Abstract
Most human tumors are heterogeneous, composed of cellular clones with different properties present at variable frequencies. Highly heterogeneous tumors have poor clinical outcomes, yet the underlying mechanism remains poorly understood. Here, we show that minor subclones of breast cancer cells expressing IL11 and FIGF (VEGFD) cooperate to promote metastatic progression and generate polyclonal metastases composed of driver and neutral subclones. Expression profiling of epithelial and stromal compartments of monoclonal and polyclonal primary and metastatic lesions revealed that this cooperation is indirect, mediated through the local and systemic microenvironments. We identified neutrophils as a leukocyte population stimulated by the IL11-expressing minor subclone and showed that depletion of neutrophils prevents metastatic outgrowth. Single-cell RNA-seq of CD45+ cell populations from primary tumors, blood, and lungs demonstrated that IL11 acts on bone-marrow-derived mesenchymal stromal cells, which induce pro-tumorigenic and pro-metastatic neutrophils. Our results indicate key roles for non-cell-autonomous drivers and minor subclones in metastasis.
Tumors are mixtures of cells with distinct characteristics1. High intratumor diversity increases the likelihood of disease progression2, as different subclones respond differently to microenvironmental cues. Treatment of heterogeneous tumors favors selection of resistant subclones, leading to therapeutic failure. Heterogeneous tumors also display phenotypes different from those of individual clones; thus, intratumor heterogeneity has a significant impact on tumor progression and therapeutic resistance.
Metastatic disease is responsible for most cancer-associated mortality; therefore, understanding drivers of metastatic progression is key for improving clinical outcomes. Cancer genome sequencing studies identified limited genetic differences between primary and metastatic tumors and demonstrated extensive subclonal heterogeneity in both primary and distant lesions3,4. However, the mechanism(s) by which polyclonal primary tumors produce polyclonal metastases remains elusive. Moreover, several recent studies implicated microenvironmental changes as key mediators of metastatic dissemination and outgrowth5,6, highlighting the role of non-cell-autonomous factors in tumor evolution.
Clonal cooperation drives polyclonal metastasis
We have been investigating the effect of subclonal interactions on tumor phenotypes using a human breast cancer cell line (MDA-MB-468)-derived xenograft model of intratumor heterogeneity. We previously established that a minor subclone can drive tumor growth through non-cell-autonomous interactions, supporting long-term subclonal heterogeneity7. Briefly, we tested 18 subclones, each expressing a secreted protein implicated in metastasis and angiogenesis, and found that polyclonal tumors with all 18 subclones grew the fastest, while in monoclonal tumors only IL11 and CCL5 were able to drive tumor growth. We also determined that a mixture of two subclones expressing IL11 (interleukin 11) and FIGF (FOS-induced growth factor, also known as VEGFD) was largely able to reproduce this phenotype. Omitting IL11+ cells from polyclonal tumors decreased tumor growth, suggesting that IL11 and FIGF may cooperate. In addition, both polyclonal tumors and tumors comprised of only IL11 and FIGF subclones were highly metastatic, but the underlying mechanism remained undefined.
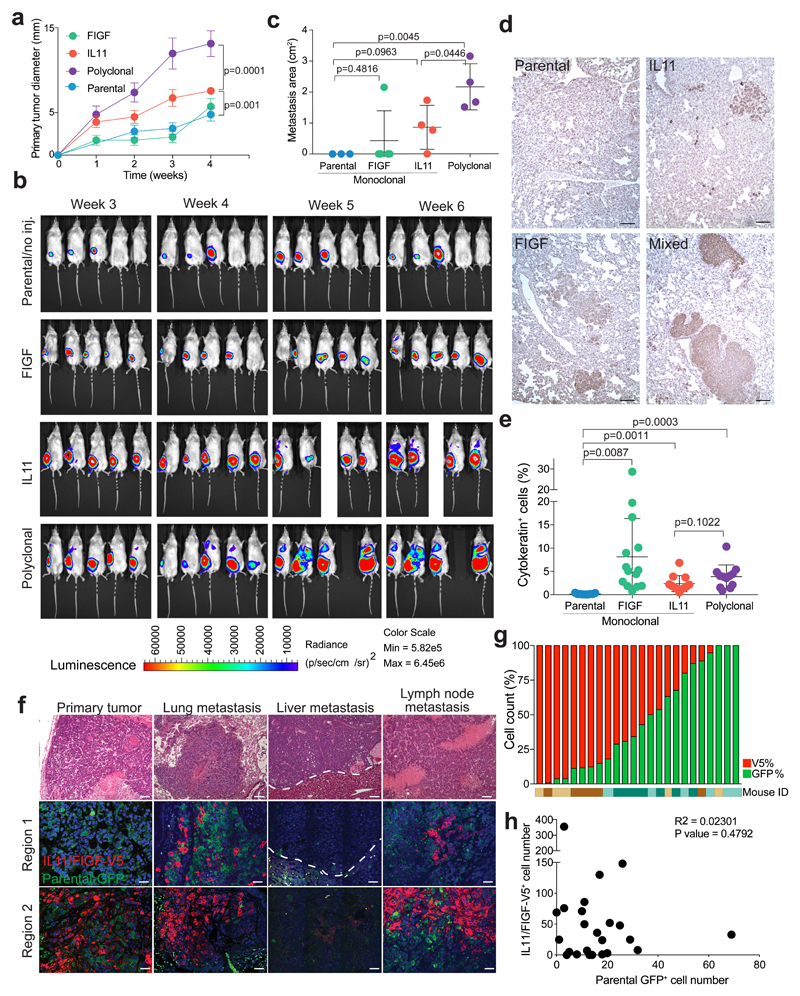
To dissect the molecular basis of this metastasis-driving subclonal cooperation, we first investigated the clonality of metastases of primary MDA-MB-468 tumors comprising IL11+ and FIGF+ driver subclones, as well as neutral subclones. Monoclonal or polyclonal mixtures of green fluorescent protein (GFP) and luciferase-expressing parental cells, red fluorescent protein (RFP) and V5-tagged IL11+ cells, and RFP+ FIGF+ cells were implanted into the mammary fat pads of immunodeficient NOG mice. We monitored primary tumor growth by weekly caliper measurements and macrometastatic lesions by weekly bioluminescence imaging. Polyclonal tumors initiated from 5% IL11+ and 5% FIGF+ RFP+ cells with 90% GFP+ parental cells grew faster and were more metastatic than monoclonal and parental tumors (Fig. 1a-c, Supplementary Table 1). Immunohistochemistry-based quantification of human cytokeratin+ (CK+) cells in the lungs revealed an increased number of metastatic lesions in mice with FIGF+ primary tumors (Fig. 1d,e) despite small primary tumors. However, most of these were micrometastases, detectable as single cells only by immunohistochemistry, while the lungs of mice with polyclonal primary tumors were filled with macrometastases emitting high bioluminescence-signal (Fig. 1b). The increased metastases by polyclonal tumors were not simply due to their faster growth, as this trend was still observed when primary tumors were surgically removed upon reaching 1 cm in diameter (Supplementary Fig. 1a, b). Furthermore, IL11+ monoclonal tumors grew faster than parental and neutral clones but were not as metastatic as polyclonal tumors. Thus, macrometastatic outgrowth is a phenotype unique to polyclonal tumors. Immunofluorescence demonstrated that metastases of polyclonal tumors were also mostly polyclonal, composed of V5-tag+ IL11 and FIGF driver and GFP+ parental subclones, the latter of which did not produce significant macrometastases on their own except for rare GFP+ monoclonal metastases (Fig. 1f,g). These results show that polyclonal tumors behave differently than monoclonal tumors and that IL11+ and FIGF+ subclones driving tumor growth and dissemination, respectively, cooperate to generate frequent large polyclonal metastases. This cooperation is likely to be indirect, as close cell contact between driver and neutral subclones was not commonly observed and clonal distribution within metastatic lesions was random (Fig. 1g,h).
Figure 1. Minor driver clones lead to polyclonal metastasis.
a, Primary tumor growth kinetics, n=10 tumors, 5 animals per group. Mean +/- SD are shown. b, Representative bioluminescence images. c, Quantification of lung macrometastases based on in vivo bioluminescence at week 6, n = 5 animals per group, except IL11 and polyclonal group: n=4 animals. Mean +/- SD is shown. All areas with detected luminescence were quantified and primary tumor area was excluded from the analysis. d, Representative images of cytokeratin staining in the lungs. Scale bar 100 μm. Staining was repeated twice with similar results. e, Quantification of human cytokeratin+ cells in the lungs. On average, 3,500 cells were counted per field, 3 fields per sample. Lungs of 5 mice per group were analyzed. Percent of positive cells per field is shown, with mean +/- SD. p values indicate statistical significance based on unpaired two-tailed t-test in all panels. f, Representative images of the histology (hematoxylin-eosin staining, upper panels) and clonal composition (GFP and V5 immunofluorescence) of primary tumors and metastases. Scale bar 100 μm. Staining was repeated twice with similar results. g, Quantification of parental GFP+ and IL11/FIGF-expressing V5+ cells in lung metastatic lesions from four polyclonal tumor- bearing animals, six lesions per animal were quantified. h, Frequency of parental GFP+ and IL11/FIGF-expressing V5+ cells per lesion, n=24 lesions in lungs of 4 animals. Pearson correlation R2 and two-tailed t-test p value are indicated. See also Supplementary Table 1 for raw data.
