Abstract
Protein prenylation is believed to be catalyzed by three heterodimeric enzymes: FTase, GGTase1, GGTase2. Here, we report the identification of a previously unknown human prenyltransferase complex consisting of an orphan prenyltransferase α subunit, PTAR1, and the catalytic β subunit of GGTase2, RabGGTB. This enzyme, which we named GGTase3, geranylgeranylates FBXL2 to allow its localization at cell membranes, where this ubiquitin ligase mediates the polyubiquitylation of membrane-anchored proteins. In cells, FBXL2 is specifically recognized by GGTase3 despite having a typical C-terminal CaaX prenylation-motif that is predicted to be recognized by GGtase1. Our crystal structure analysis of the full-length GGTase3-FBXL2-SKP1 complex reveals an extensive multivalent interface specifically formed between the leucine-rich repeat domain of FBXL2 and PTAR1, which unmasks the structural basis of the substrate-enzyme specificity. By uncovering a missing prenyltransferase and its unique mode of substrate recognition, our findings call for a revision of the “prenylation code”.
Introduction
Association with cellular membranes is a prerequisite for the function of many regulatory proteins, which can be either embedded in the lipid bilayer or located at its surface (integral vs. peripheral membrane proteins). Many peripheral proteins are targeted to biological membranes as a consequence of posttranslational modification with lipids1. Two isoprenoid lipids derived from intermediates in the cholesterol biosynthetic pathway are utilized by eukaryotic cells for such modification: the 15-carbon farnesyl lipid and the 20-carbon geranylgeranyl lipid2–4. Covalent modification of the target proteins by these lipids at a C-terminal cysteine residue, generally referred to as prenylation, is catalyzed by a group of enzymes known as prenyltransferases. In the human proteome, about 300 proteins, many involved in fundamental cellular functions, such as membrane trafficking and signal transduction, are modified by prenyltransferases5. Importantly, some oncogenic proteins, such as the activating mutant forms of H-, N-, and K-RAS, require prenylation for their transforming activities3. Inhibition of prenylation, therefore, has been proposed as a therapeutic approach for treating the ~30% of human cancers that are driven by RAS activating mutations6–8.
Three prenyltransferases have been identified in mammals, farnesyltransferase (FTase), geranylgeranyltransferase type 1 (GGTase1), and geranylgeranyltransferase type 2 (GGTase2)7,9–12. All three prenyltransferases are heterodimeric enzyme complexes, each consisting of one α and one β subunit. FTase and GGTase1 share a common α subunit, FNTA (also known as PTAR2), but contain distinct β subunits, which are encoded by FNTB and PGGT1B, respectively (Fig. 1a). The substrate specificity of FTase and GGTase1 are thought to be determined by a C-terminal CaaX sequence (C: Cysteine; a: aliphatic; X: any amino acid), which constitutes the site of lipid modification. Depending on the nature of the utmost X residue, a substrate CaaX motif is recognized by either FTase for farnesylation or GGTase1 for geranylgeranylation. The third prenyltransferase, GGTase2, is formed by RabGGTA (the α subunit, also known as PTAR3) and RabGGTB (the β subunit) (Fig. 1a). GGTase2 prenylates the substrate cysteine(s) in less defined C-termini, including XXCC, XCCX, CCXX, CCXXX, and XCXC. Unlike FTase and GGTase1, GGTase2 requires an accessory protein designated RAB escort protein (REP) that provides substrate recognition13–15. Although distinct group of substrates have been identified for FTase (e.g., RAS GTPases, pre-Lamin A and Lamin B), GGTase1 (e.g., RHO-RAC GTPases and RAP1B), and GGTase2 (e.g., RAB GTPases), several cases of cross-prenylation have been described in the literature16–20. The molecular details of cross-prenylation remain unclear, underscoring the fact that we do not completely understand how these enzymes work.
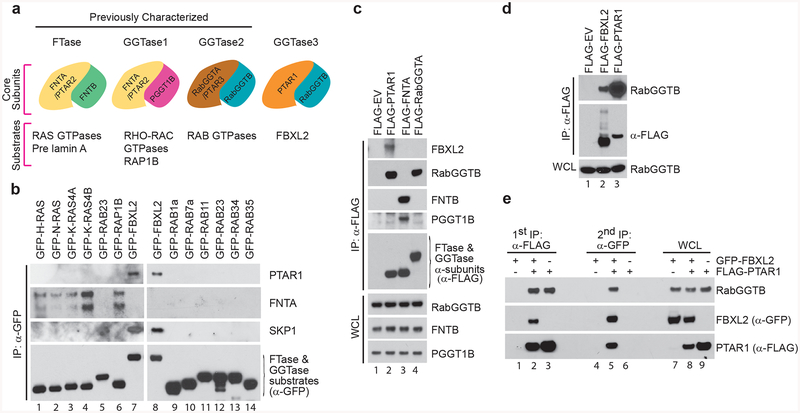
Figure 1. PTAR1, an orphan prenyltransferase α subunit, binds FBXL2 and RabGGTB.
(a) Schematic representation of the four human prenyltransferases comprised by combinations of α [PTAR1, FNTA (PTAR2), and RabGGTA (PTAR3)] and β (FNTB, PGGT1B and, RabGGTB) subunits, and their substrates. As shown in this study, PTAR1 and RabGGTB interact to form a prenyltransferase that we named GGTase3.
