may be utilized to control pituitary tumor growth and progression (28–32). However, most targeted therapies based on the above pathways have been administered to patients with aggressive pituitary tumors without success (32–34). Therefore, further research is needed to discover aggressiveness-associated biomarkers with diagnostic and therapeutic value for aggressive prolactin pituitary tumors.
miRNAs are a group of small, endogenous noncoding RNAs that can repress protein expression by cleaving mRNA or inhibiting translation (8,9). Mostly, miRNAs are recognized as having a significant role in the negative regulation of target gene expression, which contributes to tumorigenesis, invasion and metastasis in various types of tumors (10–12). Recent studies have shown that aberrant miRNA expression is involved in tumorigenesis and tumor development of pituitary adenomas, especially prolactin pituitary tumors (13–16). D'Angelo et al (35) found that miR-603, miR-34b, miR-548c-3p, miR-326, miR-570 and miR-432 were downregulated in prolactinomas, which can affect the G1-S transition process. Mussnich et al (36) found that miR-15, miR-26a, miR-196a-2, miR-16, Let-7a and miR-410 were downregulated in prolactinomas, which can negatively regulate pituitary cell proliferation. Roche et al (17) demonstrated that miR-183 was downregulated in aggressive prolactin tumors and inhibited tumor cell proliferation by directly targeting KIAA0101, which is involved in cell cycle activation and the inhibition of p53-p21-mediated cell cycle arrest. However, few studies, except for one reported by Roche et al (17) in 2015, have been performed to explore aggressiveness-associated miRNAs in aggressive prolactin pituitary tumors based on large-scale human tissue datasets. Additionally, based on the GSE46294 dataset, our study obtained different DEMs compared with those reported by Roche et al. The reasons may be due to different softwares or different algorithms when analyzing differentially expressed genes or RNAs, and due to the small sample size of the GSE46294 dataset (37).
In the present study, some aggressiveness-associated miRNAs were screened by performing a differential expression analysis based on an miRNA expression profile downloaded from the GEO database. The potential target genes of the top 3 most highly upregulated and most highly downregulated DEMs were collectively enriched for regulation of transcription from RNA polymerase II promoter, DNA-templated transcription, Wnt signaling pathway, protein binding, and transcription factor activity (sequence-specific DNA binding). Moreover, by constructing PPI networks, we identified the top 10 hub genes with the highest degree of connectivity with the top 3 most highly upregulated and downregulated DEMs. Hub genes of the upregulated DEMs were mainly enriched for proteoglycans in cancer, while hub genes of the downregulated DEMs were mainly enriched for proteoglycans in cancer, pathways in cancer, FoxO signaling pathway, and focal adhesion. Those pathways were all reported by previous studies to be associated with tumor growth, progression invasion and metastasis of various tumors (38–40). In our study, proteoglycan in cancer is the enriched pathway shared by both upregulated and downregulated DEMs. However, there is little research on proteoglycan in tumorigenesis, invasiveness and progression of pituitary tumors. Matano et al reported that endocan, a novel soluble dermatan sulfate proteoglycan, can function as a new invasion and angiogenesis marker of pituitary adenomas (40). More studies are needed to further research the functions of proteoglycan in pituitary adenomas, especially aggressive tumors.
Among the 20 hub genes, EGFR demonstrated the highest node degrees, suggesting that EGFR was a key target associated with the aggressiveness of prolactin pituitary tumors, which is consistent with previous studies (4,41). EGFR encodes a transmembrane glycoprotein that is located on the cell surface and binds to epidermal growth factor (EGF). Binding of the protein to a ligand induces receptor dimerization and tyrosine autophosphorylation, leading to cell proliferation. EGFR involvement in the tumorigenesis and invasion of pituitary tumors, especially aggressive prolactinomas, has been reported by previous studies, and mutations in this gene can be utilized as potential targets in the treatment of aggressive prolactinomas. As reported in the literature, tyrosine kinase inhibitors (TKIs), such as lapatanib, sunitinib and erlotinib, have been trialed as first- or second-line treatments based on the VEGFR pathway, but most of them have failed (4,29–32,34). In addition, in the present study, we found that EGFR may be negatively modulated by hsa-miR-520b using the miRTarBase database; furthermore, hsa-miR-520b can be regulated by EGFR due to its association with the biological process regulation of production of miRNAs involved in gene silencing by miRNA (30–32). This interesting finding may allow the use of this potential pathway for the diagnosis or treatment of aggressive prolactinomas in the future.