Indirect clonal cooperation via the microenvironment
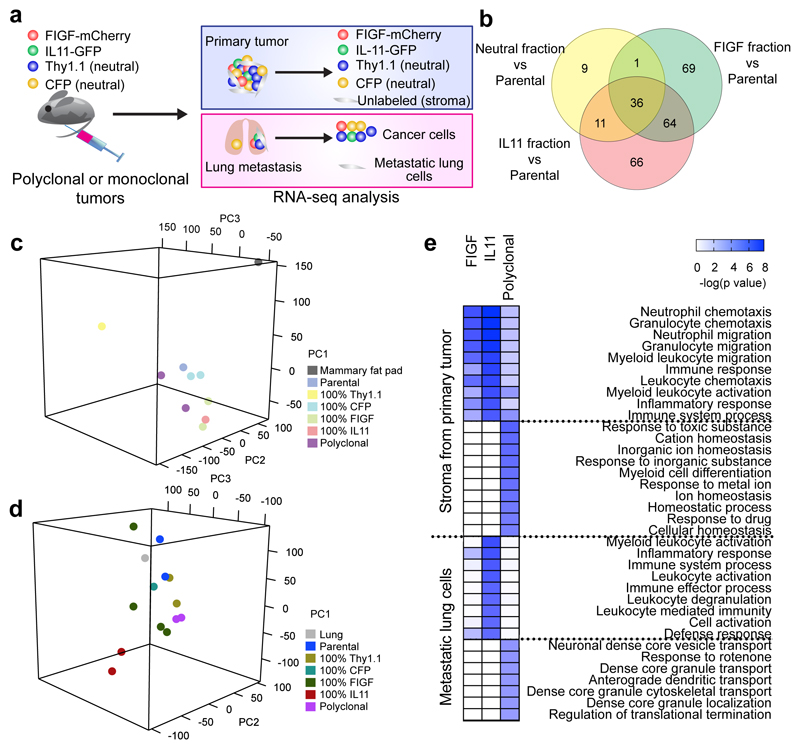
To explore mechanisms by which driver subclones cooperate to promote metastasis, we generated polyclonal tumors composed of fluorescently-labeled driver clones (10% FIGF-mCherry and 10% IL11-GFP cells) and neutral clones (40% Thy1.1 cell surface marker and 40% CFP [cyan fluorescent protein]-expressing cells). To be able to sort a sufficient number of cells of each fraction from these tumors, 10% driver clones was used in this experiment. As controls, we established tumors with 100% of each individual clone. Mice were terminated when the primary tumors reached ~2 cm. Primary and metastatic lesions were dissociated and fluorescently-labeled cancer and unlabeled murine cells were purified by FACS and analyzed by RNA-seq (Fig. 2a, Supplementary Fig. 1c, and Supplementary Table 2).
Figure 2. Driver clone-induced changes in the stroma of primary and metastatic tumors.
a, Schematic outline of the experimental design. b, Overlap of differentially expressed genes in cancer cell fractions from polyclonal primary tumors (n = 3) compared to parental cell line tumors (n = 3). c, d, Principal component analysis of RNA-seq profiles of stromal cells from mono- and polyclonal primary tumors (c) and non-tumor cells from metastatic lungs (d). n=3 independent tumors per group. e, MetaCore GO Processes overrepresented in expression profiles from stroma of primary tumors (n=3) and metastasis-bearing lung cells (n=3). Color scale corresponds to -log (p-value) of significance of enrichment, calculated by MetaCore Enrichement Analysis test.
First, we investigated whether a common transcriptional program was induced by the driver subclones in polyclonal tumors. We compared the expression profiles of each subclone purified from polyclonal tumors to that of parental tumors (Supplementary Table 3). We found 36 commonly upregulated genes in fractions from tumors with driver clones (Fig. 2b), including TMEM173 encoding STING, an immune modulator8, and several extracellular matrix (ECM) components. These expression changes suggested that driver subclones may promote metastasis by altering the tumor microenvironment. Therefore, we analyzed the differences in stromal cells (i.e., cells lacking human cancer cell markers) from monoclonal and polyclonal primary tumors and lungs. Since the parental cells did not generate metastases, we used lung tissue from mice with 100% parental cell tumors as controls in our comparisons. Principal component analysis demonstrated a trend toward clustering according to the tumor of origin, implying that tumor cells have a significant impact on stromal gene expression (Supplementary Fig. 2a,b and Fig. 2c,d). Using MetaCore9, we analyzed pathway and process network enrichment within upregulated genes in stromal cells of primary tumors capable of seeding metastases (i.e., containing a driver subclone) compared to the stroma of parental tumors (Supplementary Tables 3 and 4). Interestingly, 9 of 10 top significantly enriched pathways were immune-related (Fig. 2e and Supplementary Fig. 2c). In contrast, all 10 top pathways enriched in non-cancer cells from lungs of mice with polyclonal tumors were related to vesicular transport and cell motility (Fig. 2e). FIGF and IL11 fractions of polyclonal tumors compared to 100% FIGF+ or IL11+ tumors also showed upregulation of angiogenesis-related genes (Supplementary Table 3). Immunohistochemistry of CD31 blood and LYVE1 lymphatic vessel markers confirmed this upregulation even in primary tumors (Supplementary Fig. 2d).
We also analyzed the relative frequencies of each of the four subclones within polyclonal tumors by FACS and found that IL11+ and FIGF+ subclones remained as minor fractions (Supplementary Fig. 2e). Thus, their ability to drive tumor growth and metastasis did not require clonal expansion indicating a non-cell-autonomous mechanism. Therefore, subclonal cooperation in primary tumors promotes metastasis by modulating the microenvironment of primary and metastatic lesions.
Polyclonal tumors affect leukocytes
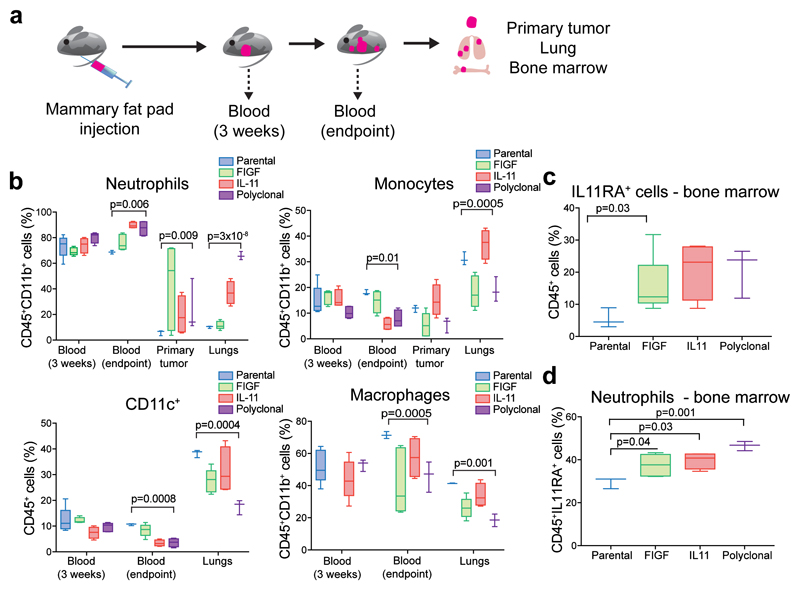
To follow up on our finding of immune-related changes in the stroma of metastatic-capable primary tumors and to analyze leukocytes, we repeated the polyclonal and monoclonal xenograft experiment using luciferase-expressing cells to monitor metastases. Immunodeficient NOG mice retain innate immune and myeloid cells (albeit not fully functional), and myeloid-related pathways were highly upregulated in the primary tumors driving metastases. We assessed qualitative changes in blood leukocytes by FACS at three weeks after tumor initiation and at the six-week experimental endpoint, when primary tumor, lungs, and bone marrow were also collected (Fig. 3a and Supplementary Fig. 3a). The most significant differences were detected in neutrophils and monocytes (Fig. 3b and Supplementary Fig. 3b). The relative fraction of neutrophils was significantly higher in the blood, primary tumors, and lungs of mice with polyclonal and IL11+ monoclonal tumors, compared to mice bearing parental cell tumors, while the fraction of CD11c+ cells was significantly lower in the blood and lungs, macrophages showed a slight decrease in primary tumors and lungs, and monocytes were decreased in the blood and primary tumors (Fig. 3b and Supplementary Fig. 3b).
Figure 3. The effect of polyclonal tumors on the immune system.
In all bar graphs: Box-and-whisker plots show mean (midline), 25th-75th percentile (box) and 5th-95th percentile (whiskers). p value of unpaired two-tailed t-test. a, Schematic outline of the experimental design. b, Analysis of myeloid cells from the blood, primary tumors, and lungs of mice bearing parental, 100% FIGF or 100% IL11 monoclonal or polyclonal tumors; n = 5 per group. c, Fraction of bone marrow CD45+ cells expressing IL11RA; n = 5 per group. d, Fraction of neutrophils among CD45+IL11RA+ bone marrow cells; n = 5 per group. See also Supplementary Table 1 for raw data.