(b) HEK-293T cells were transfected with the indicated GFP-tagged substrates of prenyltransferases for immunoprecipitations and immunoblotting.
(c,d) HEK-293T cells were transfected with the indicated plasmids for immunoprecipitations and immunoblotting. WCE: Whole cell extract; EV: empty vector.
(e) HEK-293T cells were co-transfected with FLAG-tagged PTAR1 and GFP-tagged FBXL2 as indicated. Immunoprecipitations were carried out sequentially using first an anti-FLAG antibody and then an anti-GFP antibody as described in methods. The first elution was done with a FLAG peptide and the second with 1%SDS. The two eluates were then immunoblotted as indicated.
F-box proteins are the substrate receptor subunits of SCF (Skp1, Cul1, F-box protein) ubiquitin ligase complexes21,22. In humans, there are 69 F-box proteins, each forming a different SCF ligase and promoting the polyubiquitylation of specific substrates. Distinct from most F-box proteins, FBXL2 and its close paralog FBXL20 (the former being ubiquitous and the latter being specifically expressed in neurons23–25) terminate with a prototypical CaaX motif (CVIL), which is strictly conserved across species. FBXL2 has been shown to be geranylgeranylated26 and, based on the sequence of its CaaX motif, is predicted to be a GGTase1 substrate. We have previously shown that the integrity of the CaaX motif is necessary for FBXL2 to assemble into an active SCF ubiquitin ligase complex and interact with two substrates localized at cellular membranes, p85β, a regulatory subunit of the PI3-kinase (PI3K), and IP3 (inositol 1,4,5-trisphosphate) receptor type 3 (IP3R3)27–29. Other independent studies have also reported membrane-localized substrates for both FBXL2 and FBXL2023,30–32. By promoting the degradation of p85β and IP3R3 and, consequently, regulating PI3K signaling and calcium flux from the ER to the mitochondria, FBXL2 plays a critical pro-survival role27–29.
Here we show that FBXL2 is geranylgeranylated by a previously unknown human prenyltransferases, which we named GGTase3. A combination of cellular, biochemical, and structural biology approaches has allowed us to reveal its unique mechanisms of substrate recognition.
Results
FBXL2 binds a previously unrecognized GGTase
The mammalian genome contains one single paralog of FNTA and RABGGTA encoding PTAR1 (prenyltransferase α subunit repeat-containing protein 1), an orphan protein with unknown functions. Peptides corresponding to PTAR1 and RabGGTB were specifically identified in most FBXL2 immunopurifications, we and others analyzed using mass spectrometry28,33,34. Notably, PTAR1 and RabGGTB peptides have never been found either in the 411 experiments reported by the contaminant repository for affinity purification-mass spectrometry data (www.crapome.org)35 or in some three hundred purifications of 26 distinct F-box proteins carried out in our laboratory, supporting the specificity of the interaction with FBXL2.
We sought to validate the binding of FBXL2 with PTAR1 and confirm its specificity by screening a panel of 15 proteins containing typical C-terminal prenylation motifs (i.e. CXXX, XCXC, and XXCC) for their ability to bind PTAR1 upon their expression in HEK-293T cells. Endogenous PTAR1 interacted specifically with FBXL2 (Fig. 1b, Supplementary Fig. 1a). In contrast, none of the other 14 prenyltransferase substrates (H-RAS, N-RAS, K-RAS4A, K-RAS4B, RAB23, RAP1B, RAB1a, RAB7a, RAB11, RAB23, RAB34, RAB35, M-RAS, and R-RAS,) or a distinct F-box protein used as control (FBXL15) interacted with PTAR1 (Fig. 1b, Supplementary Fig. 1b, and data not shown). As expected, RAS-GTPases and RAP1B, but not FBXL2 or RAB-GTPases, were able to co-immunoprecipitate the short and long isoforms of endogenous FNTA (Fig. 1b, Supplementary Fig. 1c). The specificity of the interaction between PTAR1 and FBXL2 was observed even when PTAR1 was co-expressed with substrates of FTase and GGTases and when FBXL2 was expressed at lower levels than the other substrates (Supplementary Fig. 1d,e). Moreover, when the three human prenyltransferase α subunits were expressed in HEK-293T cells, we found that only PTAR1, but not FNTA or RabGGTA, co-immunoprecipitated with endogenous FBXL2 (Fig. 1c).
To confirm the binding between RabGGTB and FBXL2, we expressed either FLAG-tagged FBXL2 or GFP-tagged FBXL2 in HEK-293T cells and found that endogenous RabGGTB co-immunoprecipitated with FBXL2, but not with FLAG-tagged empty vector or GFP-tagged K-RAS4B (Fig. 1d and Supplementary Fig. S1f). In line with a systematic study of the human interactome34, FLAG-tagged PTAR1 was also able to co-immunoprecipitate endogenous RabGGTB (Fig. 1d). By co-expressing GFP-tagged FBXL2 and FLAG-tagged PTAR1 in HEK-293T cells and performing sequential immunoprecipitations, we found that FLAG-tagged PTAR1 co-immunopurified endogenous RabGGTB as well as GFP-tagged FBXL2 (Fig. 1e). When the FLAG immunoprecipitations were subsequently eluted with an excess of FLAG peptide and re-immunoprecipitated with an anti-GFP antibody, the three proteins were again present in the second immunoprecipitation, indicating a FBXL2-PTAR1-RabGGTB ternary complex.