Subsequently, by constructing an miRNA-hub gene network, we found that most hub genes were potentially modulated by hsa-miR-489 and hsa-miR-520b, suggesting that these miRNAs may be the most important regulators of prolactin pituitary tumor aggressiveness. Recent studies demonstrated that hsa-miR-489 acts as a tumor suppressor in hepatocellular carcinoma (42), gastric cancer (43), breast cancer (44), glioma (45), hypopharyngeal squamous cell carcinoma (46), bladder cancer (47) and colorectal cancer (48). Downregulation of miR-489 was reported to be associated with the tumorigenesis, invasion, and metastasis of various tumors, suggesting an important role for hsa-miR-489 in predicting prognosis and acting as a drug target. However, the roles of hsa-miR-489 in pituitary tumors, especially aggressive prolactinomas, have not been previously studied. Additionally, hsa-miR-520b was reported to have a suppressive effect on tumor cell proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) in colorectal cancer (49), glioblastoma (50), hepatoma (51), head-neck cancer (52), breast cancer (53), lung cancer (54) and gastric cancer (55). Expression of hsa-miR-520b is lower in tumor tissues than in normal tissues, significantly promoting the proliferation, migration, and invasion of tumor cells. Unlike other tumors, Liang et al (56) reported that hsa-miR-520b was upregulated in nonfunctioning and gonadotropin-secreting pituitary adenomas relative to normal pituitaries, which indicated that miR-520b functions as a tumor inducer in benign pituitary adenoma (56). However, whether hsa-miR-520b acts as a promoter or suppressor in aggressive prolactin pituitary tumors has not been previously studied. According to our study, we speculate that upregulation of hsa-miR-489 suppresses aggressiveness and progression, while downregulation of hsa-miR-520b promotes the aggressiveness and progression of aggressive prolactinomas. Such ambivalent miRNA expression might be one of the reasons that aggressive prolactin pituitary tumors lie on the spectrum between ‘benign’ pituitary adenomas and ‘malignant’ pituitary carcinomas. It will be extremely meaningful to authenticate the functions of hsa-miR-489 and hsa-miR-520b and elucidate the mechanisms by which they regulate aggressive behaviors, resistance to treatments and early recurrence in aggressive prolactin pituitary tumors.
There are some limitations of the present study. First, the sample size of GSE46294 is rather small (only 12 samples), which may cause some bias when identifying the differentially expressed miRNAs. Second, the expression of the differentially expressed miRNAs was not validated by RT-qPCR analysis with our clinical pituitary samples. Further studies are needed to experimentally verify the results of this study.
In conclusion, we successfully identified one key target gene, EGFR, and two crucial miRNAs, hsa-miR-489 and hsa-miR-520b, associated with aggressiveness based on bioinformatic analysis. These findings may provide potential candidate biomarkers for the early diagnosis and individualized treatment of aggressive prolactin pituitary tumors. However, further research is needed to experimentally verify the results of this study.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- miRNAs
microRNAs
- DEMs
differentially expressed miRNAs
- PPI
protein-protein interaction
- TMZ
temozolomide
- mRNA
messenger RNA
- DE-miRNAs
differentially expressed miRNAs
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GEO
Gene Expression Omnibus
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- MCODE
Molecular Complex Detection
- BiNGO
Biological Networks Gene Oncology tool
- BP
biological process
- CC
cellular component
- MF
molecular function
- EGFR
epidermal growth factor receptor
- EGF
epidermal growth factor
- TKI
tyrosine kinase inhibitor
Funding
Not applicable.
Availability of data and materials
The GSE46294 datasets analyzed during the present study are available in the GEO repository (http://www.ncbi.nlm.nih.gov/geo/). The potential target genes of DEMs were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/). The DAVID database (http://david.ncifcrf.gov/) was used to perform functional annotation and pathway enrichment analysis for genes. The STRING database (http://string-db.org) was used to assess functional associations among genes.