Neutrophil influx has been associated with IL11 signaling and inflammatory bursts10; therefore, we analyzed bone marrow CD45+ cells for IL11Rα expression. Approximately 20-30% of CD45+ cells of mice bearing IL11+ or FIGF+ monoclonal and polyclonal tumors were IL11Rα+ (Fig. 3c), and the fraction was significantly higher in mice with IL11+ or FIGF+ monoclonal and polyclonal tumors, compared to mice with parental tumors (Fig. 3d). Hence, the presence of minor metastasis-promoting subclones within tumors alters the immune system, particularly the neutrophil population.
In breast cancer patients, distant metastases are usually recurrences after surgical removal of primary tumors, and some studies suggested that the surgery itself may induce metastatic lesions11,12. Thus, in another experiment, we performed mock or primary tumor removal surgery but did not observe any effects of surgery on metastatic burden in lungs (Supplementary Fig. 4a-c). Therefore, surgery-related wound healing and inflammation are not inherently responsible for the observed immune-related changes or increased metastasis in our polyclonal tumors.
IL11 and FIGF cooperate to induce metastasis-promoting systemic changes
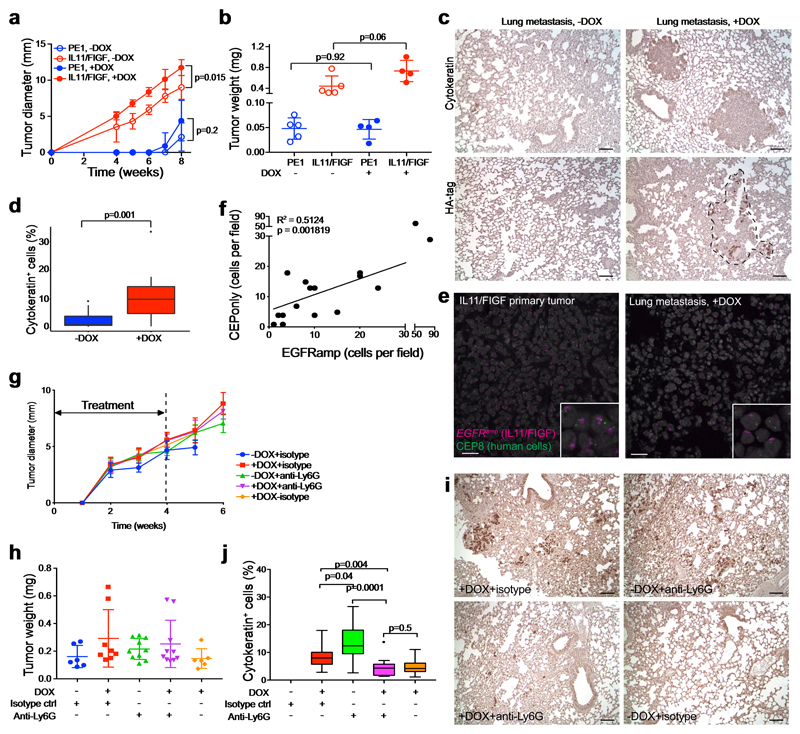
Next, we investigated whether the metastasis-promoting subclones exert their effects via local or systemic changes. We injected a 1:1 mixture of MDA-MB-468 cells expressing doxycycline (DOX)-inducible, HA-tagged IL11 and FIGF into the right mammary fat pad and a slow-growing, weakly metastatic patient-derived xenograft (PE1) into the contralateral mammary fat pad. Mice were randomized into DOX -/+ groups to induce IL11 and FIGF expression. The IL11+FIGF+ tumors grew faster than parental tumors, as expected, although the difference was not as profound as in polyclonal tumors with constitutive expression. The growth of PE1 primary tumors was not affected by the contralateral IL11+FIGF+ tumor (Fig. 4a,b), but lung metastases were significantly increased in DOX+ mice (Fig. 4c,d). Using immunohistochemistry for the HA-tag, we found that metastatic lesions were a mixture of HA+ and HA- cells, with the latter presumably representing PE1 cells (Fig. 4c). To confirm that these negative cells were not MDA-MB-468 cells that lost HA-tagged IL11 and FIGF expression, we performed fluorescence in situ hybridization (FISH) for EGFR and human-specific CEP8, since MDA-MB-468 cells have an EGFR amplification not detected in PE1, while CEP8 marks all human cells. This analysis clearly demonstrated that metastases were a mixture of PE1 andEGFRamp IL11+/FIGF+ MDA-MB-468 cells (Fig. 4e). Single-cell quantification of the FISH signal showed a significant correlation between the numbers of IL11+/FIGF+ (EGFRamp) and PE1 (CEP8only) cells, implying potential local interactions between these populations (Fig. 4f). Therefore, IL11 and FIGF promote metastasis via systemic effects, potentially mediated by neutrophils.
Figure 4. Systemic effects of metastasis-driver subclones.
a-d, Effects of driver subclones (IL11 and FIGF under a DOX-inducible promoter) on the growth and metastasis of contralaterally injected patient-derived xenografts. a-b, Primary tumor growth kinetics (a) and final tumor weight (b); n = 5 animals per group; representative of two independent experiments. PE1 – non-metastatic patient-derived xenograft. DOX – doxycycline. Mean ± SD shown. Two-tailed unpaired t-test p values are shown. c, Representative immunohistochemical images of the indicated staining. d, Fraction of cytokeratin+ cells. On average, 2,900 cells were counted per field, three fields per sample. Lungs of five mice per group were analyzed. Data shown are representative of two independent experiments. Box-and-whisker plots show mean (midline), 25th-75th percentile (box), 5th-95th percentile (whiskers) and outliers (dot). Two-tailed unpaired t-test p values are shown. e, Representative images of EGFR and CEP8 FISH on primary tumor and lung metastasis under DOX treatment. Scale bar 25 μm. Staining was repeated twice with similar results. f, Correlation between numbers of cells that are EGFRamp or have CEP8 only FISH signal per metastatic lesion. 3-4 fields of view were quantified in lungs from 4 animals; total of 16 lesions analyzed. Pearson correlation R2 and two-tailed t-test p value are indicated. g-j, Neutrophil depletion in mice bearing polyclonal tumors with minor driver subclones. g-h, Tumor growth kinetics (g) and final tumor weight (h, n = 5 per group). Dashed line marks the anti-Ly6G and isotype antibody treatment duration. Mean ± SD shown. i, Representative images of cytokeratin staining in lungs of treated mice. Scale bar 100 μm. j, Quantification of cytokeratin+ cells. On average, 2,600 cells were counted per field, three fields per sample. Lungs of five mice per group were analyzed. Box-and-whisker plots show mean (midline), 25th-75th percentile (box), 5th-95th percentile (whiskers) and outliers (dot). p values indicate statistical significance of the observed differences defined by unpaired two-tailed t-test. See also Supplementary Table 1 for raw data.
Depletion of neutrophils affects metastases
To determine whether neutrophils are required for the increased metastasis driven by IL11 and FIGF, we treated mice with a neutrophil-depleting Ly6G antibody13. To determine the optimal timing to initiate treatment, we used mathematical modeling to predict the onset of metastasis based on our bioluminescence data (see Methods for details). Our computational framework considers two types of cells – those residing in the primary tumor and those that have left the primary site (Supplementary Fig. 4d). Each time a cell leaves (at a dissemination rate q), it establishes a new metastatic colony. Growth and dissemination rates were estimated from experimental data (Supplementary Fig. 4b), dependent on the experimental condition (Supplementary Fig. 4e). Briefly, to estimate the metastatic rate q, we minimized the distance between experimental results and model predictions for the distribution of the size of the first metastasis six weeks after the start of the experiment and the primary tumor size when the first metastasis is detected. We identified the metastatic rate as 10-7 < q < 10-6 per cell per week and, using this, defined the timing of the first metastatic cell leaving the primary site as 5.18±2.45 days for IL11+ monoclonal tumors and 5.11±2.45 days for polyclonal tumors when q = 10-7, and 1.33±1.05 and 1.68±0.91 days, respectively when q = 10-6 (Supplementary Fig. 4f-k). Thus, we concluded that to prevent metastatic seeding, treatment should be initiated immediately after tumor inoculation.
We initiated polyclonal xenografts with 10% DOX-inducible IL11+ and FIGF+ and 90% parental cells and assigned animals to receive DOX with a neutrophil-blocking anti-Ly6G antibody or an isotype control, administered for four weeks (Supplementary Fig. 5a). We confirmed the efficacy of anti-Ly6G antibody treatment after two weeks by assessing blood neutrophil counts (Supplementary Fig. 5b-d). At 6 weeks, we found that neutrophil depletion did not affect primary tumor growth (Fig. 4g,h), but significantly decreased CK+ metastatic cells in the lungs of mice compared to isotype control (Fig. 4i,j). Comparing to untreated animals achieved borderline significance. Unexpectedly, anti-Ly6G treatment increased the metastatic load in -DOX mice with tumors not expressing IL11 and FIGF (Fig. 4i,j). These results demonstrate that systemic depletion of neutrophils decreases metastatic spread (or outgrowths) of polyclonal tumors driven by IL11+ and FIGF+ subclones and can have opposing effects on metastases depending on factors produced by the primary tumor. Furthermore, injection of an isotype control antibody itself can enhance metastasis. Therefore, the phenotype and functional properties of neutrophils depend on the systemic host environment.