Based on the above results and the fact that FLAG-tagged PTAR1 co-immunoprecipitated a greater amount of endogenous RabGGTB than FLAG-FBXL2, we hypothesized that RabGGTB might heterodimerize with PTAR1, serving as the β subunit of a previously unrecognized prenyltransferase. To explore this possibility, we expressed the three human prenyltransferese α subunits in HEK-293T cells and examined their binding to the three endogenous β subunits. Similar to RabGGTA, we found that PTAR1 interacted with endogenous RabGGTB, but not FNTB and PGGT1B (Fig. 1c). In contrast, FNTA interacted with FNTB and PGGT1B, but not RabGGTB. These data strongly support the identification of a previously unrecognized mammalian GGTase, which we named GGTase3, comprising PTAR1 and RabGGTB as α and β subunits, respectively (Fig. 1a).
GGTase3 geranylgeranylates FBXL2 and targets it to cellular membranes
Previous studies have shown that FBXL2 is geranylgeranylated26. We found that bacterially purified FBXL2 served as a substrate of GGTase3 in vitro (Fig. 2a,b). Notably, GGTase3 was able to in vitro geranylgeranylate FBXL2, but not K-RAS4B and FBXW7 (Fig. 2b). We obtained identical results using two bacterial purified GGTase3 preps that were made independently in our two laboratories using different constructs (Fig. 2a,b and Supplementary Fig. S2a). GGTase1 was also able to prenylate FBXL2 in vitro (Supplementary Fig. S2b), which is expected given the presence of the typical CaaX signature motif of GGtase1 substrates in FBXL2. To investigate which of the two enzymes is responsible for FBXL2 geranylgeranylation in vivo, we incubated RPE1-HTERT1 and HeLa cells with geranylgeranyl-azide and monitored the geranylgeranylation of FBXL2 using a Click-IT assay. We found that expression of PTAR1 promoted the geranylgeranylation of FBXL2, but not FBXL2(C420S), a mutant in which cysteine 420 in the CaaX motif is mutated to serine (Fig. 2c). Moreover, we found that PTAR1 silencing using two different oligos (individually) drastically decreased the prenylation of FBXL2 in vivo (Fig. 2d). This effect was reversed by expressing PTAR1 cDNA, which was siRNA-insensitive since the two oligos used targeted the 5’UTR. This experiment demonstrated that the decrease in FBXL2 prenylation is not due to an off-target effect of the oligo to PTAR1. Silencing of FNTA or PGGT1B had no effect on FBXL2 prenylation (Fig. 2d). However, when both PTAR1 and PGGT1B were silenced, the prenylation of FBXL2 became virtually undetectable. This result suggests that GGTase1 is able to compensate for GGTase3 in its absence. In agreement with this possibility, when PTAR1 was depleted, FBXL2 acquired the ability to bind endogenous FNTA (Supplementary Fig. 1f).
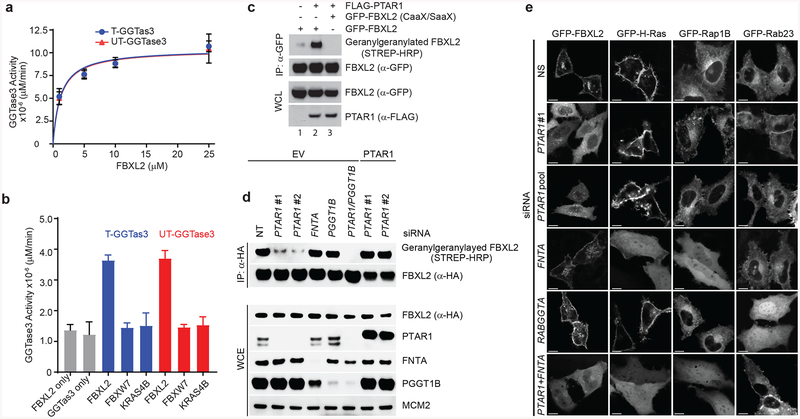
Figure 2. GGTase3 geranylgeranylates FBXL2 and is required for its localization to cellular membranes.
(a) Recombinant GGtase3 geranylgeranylates purified FBXL2. Indicated amounts of purified FBXL2 were incubated with 100 ng of purified GGTase3 (either tagged [T] or untagged [UT] versions) to carry out in vitro geranylgeranylation assay using saturating concentrations of tritiated [H3]-GGPP as described in methods. Each data point represents mean+/− SD of three biological replicates. Michaelis-Menten kinetics was used to generate an apparent Km value of 1.2μM using Prism Graphpad software.
(b) In vitro geranylgeranylation assay was carried out and measured as in (a) using 10 μM of purified FBXL2, FBXW7, or K-RAS4B and 100 ng of purified GGTase3. Bar graphs represent mean +/− SD from three biological replicates. Source data for panels a and b are available with the paper online.
(c) RPE1-HTERT cells were cotransfected with the indicated plasmids and processed for the detection of geranylgeranylated FBXL2 using a “Click-IT” assay, as described in methods. The experiment was repeated three times. Representative result is shown. Uncropped blot/gel images are shown in Supplementary Data Set 1.
(d) HeLa cells were transfected with the indicated siRNA oligos and cDNAs. Twenty-four hours post-transfection cells were incubated with geranylgeranyl-azide for 16 hours, harvested, lysed, and azide selective ligation reaction with sDIBO-Biotin was performed for one hour to label geranylgeranylated proteins via copper-free “Click-IT” reaction. After immunoprecipitation with an anti-HA antibody, immunoblots were carried out. The experiment was repeated four times. Representative result is shown. Uncropped blot/gel images are shown in Supplementary Data Set 1.