Authors' contributions
All authors conceived and designed the study. LG, XG and CF performed data curation and analysis. KD and WL analyzed and interpreted the results. ZW and BX drafted and reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kaltsas GA, Nomikos P, Kontogeorgos G, Buchfelder M, Grossman AB. Clinical review: Diagnosis and management of pituitary carcinomas. J Clin Endocrinol Metab. 2005;90:3089–3099. doi: 10.1210/jc.2004-2231. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez A, Karavitaki N, Wass JA. Prevalence of pituitary adenomas: A community-based, cross-sectional study in Banbury (Oxfordshire, UK) Clin Endocrinol (Oxf) 2010;72:377–382. doi: 10.1111/j.1365-2265.2009.03667.x. [DOI] [PubMed] [Google Scholar]
- 3.Dai C, Feng M, Liu X, Ma S, Sun B, Bao X, Yao Y, Deng K, Wang Y, Xing B, et al. Refractory pituitary adenoma: A novel classification for pituitary tumors. Oncotarget. 2016;7:83657–83668. doi: 10.18632/oncotarget.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raverot G, Burman P, McCormack A, Heaney A, Petersenn S, Popovic V, Trouillas J, Dekkers OM, European Society of Endocrinology European society of endocrinology clinical practice guidelines for the management of aggressive pituitary tumours and carcinomas. Eur J Endocrinol. 2018;178:G1–G24. doi: 10.1530/EJE-17-0796. [DOI] [PubMed] [Google Scholar]
- 5.Heaney A. Management of aggressive pituitary adenomas and pituitary carcinomas. J Neurooncol. 2014;117:459–468. doi: 10.1007/s11060-014-1413-6. [DOI] [PubMed] [Google Scholar]
- 6.Lasolle H, Cortet C, Castinetti F, Cloix L, Caron P, Delemer B, Desailloud R, Jublanc C, Lebrun-Frenay C, Sadoul JL, et al. Temozolomide treatment can improve overall survival in aggressive pituitary tumors and pituitary carcinomas. Eur J Endocrinol. 2017;176:769–777. doi: 10.1530/EJE-16-0979. [DOI] [PubMed] [Google Scholar]
- 7.Losa M, Bogazzi F, Cannavo S, Ceccato F, Curtò L, De Marinis L, Iacovazzo D, Lombardi G, Mantovani G, Mazza E, et al. Temozolomide therapy in patients with aggressive pituitary adenomas or carcinomas. J Neurooncol. 2016;126:519–525. doi: 10.1007/s11060-015-1991-y. [DOI] [PubMed] [Google Scholar]
- 8.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 9.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 10.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Mao ZG, Wang X, Du Q, Jian M, Zhu D, Xiao Z, Wang HJ, Zhu YH. MicroRNAs and target genes in pituitary adenomas. Horm Metab Res. 2018;50:179–192. doi: 10.1055/a-0585-7410. [DOI] [PubMed] [Google Scholar]
- 14.Di Ieva A, Butz H, Niamah M, Rotondo F, De Rosa S, Sav A, Yousef GM, Kovacs K, Cusimano MD. MicroRNAs as biomarkers in pituitary tumors. Neurosurgery. 2014;75:181–189. doi: 10.1227/NEU.0000000000000369. discussion 188–189. [DOI] [PubMed] [Google Scholar]
- 15.Zhang QJ, Xu C. The role of microRNAs in the pathogenesis of pituitary tumors. Front Biosci (Landmark Ed) 2016;21:1–7. doi: 10.2741/4371. [DOI] [PubMed] [Google Scholar]
- 16.Wei Z, Zhou C, Liu M, Yao Y, Sun J, Xiao J, Ma W, Zhu H, Wang R. MicroRNA involvement in a metastatic non-functioning pituitary carcinoma. Pituitary. 2015;18:710–721. doi: 10.1007/s11102-015-0648-3. [DOI] [PubMed] [Google Scholar]
- 17.Roche M, Wierinckx A, Croze S, Rey C, Legras-Lachuer C, Morel AP, Fusco A, Raverot G, Trouillas J, Lachuer J. Deregulation of miR-183 and KIAA0101 in aggressive and malignant pituitary tumors. Front Med (Lausanne) 2015;2:54. doi: 10.3389/fmed.2015.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res 41 (Database Issue) 2013:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46D. 2018:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45D. 2017:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY, Arai AE. MultiContrast delayed enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J Cardiovasc Magn Reson. 