Single-cell analysis of CD45+ cell populations
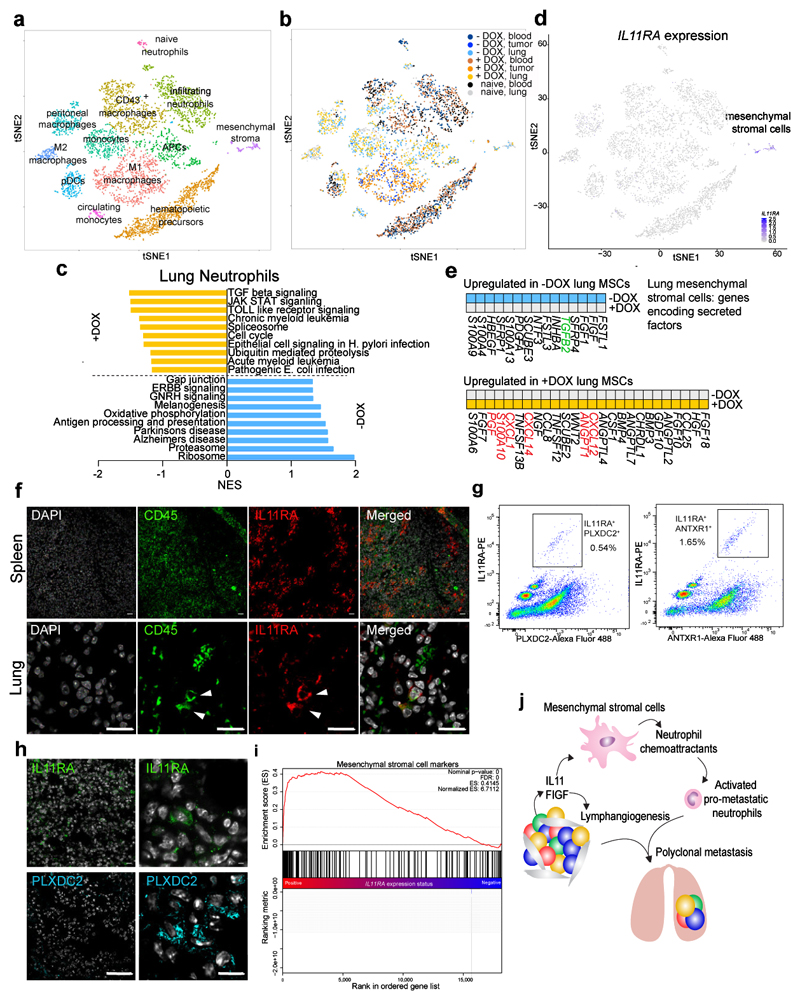
To further investigate the effects of IL11+ and FIGF+ subclones on leukocytes, we repeated the above experiment with polyclonal xenografts composed of 10% DOX-inducible IL11+ and FIGF+ and 90% parental cells. Six weeks after tumor inoculation, we purified CD45+ cells from blood, primary tumors, and lungs by FACS, and performed single-cell RNA-seq (scRNA-seq)13. Clustering analysis revealed a minor fraction of CK+EGFR+ contaminating cancer cells that were removed from subsequent analyses (Supplementary Fig. 6a-c), as well as numerous identifiable leukocyte subpopulations (Fig. 5a, Supplementary Fig. 6d-g, and Supplementary Table 5). CD45+ cells from lungs, blood, and primary tumors contributed to the distinct cell clusters to different extents (Fig. 5b). For example, M1 macrophages were found only in primary tumors, whereas M2 macrophages were only present in lung metastases. Since our previous results indicated that neutrophils contribute to metastasis, we compared the single-cell transcriptomic profiles of blood and lung neutrophils from mice with or without DOX. IL11 and FIGF induced upregulation of several pathways, including TGFβ and JAK/STAT signaling, associated with pro-tumorigenic and pro-metastatic immune signatures, in neutrophils from lungs but not from blood (Fig. 5c and Supplementary Tables 6 and 7). Despite this change in lung neutrophils, we did not detect IL11 or FIGF receptor expression in this cell population by scRNA-seq. However, cells with IL11RA transcripts were clearly present in a separate cluster (Fig. 5d), not classified as either neutrophils or any other leukocyte subpopulation. These IL11RA+ cells express IL6ST encoding GP130, a coreceptor necessary for IL11 signaling, and STAT3, a downstream IL11 effector (Supplementary Fig. 6f). Based on gene expression in this cluster (Supplementary Table 8), which also included ECM and development-related proteins, we denoted this population as IL11-responsive mesenchymal stromal cells (MStrCs). Although this population does not express the classic mesenchymal stem cells (MSC) markers, its signature is rich in stem cell-related genes, implying that it could be a previously uncharacterized MSC-like cell type. Prior studies have described interactions between MSCs and leukocytes mediated by various cytokines and chemokines14. Thus, we identified secreted factors from our MStrC-specific expression signatures using gene-enrichment analysis and analyzed them in different conditions (Fig. 5e). Comparison of lung MStrCs from -/+ DOX animals revealed that the induction of IL11/FIGF expression in the primary tumors upregulated the expression of chemoattractants for pro-metastatic neutrophils including CXCL12, CXCL14, and CXCL1 (Fig. 5e).
Figure 5. Single-cell RNA-seq data analysis of CD45+ cells.
a-b, tSNE clustering of single-cell expression profiles. Each dot represents a single cell (n=7,704), and the distance between cells is a measure of their transcriptional similarity. a, Twelve distinct clusters of immune were identified according to expression of cell type-specific marker genes or expression signatures (Supplementary Fig. 6 and Supplementary Table 5). Colors denote distinct clusters of cells. b, Contribution of cells from different samples to the distinct clusters. Cells are colored according to their sample of origin. c, Gene expression signature analysis of lung neutrophils from mice with (+DOX) or without (-DOX) IL11/FIGF-expressing tumors. NES – normalized enrichment score. d, The expression of IL11RA in the single cells analyzed (n=7,704). e, Secreted factors among cluster-specific differentially expressed genes in lung mesenchymal stromal cells. Genes highlighted in green and red are characteristic of anti-tumorigenic neutrophils and neutrophil chemoattractants, respectively. f, Immunofluorescence analysis of CD45 and IL11RA in spleen and lungs of mice with IL11/FIGF-expressing tumors. White arrows highlight rare CD45+IL11RA+ cells with mesenchymal morphology in the lungs. Staining was repeated twice with similar results. g, Flow cytometry analysis of lungs of tumor-free mice stained with AF55-conjugated IL11RA, and AF488-conjugated PLXDC2 and ANTXR1 (TEM8) antibodies. Highlighted populations show IL11RAhiPLXDC2+ as well as IL11RAhiANTXR1+ double positive cells. h, Immunofluorescence analysis depicting rare IL11RA+ and PLXDC2+ cells with mesenchymal morphology in the lungs of +DOX-isotype control-treated mice. Staining was repeated twice with similar results. i, Mesenchymal stromal cell signature enrichment in lung cells FACS-sorted for IL11RA expression from tumor-bearing mice (n=3 animals). P values of the Gene Set Enrichment Analysis (Wald test with Benjamini-Hochberg multiple comparison correction) are shown. j, Model of metastatic cascade driven by IL11/FIGF-expressing minor primary tumor subclones.
To validate the presence of CD45+IL11RA+ cells in lungs and to assess co-expression of MStrC markers with IL11RA we performed immunofluorescence and FACS. CD45+IL11RA+ cells were relatively common in the spleen and present as a rare subpopulation with mesenchymal morphology in the lungs (Fig. 5f). Rare IL11RAhigh cells co-expressing PLXDC2 and ANTRXR1, markers of these cells based on scRNA-seq (Supplementary Table 8), were also identified in the lungs by FACS representing 0.5-1% of cells (Fig. 5g and Supplementary Fig. 6h-l). Immunofluorescence analysis of lungs demonstrated the presence of rare IL11RA+PLXDC2+ cells with mesenchymal morphology (Fig. 5h). Lastly, we sorted cells from lungs of mice with IL11+/FIGF+ tumors by IL11RA expression and analyzed their gene expression profiles. Our MStrC signature was significantly enriched in IL11RAhigh samples (Fig. 5i) and enrichment for cell adhesion and ECM remodeling-related process networks was noted (Supplementary Table 9).
In summary, our scRNA-seq data and its subsequent validation by FACS, immunofluorescence, and bulk RNA-seq uncovered an interaction loop among different subpopulations of bone marrow-derived cells that mediates the pro-metastatic effects of tumor cell-secreted IL11 and FIGF (Fig. 5j). The expression of IL11 and FIGF in a minor subpopulation of cancer cells in polyclonal tumors alters these MStrCs in the lungs, leading to the activation of “effector” neutrophils, which are crucial for metastatic progression.