(e) HeLa cells were transfected first with the indicated siRNA oligos and then with the indicated GFP-tagged proteins. Live cell confocal imaging was carried out as described in methods. Images show representative frames of three independent experiments. Bar size: 10 μm.
Prenylation of proteins is required for their proper subcellular trafficking and localization. Therefore, we investigated the localization of GFP-tagged FBXL2 with and without geranylgeranylation. Using live-cell imaging confocal microscopy, we observed that GFP-FBXL2 decorated cellular membranes, including the plasma membrane and perinuclear vesicles (Fig. 2e). In contrast, GFP-FBXL2(C420S) was observed in a homogeneous pattern in the cytoplasm (with negatively imaged organelles) and the nucleoplasm (Supplementary Fig. S2c). Thus, the CVIL sequence in FBXL2 is required for its membrane localization. Moreover, whereas silencing FNTA or RABGGTA did not affect the subcellular localization of GFP-FBXL2, PTAR1 depletion using either a pool of four different siRNA oligos or a single one resulted in the loss of FBXL2 from the plasma membrane and its accumulation in the cytoplasm (Fig. 2e). In contrast, PTAR1 silencing had no effect on the localization of H-RAS, RAP1B, or RAB23. As expected, FNTA depletion resulted in the delocalization of both H-RAS and RAP1B, but not RAB23, and depletion of RabGGTA resulted in the delocalization of RAB23, but had no effect on the localization of H-RAS or RAP1B. Together, these results indicate that GGTase3 specifically geranylgeranylates FBXL2 and controls its cellular localization.
The relocalization of GFP-FBXL2 from the plasma membrane to the cytoplasm upon knockdown of PTAR1 was unequivocal, however GFP-FBXL2 still persisted on some vesicles (Fig. 2e), suggesting that it has the capability to associate with membranes through a different and/or a compensatory mechanism. When both PTAR1 and FNTA were silenced, GFP-FBXL2 lost its residual vesicular localization and became homogenously distributed in cytoplasm and nucleus (Fig. 2e), in agreement with the notion that, in the absence of PTAR1, FBXL2 becomes a ‘neo-substrate’ of GGTase1, as suggested by its binding to FNTA (Supplementary Fig. 1f) and prenylation in vitro (Supplementary Fig. 2b).
Overall structure of GGTase3-FBXL2-SKP1
Previous studies have shown that high affinity binding of substrates to FTase and GGTase1 requires preloading of the enzymes with the farnesyl and geranylgeranyl diphosphate (FPP or GGPP) substrate, respectively36–38. Substrate recognition by GGTase2 also depends on an accessory protein that bridges RAB GTPases and the enzyme13,15. Unexpectedly, we detected robust high-affinity (~70 nM) interaction between GGTase3 and FBXL2-SKP1, both purified from E. coli, without the addition of GGPP or a GGPP analog (Fig. 3a). To further validate their strong interaction, we co-expressed the hetero-dimeric GGTase3 with FBXL2-SKP1 and successfully isolated GGTase3-FBXL2-SKP1 as a tetrameric complex in equal stoichiometry (Fig. 3b). To understand how GGTase3 assembles and recognizes FBXL2, we crystallized the GGTase3-FBXL2-SKP1 complex and determined its structure at a 2.5 Å resolution (Table 1).
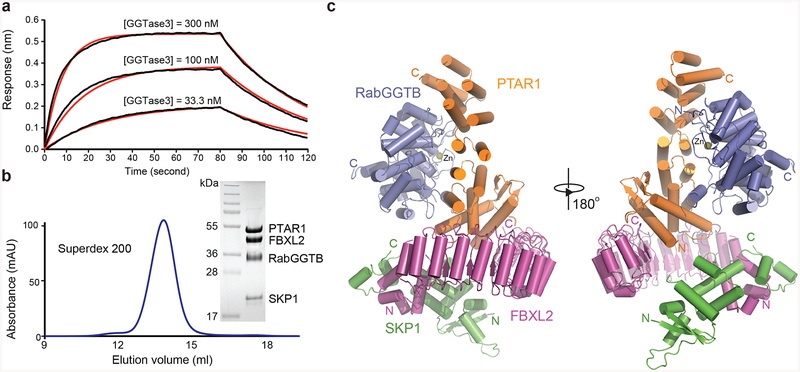
Figure 3. Overall structure of GGTase3-FBXL2-SKP1 complex.
(a) GGTase3 binding to FBXL2-SKP1 monitored by Octet BioLayer Interferometry analysis in the absence of GGPP or its analog. Sensorgram traces (black lines) of the interactions of His-FBXL2-SKP1 with GGTase3 at three concentrations (300 nM, 100 nM, and 33.3 nM) overlaid with curve fits (red lines). The global fit yields a calculated KD of 69.0 nM ± 1.1 nM.
(b) Size exclusion chromatography analysis of the purified GGTase3-FBXL2-SKP1 protein complex with sodium dodecylsulfate-polyacrylamide gel electrophoresis analysis of the central fraction containing the protein complex with all four subunits in equal stoichiometry.
(c) Two orthogonal views of the tetrameric complex containing PTAR1 (orange), RabGGTB (slate), FBXL2 (magenta) and SKP1 (green). The zinc ion at the active site of RabGGTB is shown in yellow sphere. The N and C termini of different proteins are labeled N and C in corresponding colors.