2012;14:83. doi: 10.1186/1532-429X-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maere S, Heymans K, Kuiper M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 28.Dworakowska D, Grossman AB. The pathophysiology of pituitary adenomas. Best Pract Res Clin Endocrinol Metab. 2009;23:525–541. doi: 10.1016/j.beem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Fukuoka H, Cooper O, Ben-Shlomo A, Mamelak A, Ren SG, Bruyette D, Melmed S. EGFR as a therapeutic target for human, canine, and mouse ACTH-secreting pituitary adenomas. J Clin Invest. 2011;121:4712–4721. doi: 10.1172/JCI60417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuoka H, Cooper O, Mizutani J, Tong Y, Ren SG, Bannykh S, Melmed S. HER2/ErbB2 receptor signaling in rat and human prolactinoma cells: Strategy for targeted prolactinoma therapy. Mol Endocrinol. 2011;25:92–103. doi: 10.1210/me.2010-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlotides G, Siegel E, Donangelo I, Gutman S, Ren SG, Melmed S. Rat prolactinoma cell growth regulation by epidermal growth factor receptor ligands. Cancer Res. 2008;68:6377–6386. doi: 10.1158/0008-5472.CAN-08-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper O, Mamelak A, Bannykh S, Carmichael J, Bonert V, Lim S, Cook-Wiens G, Ben-Shlomo A. Prolactinoma ErbB receptor expression and targeted therapy for aggressive tumors. Endocrine. 2014;46:318–327. doi: 10.1007/s12020-013-0093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donovan LE, Arnal AV, Wang SH, Odia Y. Widely metastatic atypical pituitary adenoma with mTOR pathway STK11(F298L) mutation treated with everolimus therapy. CNS Oncol. 2016;5:203–209. doi: 10.2217/cns-2016-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz LD, Syro LV, Scheithauer BW, Ersen A, Uribe H, Fadul CE, Rotondo F, Horvath E, Kovacs K. Anti-VEGF therapy in pituitary carcinoma. Pituitary. 2012;15:445–449. doi: 10.1007/s11102-011-0346-8. [DOI] [PubMed] [Google Scholar]
- 35.D'Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, Raverot G, Fedele M, Croce CM, Trouillas J, Fusco A. Altered microRNA expression profile in human pituitary GH adenomas: Down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab. 2012;97:E1128–E1138. doi: 10.1210/jc.2011-3482. [DOI] [PubMed] [Google Scholar]
- 36.Mussnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Wierinckx A, Trouillas J, Fusco A, D'Angelo D. Downregulation of miR-410 targeting the cyclin B1 gene plays a role in pituitary gonadotroph tumors. Cell Cycle. 2015;14:2590–2597. doi: 10.1080/15384101.2015.1064207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finotello F, Di Camillo B. Measuring differential gene expression with RNA-seq: Challenges and strategies for data analysis. Brief Funct Genomics. 2015;14:130–142. doi: 10.1093/bfgp/elu035. [DOI] [PubMed] [Google Scholar]
- 38.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu F, Zhang J, Hu G, Liu L, Liang W. Hypoxia and TGF-β1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 2017;17:54. doi: 10.1186/s12935-017-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matano F, Yoshida D, Ishii Y, Tahara S, Teramoto A, Morita A. Endocan, a new invasion and angiogenesis marker of pituitary adenomas. J Neurooncol. 2014;117:485–491. doi: 10.1007/s11060-014-1377-6. [DOI] [PubMed] [Google Scholar]
- 41.Cooper O, Vlotides G, Fukuoka H, Greene MI, Melmed S. Expression and function of ErbB receptors and ligands in the pituitary. Endocr Relat Cancer. 2011;18:R197–R211. doi: 10.1530/ERC-11-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y, Liu J, Huang Y, Liu D, Zhang G, Kan H. microRNA-489 plays an anti-metastatic role in human hepatocellular carcinoma by targeting matrix metalloproteinase-7. Transl Oncol. 2017;10:211–220. doi: 10.1016/j.tranon.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, Ji S, Ma F, Ma Q, Lu X, Chen X. miR-489 acts as a tumor suppressor in human gastric cancer by targeting PROX1. Am J Cancer Res. 2016;6:2021–2030. [PMC free article] [PubMed] [Google Scholar]