IL11-driven signatures in human metastatic breast cancer
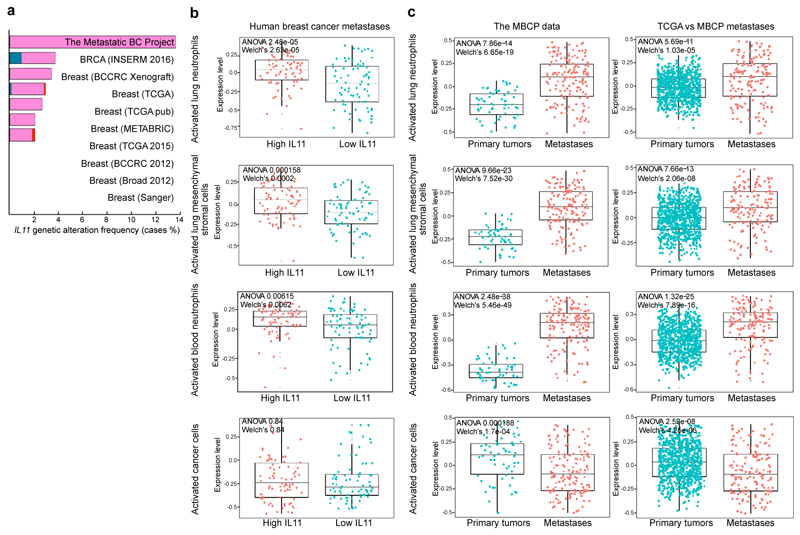
To investigate the clinical relevance of our findings, we analyzed the expression of IL11 and IL11-driven neutrophil and MStrC signatures in breast cancer metastases. IL11 is amplified in a subset of primary breast tumors, and this amplification is more commonly observed in metastases (cBioPortal15,16, Fig. 6a). Based on our data, IL11 and FIGF can drive significant systemic changes even when expressed by minor subclones making them difficult to detect. Thus, we explored whether gene signatures of IL11/FIGF-activated neutrophils and MStrCs, defined by our single-cell RNA data, are present in distant metastases in the Metastatic Breast Cancer Project (MBCP) dataset17, which contains RNA expression data of 156 distant metastatic lesions. Since most of the systemic changes were IL11-driven in our experimental system, we classified metastatic lesions into IL11high and IL11low groups based on mRNA levels. We then compared the IL11high and IL11low metastases for the expression of gene signatures activated by IL11/FIGF in lung and blood neutrophils and in MStrCs (Supplementary Table 6). Signatures for activated lung and blood neutrophils and lung MStrCs were significantly higher in IL11high than IL11low metastases (Fig. 6b). In contrast, gene signature of IL11/FIGF-activated cancer cells, derived from genes expressed by neutral clones in polyclonal tumors, did not correlate with IL11 expression in metastases (Fig. 6b, bottom panel). Analyzing the expression of these gene signatures in matched primary and metastatic lesions using the MBCP dataset showed that activated neutrophil and MStrC signatures were significantly higher in metastases, while cancer cell signatures did not show a difference (Fig. 6c, left). To eliminate potential bias due to differences in tissue type used for RNA extraction (FFPE for primary tumors and frozen for metastases), we also compared the MBCP metastases to ER+ tumors in the TCGA data with essentially the same findings (Fig. 6c, right). Therefore, IL11/FIGF-driven microenvironmental changes are also detected in human breast cancer patients and are associated with distant metastases.
Figure 6. IL11 is associated with metastasis in breast cancer patients.
a, Frequency of genetic alterations of the IL11 locus in cBioPortal breast cancer dataset. b, Expression of IL11/FIGF-activated signatures of lung and blood neutrophils, mesenchymal stromal cells and primary cancer cells in human breast cancer metastases with high and low IL11 levels. n=156. Cancer cell signature was derived from neutral clone isolated from polyclonal primary tumors. c, Gene expression signatures of IL11/FIGF-activated cells in the Metastatic Breast Cancer Project dataset (matched primary (n=58) and metastatic lesions (n=156); left) and in the ER+ TCGA primary tumors (n=868) versus MBCP metastatic lesions (n=156; right). Box-and-whisker plots show mean (midline), 25th-75th percentile (box) and 5th-95th percentile (whiskers). ANOVA p values and two-sided Welch’s test p values are shown. See also Supplementary Table 1 for raw data.
Discussion
Polyclonal metastasis of breast cancer has been described in several recent studies18,19. Here, we provide evidence that in polyclonal tumors, minor subpopulations can drive metastatic progression and outgrowth of polyclonal metastases that also contain neutral subclones (non-metastatic on their own) by modulating local and systemic microenvironments.
In our study, we focused on two secreted factors expressed by minor subclones that cooperate to drive metastasis, selected based on our prior data7. IL11 belongs to the IL6 family of cytokines and has been shown to play a role in progression and resistance in multiple cancer types including prostate and colon cancer20,21. In breast cancer, IL11 has been implicated in therapeutic resistance22 and bone metastasis23, as well as a marker of poor outcomes24. FIGF is a ligand for VEGFR2 and VEGFR3 and can stimulate both angiogenesis and lymphangiogenesis25,26. Our results show that FIGF enhances dissemination, as evidenced by increased numbers of CK+ cells in the lungs, but is not sufficient to drive macrometastatic outgrowths. In our model, IL11 and FIGF secreted by minor subclones have profound effects on the metastatic ability of breast cancer cells. While IL11 acts on leukocytes to induce metastasis-permissive changes in the microenvironment of distant organs, FIGF acts more locally by increasing lymphangiogenesis and vascular permeability. However, because these subclones represent only 1-5% of tumor cells, their presence and their secreted factors may be difficult to detect in clinical samples, making it challenging to predict their metastasis-promoting effects in human tumors. Nevertheless, our analysis of human metastatic breast tumors suggests that the systemic changes in the immune and other bone marrow-derived cells induced by even low levels of these secreted factors can be detected and potentially exploited for patient stratification and treatment.
We identified neutrophils as the leukocyte population required for IL11- and FIGF-induced polyclonal metastasis. A caveat of our approach is the use of immunocompromised animals, which did not allow us to study the roles of lymphoid cells in metastatic cooperation. Thus, it is possible that other immune cells may also play a role in the metastatic process. Neutrophils have been implicated in metastatic progression by promoting the premetastatic niche27–31. Furthermore, high pre-operative neutrophil-to-lymphocyte ratio is associated with poor prognosis in breast cancer patients32. Neutrophils have also been shown to have antitumor effects and inhibit seeding of cancer cells in the lungs33,34. In our experiments, depending on whether or not the tumors expressed IL11 and FIGF, depleting neutrophils could either decrease or enhance lung metastasis. These seemingly contradictory roles for neutrophils could potentially be explained by our observation, based on scRNA-seq data, that there are multiple different types of neutrophils, potentially pro- and anti-tumorigenic, depending on anatomical location and gene expression patterns. Our findings are therefore in agreement with reports that the context and temporal infiltration of monocytes and neutrophils is critical to outcome31,35,36 and that heterogeneity of not only tumor cells but also immune cells must be considered. Another point to consider is that injection of isotype control antibody unexpectedly increased metastatic burden potentially by triggering an inflammatory response. The magnitude of neutrophil-depletion effect on metastasis may therefore be very sensitive to the systemic environment of the host.
The use of single-cell expression profiling of CD45+ cells in our study allowed us to uncover interactions between different leukocytes subpopulations. We found that leukocytes may not be the direct cellular targets of IL11, but indirectly affected via factors secreted by MStrCs responsive to IL11. Interestingly, these cells also express PLXDC2 and ANTXR1, which are highly expressed in tumor-associated endothelial cells37,38. Thus, these IL11RA+ mesenchymal cells may represent progenitors that can give rise to multiple cell types. Further characterization of these cells and their effector leukocytes will shed new light on the biology of pro-tumorigenic neutrophils. An improved understanding of neutrophil subtypes and the development of tools to target pro-metastatic neutrophils, in combination with cancer cell-targeting drugs, could potentially be used to prevent breast cancer metastasis.
Methods
Cell lines and tissue culture conditions
Parental MDA-MB-468 cell line was used to generate IL11- and FIGF-overexpressing subpopulations using lentiviral vectors, as previously described7. Inducible IL11 and FIGF expression was achieved by cloning the respective cDNAs into pLentiX2 vectors, followed by viral transduction of respective constructs into parental MDA-MB-468 cells7, and selection with 2μg/ml puromycin. All cell lines were cultured in McCoy’s medium (Cell Grow) with 10% fetal bovine serum (HyClone) and 10 μg/ml insulin (Life Technologies), at 37°C with 4% CO2. Cell line identity was confirmed by short tandem repeats (STR) analysis and the cells were regularly tested for mycoplasma (Venor GeM Mycoplasma Detection Kit, Sigma).
PE1 was a primary cell line from pleural effusion of a patient with triple-negative (ER-PR-HER2-) breast cancer. The pleural effusion was collected under Dana-Farber Harvard Cancer Center (DF/HCC) Institutional Review Board (IRB) protocol #93-085 following written informed consent and used in the lab in compliance with DF/HCC IRB protocol #14-400 approved for the use of de-identified tissue samples. The study is compliant with all relevant ethical regulations regarding research involving human participants.
Xenograft experiments
All animal procedures were approved by DFCI IACUC and performed according to DFCI protocol #11-023. The study is compliant with all relevant ethical regulations regarding animal research. Female NOG (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) mice (Taconic) were used at 4-6 weeks of age. For tumor growth assays, 2 million cells in 50% Matrigel (BD Biosciences) in DMEM media (Gibco), total volume 50μl, were injected per mammary fat pad in a total volume 50 μl. Tumor growth was measured weekly with a caliper. In some experiments, metastatic growth was followed by weekly bioluminescence imaging measurements of luciferase activity using an IVIS imaging system. The size of macrometastasis was determined by Living Image software (Perkin Elmer) analysis of the areas of bioluminescence in the images. Regions of interest (ROI) were selected automatically by the software and the area of each individual ROI was measured. Parental tumor ROI was excluded from the analysis. Blood samples at early time-points were collected via retroorbital bleeding. For in vivo inducible expression of IL11 and FIGF, mice were switched to doxycycline diet 24 h after mammary fat pad injection.