Table 1.
Data collection and refinement statistics
| GGTase3-FBXL2-SKP1 (PDB 6O60)a | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 86.8, 99.3, 151.2 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.00–2.50 (2.54–2.50)b |
| Rmerge | 0.098 (0.890) |
| Rmeas | 0.109 (0.986) |
| I/σ(I) | 24.3 (2.3) |
| CC1/2 | 0.991 (0.754) |
| Completeness (%) | 90.5 (93.6) |
| Redundancy | 4.8 (4.7) |
| Refinement | |
| Resolution (Å) | 49.5–2.5 |
| No. reflections | 41320 |
| Rwork / Rfree | 0.194/0.247 |
| No. atoms | 9299 |
| Protein | 9228 |
| Znion | 1 |
| Water | 70 |
| B factors | |
| Protein | 55 |
| Znion | 41 |
| Water | 51 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.025 |
A single crystal was used for diffraction data collection and structure determination.
Values in parentheses are for highest-resolution shell.
The PTAR1-RabGGTB-FBXL2-SKP1 complex adopts a torch-shaped architecture with GGTase3 representing the flame emitting from the FBXL2-SKP1 handle (Fig. 3c). Similar to the subunit of other prenyltransferase complexes15,39, PTAR1 folds into an elongated crescent, mainly consisting of linearly packed α-helical hairpins. It closely wraps around the globular RabGGTB subunit, which is characterized by a zinc ion at its active site. The FBXL2 protein possesses a typical N-terminal helical F-box motif that interacts with the SCF adaptor subunit SKP140,41. Its C-terminal domain is constructed by leucine-rich-repeats (LRRs) in a remarkably regular curved solenoid structure encompassing half a circle. GGTase3 recruits its substrate FBXL2 predominantly via a unique N-terminal extension (NTE) of PTAR1, which folds into an α/β sub-domain and anchors itself onto the concave surface of FBXL2 LRR domain. Interestingly, no electron density was found for the utmost CaaX motif-containing FBXL2 C-terminal tail, which is most likely to be disordered in the crystal (Supplementary Note 1). This observation reinforces the notion that recognition of FBXL2 by GGTase3 is mediated by interacting regions outside the C-terminal CaaX motif of the substrate. The GGTase3-FBXL2 complex buries over 2,500Å2 solvent accessible surface area, more extensive than that of any other prenyltransferase-substrate complexes. The high affinity binding between GGTase3 and FBXL2 is most likely attributable to this large intermolecular interface.
GGTase3 contains a unique N-terminal extension in PTAR1
PTAR1, the α subunit of the newly identified GGTase, harbors a series of right-handed antiparallel coiled-coils that fold into six successively packed α-helical hairpins, which are highly comparable to those of FNTA (PTAR2) and RabGGTA (PTAR3), the α subunits of known prenyltransferases (Fig. 4a,b)15,39. Despite missing an additional α-helical hairpin at the C-terminus, PTAR1 adopts the same crescent-shaped fold that envelops the RabGGTB catalytic subunit through an extensive and conserved interface found in all three previously characterized heterodimeric prenyltransferases. In complex with PTAR1, RabGGTB displays the identical α-α barrel fold as found in GGTase2, consisting of a core of six parallel helices and six peripheral helices15. At a funnel-shaped cavity in the center of α-α barrel, the active site of the enzyme is lined with conserved hydrophobic residues with an intrinsic zinc ion bound at the top. Overall, the architecture of GGTase3 is very similar to the known prenyltransferase complexes.
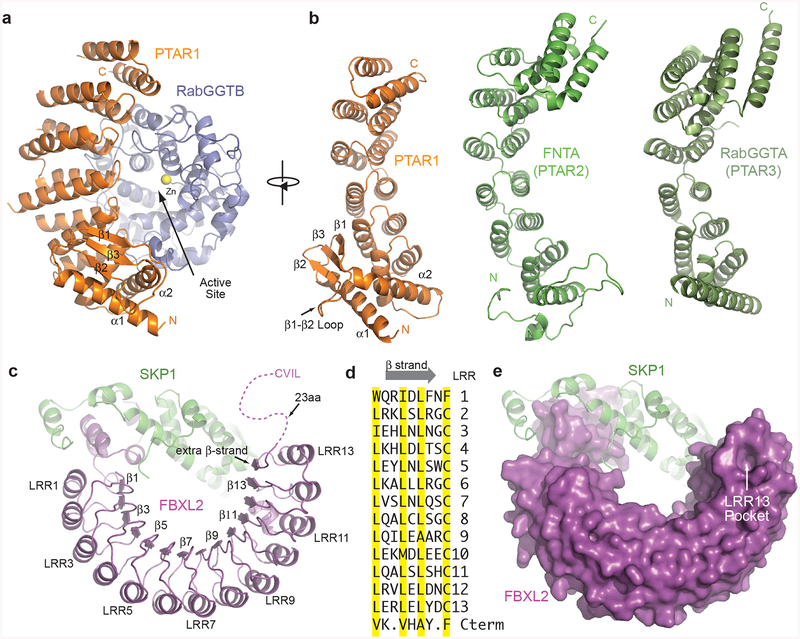
Figure 4. Structural features of GGTase3 and FBXL2.