- 44.Chai P, Tian J, Zhao D, Zhang H, Cui J, Ding K, Liu B. GSE1 negative regulation by miR-489-5p promotes breast cancer cell proliferation and invasion. Biochem Biophys Res Commun. 2016;471:123–128. doi: 10.1016/j.bbrc.2016.01.168. [DOI] [PubMed] [Google Scholar]
- 45.Xu D, Liu R, Meng L, Zhang Y, Lu G, Ma P. Long non-coding RNA ENST01108 promotes carcinogenesis of glioma by acting as a molecular sponge to modulate miR-489. Biomed Pharmacother. 2018;100:20–28. doi: 10.1016/j.biopha.2018.01.126. [DOI] [PubMed] [Google Scholar]
- 46.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Qu W, Jiang Y, Sun Y, Cheng Y, Zou T, Du S. miR-489 suppresses proliferation and invasion of human bladder cancer cells. Oncol Res. 2016;24:391–398. doi: 10.3727/096504016X14666990347518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao S, Liu H, Hou S, Wu L, Yang Z, Shen J, Zhou L, Zheng SS, Jiang B. miR-489 suppresses tumor growth and invasion by targeting HDAC7 in colorectal cancer. Clin Transl Oncol. 2018;20:703–712. doi: 10.1007/s12094-017-1770-7. [DOI] [PubMed] [Google Scholar]
- 49.Xiao J, Li G, Zhou J, Wang S, Liu D, Shu G, Zhou J, Ren F. MicroRNA-520b functions as a tumor suppressor in colorectal cancer by inhibiting defective in cullin neddylation 1 domain containing 1 (DCUN1D1) Oncol Res. 2018;26:593–604. doi: 10.3727/096504017X14920318811712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Wang F, Tian L, Wang T, Zhang W, Li B, Bai YA. MicroRNA-520b affects the proliferation of human glioblastoma cells by directly targeting cyclin D1. Tumour Biol. 2016;37:7921–7928. doi: 10.1007/s13277-015-4666-6. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Kong G, Zhang J, Wang T, Ye L, Zhang X. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One. 2012;7:e31450. doi: 10.1371/journal.pone.0031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu YC, Cheng AJ, Lee LY, You GR, Li YL, Chen HY, Chang JT. miR-520b as a novel molecular target for suppressing stemness phenotype of head-neck cancer by inhibiting CD44. Sci Rep. 2017;7:2042. doi: 10.1038/s41598-017-02058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, Bai X, Zhang Z, Zhang W, Zhang X, Ye L. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J Biol Chem. 2011;286:13714–13722. doi: 10.1074/jbc.M110.204131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin K, Zhao W, Xie X, Pan Y, Wang K, Zhang H. miR-520b restrains cell growth by targeting HDAC4 in lung cancer. Thorac Cancer. 2018;9:1249–1254. doi: 10.1111/1759-7714.12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Zhang H, Ning T, Wang X, Liu R, Yang H, Han Y, Deng T, Zhou L, Zhang L, et al. miR-520b/e regulates proliferation and migration by simultaneously targeting EGFR in gastric cancer. Cell Physiol Biochem. 2016;40:1303–1315. doi: 10.1159/000453183. [DOI] [PubMed] [Google Scholar]
- 56.Liang S, Chen L, Huang H, Zhi D. The experimental study of miRNA in pituitary adenomas. Turk Neurosurg. 2013;23:721–727. doi: 10.5137/1019-5149.JTN.7425-12.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GSE46294 datasets analyzed during the present study are available in the GEO repository (http://www.ncbi.nlm.nih.gov/geo/). The potential target genes of DEMs were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/). The DAVID database (http://david.ncifcrf.gov/) was used to perform functional annotation and pathway enrichment analysis for genes. The STRING database (http://string-db.org) was used to assess functional associations among genes.