FACS analysis
For all experiments, blood was freshly collected and processed immediately, while bone marrow was viably frozen after erythrolysis. Tumor and lung samples were digested to single-cell suspension with collagenase type IV (Worthington, cat# LS004189)/DNase I digestion buffer (2 mg/ml and 0.02 mg/ml, respectively, in DMEM with 10% FBS, 1% PenStrep). The digestion was performed at 37°C for 1 h for tumor tissues and for 0.5 h for lung tissues, with stirring. Digestion mix was terminated with DMEM with 10% FBS, and cells were washed and stained immediately. Red blood cells were lysed with lysis buffer (BD Pharm Lyse, BD Biosciences). After washing with FACS buffer (2% FBS in PBS and 2mM EDTA), cells were stained with antibody cocktail 1 or 2 (Supplementary Table 2) for 20-30 min on ice and washed twice. The final cell pellet was resuspended in FACS buffer with or without 7AAD for viability detection. The analysis was performed on a BD FACS Canto system (BD Biosciences). Gating and PBMC subpopulation analysis were performed using FlowJo software.
In vivo neutrophil depletion
Twenty-four hours post-mammary fat pad injection, the mice were treated with 12.5 μg anti-Ly6G (BioCell) or isotype control antibody (BioCell) in a 100 μl volume via intraperitoneal injection. The treatment was administered daily for 28 days. On-target effect was measured after two weeks of treatment by peripheral blood mononuclear cell (PBMC) FACS analysis of mouse blood.
Immunofluorescence and immunohistochemistry
Xenograft samples and mouse organs were fixed in buffered formalin overnight after collection and stored in 70% ethanol before being processed into formalin-fixed paraffin-embedded blocks by the DFCI Pathology Core. Staining was performed on 5-μm sections. Before staining, the slides were baked at 65°C and standard deparaffinization protocol using xylene and ethanol washes was used. If required, antigen retrieval was performed using citrate buffer (pH 6; Invitrogen) or Target Retrieval solution (pH 9; Dako) for 20 min in a steamer. Some antibodies then performed better after a 10-min permeabilization step using 0.5% Triton X-100 (Sigma Aldrich) in PBS. The specific antibodies and dilutions used are listed in Supplementary Table 2. Incubation with primary antibodies diluted with PBST buffer (0.05% Tween-20 in PBS) in 5% goat serum was performed overnight at 4°C. After three washes with PBST, secondary antibody was applied to the slides and incubated for 1 h at room temperature. For immunohistochemistry, the ABC peroxidase system (Vectastain, ABC System Vector Laboratories) was used with 3,3’-diaminobenzidine (DAB) was used as the colorimetric substrate. Counterstaining was performed using Harris hematoxylin and Scott’s tap water substitute (Leica). Automated quantification of individual positive cells was performed using ImageJ macros (available upon request) by scoring total cell numbers based on nuclear stain and strongly positive cells across five different areas of a given specimen. Following immunofluorescent staining slides were counterstained with DAPI (Vectashield Hard Set Mounting Medium with DAPI) and stored for at least 24 h at -20°C to reduce autofluorescence. Images from multiple areas of each sample were acquired using a Nikon Ti microscope attached to a Yokogawa spinning-disk confocal unit using a 603 plain apo objective, and OrcaER camera controlled by Andor iQ software; or a Leica SP5 confocal scanning microscope.
Stroma and epithelial cell fractionation and RNA-seq
Cell fractionation
To separate subpopulations from polyclonal tumors, the following MDA-MB-468 cell line derivatives with fluorescent tags were used: neutral clones (expressing CFP and Thy1.1) and with IL11-mCherry and FIGF-GFP clones. Polyclonal tumors were dissociated to single-cell suspension by digestion in collagenase/hyaluronidase digestion buffer (2 mg/ml each, in DMEM with 10% FBS, 1% Pen Strep). After washes with PBS, the samples were resuspended in FACS buffer (2% FBS in PBS) and the populations expressing CFP, mCherry, GFP, and Thy1.1 were sorted into FACS buffer, pelleted and stored at -80°C. Negative cell population was also collected, as this constitutes the tumor stroma. The same procedure was performed on metastatic lungs, except all labeled cell were collected as a group, due to their overall small number within metastatic lungs.
mRNA extraction and library preparation
mRNA selection and whole transcriptome amplification were performed as previously described with some modifications39,40. A primer 5’-phosphorylated oligo-dT24 (pdT24) was used to select mRNA from total RNA. For denaturation and primer annealing, total RNA was mixed with dNTPs (25 mM) and pdT24 and put in a preheated 68°C PCR machine for 5 min. and then immediately placed on ice. Reverse transcription was performed with the addition of the First-Strand buffer, DTT, RNase Inhibitor, and 10 U/μl Superscript Reverse Transcriptase III. Double-stranded cDNA was generated by the addition of 30 μl mix containing 10x Second Strand buffer, dNTPs (25 mM), E. coli DNA ligase (10 U/μl), RNaseH, and DNA Polymerase (10U/μl), followed by a 3 hr incubation at 16°C and 5 min. at 70°C. The product was purified with the Genomic DNA Clean and Concentrator kit (Zymo Research). Then, DNA blunt-ending, 5’-end phosphorylation, and ligation were performed with End-It DNA End-Repair Kit (Epicentre) and T4 DNA ligase (Epicentre). The product was directly amplified by adding a mixture of Reaction Buffer and DNA polymerase from REPLI-g UltraFast Kit (Qiagen) at a 15:1 ratio. The amplification reaction was carried out at 30°C for 2 hrs. The amplified product was purified using the same Genomic DNA Clean and Concentrator kit (Zymo Research). When eluted in 50 μl elution buffer, typically 5-10 μg of mRNA-derived cDNA amplicons were obtained. The amplicons were then evaluated for quantity, coverage, and purity by PCR using a set of primers for housekeeping genes (GAPDH: Forward, 5’-GTCTCCTCTGACTTCAACAGCG-3’, Reverse, 5’-ACCACCCTGTTGCTGTAGCCAA-3’; B2M: Forward, 5’-CCACTGAAAAAGATGAGTATGCCT-3’, Reverse, 5’-CCAATCCAAATGCGGCATCTTCA-3’), cDNA 5’ end (CERB5: Forward, 5’-GTCAGTGAACTCCAGCATCATGG-3’, Reverse, 5’-GTGGTGAGTCAATGCAGCCTTC-3’), and gDNA (10p Forward, 5’-GTTCTGCTGCCTCTACACAGG-3’, Reverse, 5’-ATCCTTCTGTGAACTCTCAAATTC-3’). cDNA libraries were constructed using Nextera XT DNA Sample Preparation kit (Illumina) according to the manufacturer’s recommendations with minor modifications. Briefly, cDNA amplicons were fragmented and barcoded by tagmentation using Tn5 DNA transposase. Tagmented cDNA was amplified with a limited-cycle PCR program. During the PCR step, indices 1 and 2 were added to enable cluster generation and multiplexed sequencing. The extension time was increased from 30s to 60s. The amplified cDNA libraries were purified and cleaned using 0.6x Agencourt AMPure XP (Beckman Coulter) and fresh ethanol and eluted in TE buffer. The quality of sequencing libraries was assessed using a high-sensitivity DNA chip (Agilent). Finally, the libraries were deep sequenced using a HiSeq2000 sequencer (Illumina).
Clustering based on differentially expressed genes
We analyzed stroma from primary tumors and metastatic lungs separately. First, we normalized for sequencing depth and applied the variance stabilizing transformation (VST), both implemented in DESeq2. The top 1000 most variable genes were used to cluster the samples based on the expression of these genes (Euclidean distance and complete clustering). The heatmap itself depicts relative VST-transformed values across samples (the difference between the sample’s value for a given gene and the mean expression of the given gene across all samples), hence the negative values. Note that this way of displaying the expression does not affect the sample clustering when compared to clustering based on expression correlation values.
RNA-seq of lungs
Lungs from tumor-bearing mice with and without doxycycline were isolated and dissociated into single cell suspension as described above. Lungs from 5 mice were pooled per group with two replicates. Staining for IL11R and PLX DC was performed as described in FACS analysis. IL11R-, IL11R+PLX DC- and IL11R+PLX DC+ populations were FACS-sorted into RLT buffer with β-mercaptoethanol (Qiagen). Then, total RNA was isolated as described by the manufacturer using the RNeasy mini kit (Qiagen). mRNA extraction and library preparation were performed as described above.