(a) Ribbon diagrams of GGTase3 containing PTAR1 (orange) as the α subunit, and RabGGTB (slate) as the β subunit. The catalytic zinc ion is shown in yellow sphere. The unique PTAR1 NTE α/β fold comprising two α helices and three β strands are labeled and numbered.
(b) Structural comparison of PTAR1 (orange) to the α subunits of known prenyltransferases, including FNTA (PTAR2) (green) and RabGGTA (PTAR3) (dark green). The N and C termini of different proteins are labeled N and C in corresponding colors. For clarity, the C-terminal IGG and LRR domain of RabGGTA (PTAR3) are not shown. All the α subunits are displayed in a view orthogonal to the PTAR1 shown in (a).
(c) Ribbon diagrams of FBXL2 (magenta) in complex with SKP1 (green). Select LRRs are labeled and numbered at their helices and β strands. The disordered C-terminal tail of FBXL2 containing the CVIL motif is shown as dashed magenta line.
(d) Sequence alignment of β strands in all the LRRs of FBXL2 and its additional short C-terminal β strands. The characteristic residues in typical LRRs are highlighted in yellow.
(e) The unusual LRR13 pocket on the apical ridge of the LRR domain of FBXL2 shown in surface representation.
A striking structural feature that distinguishes PTAR1 from the other two prenyltransferase α subunits is its unique N-terminal extension (NTE), which is highly conserved in animals (Supplementary Note 2). Unlike the NTEs of the other two prenyltransferase α subunits that are largely disordered, PTAR1-NTE is unexpectedly well structured. The PTAR1 NTE adopts an α/β fold starting with α1 helix followed by a β-sheet comprising three β-stands (β1-β3) (Fig. 4a,b). It terminates with α2 helix, which packs orthogonally against α1 and couples NTE to the helical-hairpin domain. Stabilized by a hydrophobic core, the PTAR1 NTE is highlighted by a 10 amino acids loop, which is flanked by the β1 and β2 strands (hereafter referred to as β1-β2 loop). As described below, this structural loop, together with the rest of the PTAR1 NTE, plays a pivotal role in FBXL2 recruitment.
The LRR domain of FBXL2 contains an unusual pocket
The FBXL2 LRR domain contains 13 complete LRRs (LRR1-LRR13) and an additional β-strand followed by a disordered C-terminal tail with the CaaX motif (Fig. 4c). The FBXL2 LRRs pack in tandem and produce a curved solenoid architecture resembling a semicircular arch. The concave surface of FBXL2 LRRs is formed by the interior parallel β-strands, while its convex surface is constructed by the exterior array of α-helices. In comparison to the LRR domains of other LRR-type F-box proteins40,42,43, the topology of FBXL2-LRRs displays a remarkable regularity with no atypical repeat, loop insert, abrupt kink in curvature, or repeat offset. The peculiar feature of the FBXL2-LRRs, interestingly, is found at the very C-terminal end of the solenoid, where the extra C-terminal β-strand forms a continuous parallel β-sheet with LRRs (Fig. 4c). In sequence, this additional β-strand and the short loop following it are two amino acids shorter than the corresponding parts of all LRRs (Fig. 4d). By packing against the last LRR (LRR13), it creates an unusual surface pocket, which we named “LRR13 pocket”, on the apical ridge of the LRR domain (Fig. 4e). As shown below, this FBXL2 pocket serves as the key docking site for GGTase3 binding.
FBXL2 and GGTase3 form a multivalent interface
Recognition of FBXL2 by GGTase3 is exclusively mediated by the α subunit PTAR1, which occupies the entire concave side of the F-box protein. A top view of the PTAR1-FBXL2 interface reveals that many FBXL2 LRRs from both the N- and C-terminal halves of the protein participate in PTAR1 binding, leaving only a small gap between the two proteins near the middle region of the FBXL2 solenoid (Fig. 5a).
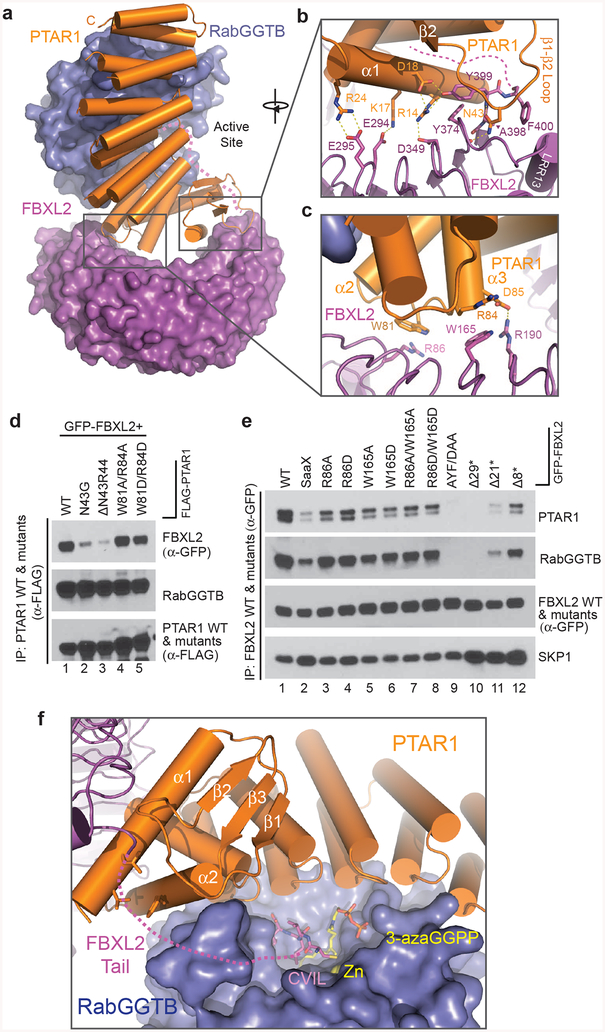
Figure 5. Multivalent FBXL2-GGTase3 interface.
(a) A top view of the overall FBXL2-GGTase3 interface formed by PTAR1 (orange ribbons) and FBXL2 (magenta surface).
(b,c) Close-up views of the PTAR1-FBXL2 interface made by the C-terminal and N-terminal FBXL2 LRRs. Select interface residues are shown in sticks. Dashed yellow lines represent a network of hydrogen bonds and polar interactions. The disordered C-terminal tail of FBXL2 is shown as dashed magenta line.
(d) HEK-293T cells were co-transfected with GFP-tagged FBXL2 and either wild-type (WT) FLAG-tagged PTAR1 or the indicated mutants. Twenty-four hours post-transfection, cells were harvested for immunoprecipitations and immunoblotting. The experiment was repeated three times. Representative result is shown. Uncropped blot/gel images are shown in Supplementary Data Set 1.
(e) HEK-293T cells were transfected with either GFP-tagged WT FBXL2 or the indicated GFP-tagged mutants. Twenty-four hours post-transfection, cells were harvested for immunoprecipitations and immunoblotting. The experiment was repeated three times. Representative result is shown. Uncropped blot/gel images are shown in Supplementary Data Set 1.
(f) A model of FBXL2 C-terminal CVIL motif at the active site of GGTase3 in presence of a GGPP analog. The zinc ion is shown in yellow sphere and is buried in the catalytic pocket of RabGGTB (slate surface). The disordered FBXL2 C-tail is shown as magenta dashed line with modeled CVIL motif as magenta sticks. A GGPP analog, 3-azaGGPP, is shown as sticks with the geranylgeranyl group in yellow and the pyrophosphate in orange. The residues in the α1 and α2 helices of PTAR1-NTE forming the hydrophobic patch possibly interacting with the FBXL2 tail are shown in orange sticks.
At the C-terminal half of the FBXL2 LRR domain, PTAR1-NTE forms a continuous and highly complementary interface with the F-box protein. The hallmark of this interface is the interlocking engagement between the LRR13 pocket of FBXL2 and the α1 helix and the β1-β2 loop of PTAR1-NTE (Fig. 5a,b). The tip of the PTAR1 β1-β2 loop harbors an asparagine residue, Asn43, which is strictly conserved among all PTAR1 orthologs (Supplementary Note 2). Acting as a claw, the PTAR1 β1-β2 loop hooks to the FBXL2 pocket with its Asn43 residue reaching to the bottom of the pocket and forming two hydrogen bonds with FBXL2 backbone groups (Fig. 5b). In doing so, the PTAR1 β1-β2 loop and its nearby α1 helix clamps down the short FBXL2 loop terminating the extra C-terminal β-strand, creating an inter-digited molecular interface. Similar to the tip of the PTAR1 β1-β2 loop, the short loop at the end of the FBXL2 LRR domain, which consists of three residues, Ala398, Tyr399, and Phe400, also has an amino acid sequence invariant among different FBXL2 orthologs (Supplementary Note 1). Phe400, which is the last traceable residue of FBXL2 in the crystal structure, encloses the LRR13 pocket and secures the docking of PTAR1 Asn43 inside. Remarkably, this portion of the FBXL2-PTAR1 interface is further reinforced by a network of polar interactions taking place between the PTAR1 α1 helix and the three FBXL2 LRRs preceding LRR13 (Fig. 5b). Together, these features of the interface strongly suggest a critical role in mediating FBXL2 binding to PTAR1.
In addition to its C-terminal LRRs, FBXL2 N-terminal LRRs are also involved in cradling PTAR1 through a relatively flat interface. The majority of the intermolecular contacts in this region is made between the apical ridge of the first five LRRs in FBXL2 and the PTAR1 loop connecting α2 and α3 helices (Fig. 5c). Distinct from the C-terminal interface, this N-terminal interface is characterized by two pairs of reciprocal Trp-Arg interactions, in which the aliphatic side chain of an arginine residue (Arg86 of FBXL2 and Arg84 of PTAR1) packs against the indole ring of a tryptophan residue (Trp81 of PTAR1 and Trp165 of FBXL2). These interactions are further stabilized by a salt bridge formed between FBXL2 Arg190 and PTAR1 Asp85.
Molecular determinants of FBXL2-GGTase3 interaction
To map the important structural elements supporting FBXL2-GGTase3 interaction, we designed and tested the binding activities of a series of PTAR1 and FBXL2 mutants by co-immunoprecipitation from HEK293T cells. Single or double-point mutations designed to disrupt the two Trp-Arg pairs located at the N-terminal interface only modestly compromised complex association (Fig. 5d, lane 4 and 5; Fig. 5e, lane 3–8), indicating that FBXL2-PTAR1 interactions at this region only play an accessary role in supporting FBXL2 recruitment to GGTase3. By contrast, removing the side chain of Asn43 or deleting two amino acids at the tip of the PTAR1 β1-β2 loop severally impaired the binding of FBXL2 to PTAR1 (Fig. 5d, lane 2 & 3). This effect can also be achieved, to a more complete degree, by both a triple-point mutation of the short FBXL2 loop terminating the extra C-terminal β-strand, or a truncation mutation of the F-box protein with C-terminal β-strand entirely eliminated (Fig. 5e, lane 9 & 10). Together, these results definitively establish the interface at the C-terminal end of the FBXL2 LRR domain as a critical “hotspot” for FBXL2 recognition by GGTase3.