RNA-seq analysis
Raw sequencing data were de-multiplexed into fastq files with bcl2fastq software (Illumina) based on unique base pairs. Quality check of the called bases of the reads were performed using the FastQC tool (Babraham Bioinformatics). Tumor sample fastq files were trimmed to 40 bases. Datasets of tumor origin were aligned to the human reference GRCh37/hg19 genome and datasets of stroma origin were aligned to the mouse reference GRCm38/mm10 genome, both using the STAR RNA-Seq aligner (version STAR_2.5.1b). Two-pass mapping was performed using the following modified parameters: --outSAMstrandField intronMotif, --outFilterMultimapNmax 20, --alignSJoverhangMin 8, --alignSJDBoverhangMin 1, --outFilterMismatchNmax 999, --outFilterMismatchNoverLmax 0.1, --alignIntronMin 20, --alignIntronMax 1000000, --alignMatesGapMax 1000000, --outFilterType BySJout, --outFilterScoreMinOverLread 0.33, --outFilterMatchNminOverLread 0.33, --limitSjdbInsertNsj 1200000, --chimSegmentMin 15, --chimJunctionOverhangMin 15, --twopassMode Basic. Samples were filtered to remove contamination (mouse contamination in tumor samples and human contamination in stroma samples). Reads uniquely mapped only to the target genome were kept, along with uniquely mapped reads that had significantly better alignment scores in the target genome compared to the contamination genome. Read counts for individual genes were generated using the htseq-count script of the HTSeq framework (version 0.6.1p1) using modified parameters (--stranded no) and the hg19/mm10 refGene annotation file available at the UCSC Genome. For stroma samples, reads for technical duplicates were added together. DESeq2 R package version 1.16.1 was used to generate differential expression gene lists, with fold-change value >=2 and padj value <=0.05.
Single-cell RNA-seq
Blood, tumors, and lungs were isolated from test groups and tumor-naïve animals. Tissues were dissociated into single cell suspension as described above. Staining for CD45 was performed as for PBMC FACS analysis. Samples were pooled across 3 animals per group, and CD45+ populations were FACS-sorted into PBS with 2% RNase-free BSA (Ambion). Then, the cells were processed according to the 10xGenomics sample preparation protocol (Chromium Single Cell 3’ v2 Reagent Kit, 10xGenomics). Two thousand cells were targeted for each sample.
Single-cell RNA-seq data analysis
Generating counts
Cell Ranger v. 1.2 was used to preprocess the Chromium single cell 3’ RNA-Seq output. Specifically, cellranger mkfastq was used to demultiplex the samples and produce FASTQ files, cellranger count was used to align and filter the FASTQ files individually, generating gene-barcode matrices, and cellranger aggr was used to aggregate all samples into a single combined matrix, normalizing the sequencing depth across runs. The resulting data was analyzed with the R (v.3.4.) package Seurat v. 2.1.041.
Quality control and normalization
The data was filtered by selecting all genes expressed in at least 1 single cell, and all cells with at least 100 and at most 5,000 expressed genes, and with a maximum percent of mitochondrial DNA of 0.1. This amounted to keeping a total of 18,203 genes and 7,704 cells. Gene expression measurements for each cell were normalized by the total expression, further multiplying by a factor of 10,000 and log-transforming the result (normalization method LogNormalize in Seurat).
Variable genes across cells
Highly variable genes across cells were identified with the Seurat function FindVariableGenes with parameters mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.0125, x.high.cutoff = 3, and y.cutoff = 0.5. This amounted to selecting 2,571 genes for downstream dimensionality reduction and clustering. The number of UMIs and the percentage of mitochondrial DNA across all cells were regressed against each gene, and the resulting residuals were further scaled and centered.
Dimensionality reduction
Principal component analysis (PCA) was performed on the scaled data. After regressing out unwanted sources of variation, the PCA component explaining the highest percentage of variation was uncorrelated with the number of genes expressed (Pearson correlation 0.01). Cells were clustered based on the PCA scores of the first 20 principal components, selected as strongly significant after running the resampling test implemented in the Seurat function JackStraw. The clustering of the data was based on the graph-based procedure implemented in the Seurat function FindClusters, for which a resolution parameter of 0.3 was chosen. Briefly, the underlying clustering methodology includes constructing a K-nearest neighbor on the Euclidean distances among cells in the PCA space, and refining edge weights based on the Jaccard distance between any two cells. tSNE was used for visualizing the data, which clustered into 14 clusters. Among those, one cluster consisting of 712 cells was identified to represent tumor cells, by its high expression of EGFR and numerous keratins, among which KRT14 and KRT18 (Supplementary Fig. 6c). The 712 cells were removed from the dataset. The new data was clustered again, using a resolution parameter of 0.2, which resulted in 12 clusters.
Differential expression
Differentially expressed genes between clustered were identified using the Wilcoxon rank sum test, as implemented in the Seurat function FindAllMarkers, with a minimum fraction of expression across cells of 0 (min.pct) and an expression threshold on the differences in log fold changes of 0.25 (logfc.threshold).
Cell type identification
Cell clusters were assigned to cell types based on expression of classical cell type-specific markers and based on similarity between top 30 cluster-specific genes and the ImmGen consortium datasets (http://rstats.immgen.org/MyGeneSet/)42.
Secreted factors identification
We used the lists of top 1000 markers differentially expressed between MSC cluster and rest of the cells from +DOX mice or -DOX mice. NABA_SECRETED_FACTORS GeneSet from GSEA database (http://software.broadinstitute.org/gsea/msigdb/index.jsp) was used to select for genes encoding for secreted factors in those lists.
Mathematical modeling of metastasis onset
To estimate the timing of the first metastatic cell emigrating from the primary tumor, we created a birth-death process model of cell growth in which an individual cell lives for an exponentially distributed amount of time before splitting into two daughters43,44. In this model, the initiating population seeded within the primary site consists entirely of so-called type-0 cells. We considered situations in which type-0 cells have already evolved the potential to disseminate to other organs such as lymph nodes, lung, and bone based on the fact that no metastatic driver genes have been reported. Type-0 cells divide at rate r0 and die at rate d0 per week, and may leave the primary site to found a new metastasis at a distant site at rate q per week. Each export event results in a new metastatic colony. Metastasized cells are referred to as a type-1 cells and may go extinct or grow with a division and death rate of r1 and d1 per week, respectively. The total number of metastatic sites until time t is denoted by I(t). During each elementary time step of the stochastic process, cells may divide, die, or leave the primary site to initiate a colony elsewhere.
Numerical simulations
We performed numerical simulations of the stochastic process. Here, w and xi denote the number of type-0 cells and type-1 cells of the i-th metastatic site, respectively. A change in w, xi, can occur by cell division, cell death, or departure of a cell from a primary site. Initially, the population of cancer cells is initialized with three million type-0 cells and no type-1 cells, corresponding to three million cells injected into mice for the in vivo experiment. Our model considers all possible events in the stochastic simulation —production and death of a type-0, or -1 cell, and metastasis of a type-0 cell to a new site. The probability of each event occurring is its rate of occurrence, normalized by the sum of the rates of all possible events, given by Γ= (r0+d0+q)w+Σi(r1+d1)xi. The timing of one event is exponentially distributed with mean 1/Γ. The transition probabilities between states in the stochastic simulation are determined as follows: the probabilities that the number of each type cell increase by one are:
| (1) |
Export of a type-0 cell to a new metastatic site increases the number of metastatic cells by one at the new site I+1 and decreases the number of type-0 cells by one; the probability of this event occurring is
| (2) |
The probabilities that the numbers of type-0, -1, and -2 cells in the i-th metastatic site decrease by one are
| (3) |
Based on the framework described in equations (1)–(3), we performed independent runs of the stochastic process, either until all cells go extinct, w = x = Σiyi = 0, or for a total time of six weeks after cancer cells were seeded. Our main goal was to determine the distribution of the size of the first metastasis at week six and the size of primary site where the first metastasis was detected.
Parameter estimation
We explored the use of mixed effects models for the datasets where repeated tumor volume measurements were made over time in each individual mouse. For both the primary site and metastatic sites in each group of IL11 and polyclonal tumor-bearing mice, the growth rates were determined by analyzing the number of cancer cells to using a log-transformed linear regression. Given that three million cells were injected into mice at the beginning of the experiment, we set the number of cancer cell in the primary site at week zero to be three million. Then the estimated slope of the regression provides the estimated growth rate. Tumor sizes utilizing two measurement methods were available for the primary site: tumor diameter by caliper measurement and the largest cross-sectional area of a tumor by bioluminescence detection. For metastatic sites, caliper measurement was not available because the tumor sizes were too small to be measured; therefore, only cross-sectional area measurements by bioluminescence detection were performed. Based on the notion that growth estimation based on caliper measurements is more accurate than bioluminescence detection methods due to oversaturation of large lesions, we calculated the growth rates of metastatic sites by converting bioluminescence to caliper measurements using primary site data. Specifically, we calculated the ratio of the estimated growth rates of primary site by the two aforementioned measurement methods: caliper measurement and bioluminescence detection; and multiplied it by growth rates of metastatic sites estimated by bioluminescence. We converted the tumor volume to cell numbers with the assumption of a spherical shape (109 cells occupy a volume of 1 cm3). We performed 100 simulations per metastatic rate q (q in [10-10, 10-5]) to estimate the parameters of our model.
Metastatic Breast Cancer (MBC) Project cohort analysis
Prior to any study procedures, all patients for whom we analyzed tumor tissue provided written informed consent for research biopsies and transcriptome sequencing of tumor RNA, as approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DF/HCC Protocol 05-246). The study is compliant with all relevant ethical regulations regarding research involving human participants. The cohort contains of 51 primary tumors (FFPE samples) and matching 155 metastatic lesions (mostly frozen tissue), enriched in ER+ subtype. Because of the difference in tissue preservation between primary and metastatic lesions, batch effects cannot be ruled out when comparing primary and metastatic lesions45. Total RNA was extracted with RNEasy Mini Kit (Qiagen) and libraries for RNA-seq were prepared using TrueSeq RNA Sample Preparation Kit protocol adapted for Sciclone liquid handler (Perkin-Elmer). Sequencing was performed on Illumina Nextseq500. Reads were aligned to the hg19 human genome using STAR v2.5.146 followed by Transcript assembly using cufflinks v2.2.147. Quality control steps were done using STAR v2.5.1 and RseQC v2.6.248. We assessed each sample on metrics of mappable reads, percentage of rRNA reads, gene body coverage, and junction saturation and insert_size for paired end, to determine samples that were of adequate quality. Signatures of IL11/FIGF-activated neutrophils and MSCs from blood and lungs were derived from differentially expressed gene lists from our single-cell RNAseq analysis. We aimed to construct the signatures using 150 top differentially expressed genes, unless fewer genes presented with adj p value < 0.05. Signature of cancer cells was derived from top 150 genes differentially expressed between Thy1.1 neutral clone sorted from polyclonal tumor vs parental cell line tumor. All signatures are presented in Supplementary Table 6. Signature analysis on the MBC and TCGA data was performed using GSVA49.
Statistics and reproducibility
Sample size for mouse experiments was determined based on prior studies and pilot experiments to ensure that sufficient power could be obtained7. Unless otherwise stated, statistical analyses were performed with Graph Pad Prism software or R. Box-and-whisker plots show mean (midline), 25th-75th percentile (box) and 5th-95th percentile (whiskers). Most experiments in this study were performed independently at least twice and all attempts at replication of the results presented in this study were successful. Biological replicates were included in all experiments.
Supplementary Material
Acknowledgements
We thank members of the Polyak and Michor laboratories for their critical reading of this manuscript and useful discussions. We thank Lisa Cameron from the DFCI Confocal Microscopy and Zach Herbert from the DFCI Molecular Biology Core Facility for their dedication and technical expertise. We also thank the DFCI Animal Facility staff for their help with imaging studies. This work was supported by the Dana-Farber Cancer Institute Physical Sciences-Oncology Center (U54CA143798 to F.M. and K.P.) and Center for Cancer Evolution (F.M. and K.P.), CDRMP Breast Cancer Research Program W81XWH-09-1-0561 (A.M.) and W81XWH-14-1-0191 (S.S.M), Swiss National Science Foundation project number P2EZP2 175139 (S.C.), NIH K99/R00 CA201606-01A1 (M.J.), R35CA197623 (K.P.), the Ludwig Center at Harvard (F.M. and K.P.), Novartis Oncology (K.P.), and the Breast Cancer Research Foundation (K.P.).
Footnotes
Code availability
Mathematical modeling code is available from the corresponding author upon request.
Data availability
RNA-seq and scRNA-seq data have been deposited in the NCBI GEO database with the accession number GSE109281. Publicly available subset of the MBC Project RNA-seq data can be found in GEO database with the accession number GSE121411. Primary breast cancer data, were indicated, were derived from TCGA dataset (http://cancergenome.nih.gov/). Source data for Fig. 1, 3, 4, 6 and Supplementary Fig. 1-6 have been provided as Supplementary Table 1. All data supporting the findings in this study are available from the corresponding author upon request.
Author Contributions
D.P.T. and M.J. performed xenograft, molecular profiling, and immunohistochemical experiments, and data analyses. M.B.E. and N.W.H. analyzed RNA-seq data. A.M. helped with study conception and xenograft experiments. N.L.K. and K.C.M. assisted with immunohistochemical staining. M.K. generated low-input RNA-seq libraries. Y.Q., Z.C., M.A., and C.G.A. performed FACS analyses. T.L. and S.S. assisted with animal experiments. K.N.Y. carried out mathematical modeling. A.L. and K.W.W. assisted with single-cell RNA-seq libraries generation. S.C. analyzed single-cell RNA-seq data. O.C. and N.W. provided MBCP cohort data and performed analyses. K.P. supervised with help from F.M., S.S.M., and R.F. All authors helped to design the study and write the manuscript.
Author Information
RNA-seq and scRNA-seq data have been deposited in the NCBI GEO database with the accession number GSE109281. The following secure token has been created to allow review of record GSE109281 while it remains in private status: cnehiokwzjaflmn
Competing Financial Interests
The authors declare competing financial interests: K.P. received research support from and was a consultant to Novartis Oncology during the execution of this study. K.P. also serves on the Scientific Advisory Board of Mitra Biotech.
References
- 1.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 2.Burrell RA, Swanton C. Re-Evaluating Clonal Dominance in Cancer Evolution. Trends Cancer. 2016;2:263–276. doi: 10.1016/j.trecan.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Yates LR, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell. 2017;32:169–184 e167. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brastianos PK, et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peinado H, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 6.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 9.Nikolsky Y, Nikolskaya T, Bugrim A. Biological networks and analysis of experimental data in drug discovery. Drug Discov Today. 2005;10:653–662. doi: 10.1016/S1359-6446(05)03420-3. [DOI] [PubMed] [Google Scholar]
- 10.Ernst M, Putoczki TL. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin Cancer Res. 2014;20:5579–5588. doi: 10.1158/1078-0432.CCR-13-2492. [DOI] [PubMed] [Google Scholar]
- 11.Retsky M, Demicheli R, Hrushesky W, Baum M, Gukas I. Surgery triggers outgrowth of latent distant disease in breast cancer: an inconvenient truth? Cancers (Basel) 2010;2:305–337. doi: 10.3390/cancers2020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tohme S, Simmons RL, Tsung A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017;77:1548–1552. doi: 10.1158/0008-5472.CAN-16-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 14.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 15.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeselsohn R, et al. Allele-Specific Chromatin Recruitment and Therapeutic Vulnerabilities of ESR1 Activating Mutations. Cancer Cell. 2018;33:173–186 e175. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel MB, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest. 2018;128:1371–1383. doi: 10.1172/JCI96153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoadley KA, et al. Tumor Evolution in Two Patients with Basal-like Breast Cancer: A Retrospective Genomics Study of Multiple Metastases. PLoS Med. 2016;13:e1002174. doi: 10.1371/journal.pmed.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onnis B, Fer N, Rapisarda A, Perez VS, Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J Clin Invest. 2013;123:1615–1629. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putoczki TL, et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. 2013;24:257–271. doi: 10.1016/j.ccr.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Bockhorn J, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 24.Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann Surg Oncol. 2006;13:802–808. doi: 10.1245/ASO.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Bower NI, et al. Vegfd modulates both angiogenesis and lymphangiogenesis during zebrafish embryonic development. Development. 2017;144:507–518. doi: 10.1242/dev.146969. [DOI] [PubMed] [Google Scholar]
- 26.Van den Eynden GG, et al. Comparison of molecular determinants of angiogenesis and lymphangiogenesis in lymph node metastases and in primary tumours of patients with breast cancer. J Pathol. 2007;213:56–64. doi: 10.1002/path.2211. [DOI] [PubMed] [Google Scholar]
- 27.Leach J, Morton JP, Sansom OJ. Neutrophils: Homing in on the myeloid mechanisms of metastasis. Mol Immunol. 2017 doi: 10.1016/j.molimm.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffelt SB, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fridlender ZG, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castano Z, et al. IL-1beta inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol. 2018;20:1084–1097. doi: 10.1038/s41556-018-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krenn-Pilko S, et al. The elevated preoperative derived neutrophil-to-lymphocyte ratio predicts poor clinical outcome in breast cancer patients. Tumour Biol. 2016;37:361–368. doi: 10.1007/s13277-015-3805-4. [DOI] [PubMed] [Google Scholar]
- 33.Granot Z, et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finisguerra V, et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature. 2015;522:349–353. doi: 10.1038/nature14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voloshin T, et al. Blocking IL1beta Pathway Following Paclitaxel Chemotherapy Slightly Inhibits Primary Tumor Growth but Promotes Spontaneous Metastasis. Mol Cancer Ther. 2015;14:1385–1394. doi: 10.1158/1535-7163.MCT-14-0969. [DOI] [PubMed] [Google Scholar]
- 36.Kersten K, et al. Mammary tumor-derived CCL2 enhances pro-metastatic systemic inflammation through upregulation of IL1beta in tumor-associated macrophages. Oncoimmunology. 2017;6:e1334744. doi: 10.1080/2162402X.2017.1334744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St Croix B, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 38.Carson-Walter EB, et al. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- 39.Pan X, et al. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc Natl Acad Sci U S A. 2013;110:594–599. doi: 10.1073/pnas.1217322109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo S, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell. 2014;156:649–662. doi: 10.1016/j.cell.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heng TS, Painter MW, Immunological Genome Project, C The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 43.McDonald TO, Michor F. SIApopr: a computational method to simulate evolutionary branching trees for analysis of tumor clonal evolution. Bioinformatics. 2017;33:2221–2223. doi: 10.1093/bioinformatics/btx146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haeno H, et al. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell. 2012;148:362–375. doi: 10.1016/j.cell.2011.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen R, et al. Robust transcriptional tumor signatures applicable to both formalin-fixed paraffin-embedded and fresh-frozen samples. Oncotarget. 2017;8:6652–6662. doi: 10.18632/oncotarget.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 49.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.