Human FBXL2 is 423 amino acids long. The last 23 residues following Phe400 are in a disordered conformation with no clear electron density in the crystal. Based on previous structural studies of FTase, GGTase1, and GGTase215,37,44,45, we model the C-terminal CVIL motif of FBXL2 into the catalytic pocket of GGTase3 together with a GGPP analog (Fig. 5f). The Cα atoms of Phe400 and the cysteine residue in the CVIL motif are separated by a distance of ~36 Å, which can be comfortably bridged by the missing 19 residues in an extended conformation. Consistent with the role of the FBXL2 LRR domain in mediating PTAR1 binding, the CVIL motif fused to GFP showed little binding to GGTase3 (data not shown). Nonetheless, mutating the cysteine residue in the FBXL2 CVIL motif weakened complex formation (Fig. 5e, lane 2), suggesting that the integrity of the C-terminal four amino acids motif contributes to the association between the F-box protein and the enzymatic complex. Interestingly, truncating the last 8 vs. 21 amino acids tail of FBXL2 showed differential impact to GGTase3 binding (Fig. 5e, lane 11 & 12). Therefore, it is most likely that select amino acids in the 19 residues region presumably disordered in the crystal structure also make contact to GGTase3. In agreement to this postulation, the α1 and α2 helices of PTAR1-NTE present a hydrophobic patch in close vicinity to FBXL2 Phe400 (Fig. 5f), poised to interact with its following ~10 residues, which are highly conserved among FBXL2 orthologs (Supplementary Note 1). Taken together, our mutational analysis reveals an unexpected multi-valency in FBXL2-GGTase3 interaction, which spans the entire LRR domain and the C-terminal tail of the F-box protein.
Discussion
Since the 1980s, the biomedical literature has stated that mammals possess three prenyltransferases. In this study, we have identified and characterized a fourth family member, GGTase 3, which consists of PTAR1, an orphan α subunit, and the β subunit RabGGTB. Prenyltransferases have been previously shown to share α subunits. Our studies reveal that this important enzyme family has also evolutionarily expanded by sharing β subunits. Notably, our structural and biochemical analyses unveil that the CaaX motif of FBXL2 contributes to, but is not sufficient for, the formation of a stable complex with GGTase3. PTAR1, instead, plays a critical role in FBXL2 recruitment through its uniquely extensive interaction with the LRR domain of the F-box protein. Therefore, in contrast to classical prenyltransferases, our results indicate that additional multivalent structural elements outside of the CaaX motif contribute to the FBXL2 specificity of GGTase3. Moreover, our study suggests that GGTase1 can prenylate FBXL2, albeit with less efficiency, when GGTase3 is absent in the cell. This is yet another example that prenyltransferases are able to cross-prenylate substrates, enlightening functional compensation under specific conditions.
By establishing FBXL2 as a substrate of GGTase3, we have also uncovered the first prenyltransferase that physically interacts with and modifies one of more than 600 ubiquitin ligases present in mammals to allow its localization to cell membranes, where FBXL2 has been shown to bind and ubiquitylate its substrates for subsequent proteasomal degradation.
PTAR1 has long been neglected until it was identified as a gene product required for optimal fitness of cultured cells in a genome-scale screen aimed at defining essential human genes46. FBXL2 is a pro-survival gene, whose product, in response to mitogens, inhibits apoptosis by promoting the activation of the PI3K-AKT pathway and by inhibiting mitochondrial calcium overloading27–29. Herein, we have shown physical and functional connections between PTAR1 and FBXL2, thereby delineating a post-translational modification cascade critical for cell survival. Together with the detailed structural basis of enzyme assembly and substrate recognition by GGTase3, our studies elucidated the structure and function of a previously unrecognized prenyltransferase. Finally, by shedding light on the complexity of substrate recognition by this family of enzymes, our work provides additional tools for developing therapeutics targeting prenyltransferases, which, despite initial obstacles, are widely pursued for treating cancer and other human diseases.
Methods
Click-IT®-Geranylgeranylation assay
RPE1-HTERT (retinal pigmented epithelium cells immortalized with human telomerase reverse transcriptase) and HeLa cells (~ 3–5 × 106) were plated in 10 cm dishes overnight. Cells were either with siRNA oligos and/or plasmids for the times indicated in figure legends using Lipofactemine®3000 reagent for 1 hour at 37°C prior to metabolic labeling with geranylgeranly azide (30uM, Thermo Scientific # C10249) for 24 hours in DMEM medium supplemented with 10% dialyzed FBS and 5 mM sodium pyruvate. Labeled cells were harvested, frozen and the subsequent lysis of the frozen pellets was carried out with NP-40(0.2%) supplemented with protease inhibitor cocktail (Roche# 04693132001), sodium vanadate (1 μM) and okadaic acid (30 nM) in PBS buffer. Soluble protein lysates (1 mg/ml) were used for the copper free “Click-IT reaction” with either 20 μM DIBO-Biotin (Fig. 2c) or 20 μM sDIBO-Biotin (Fig. 2d) for 1 hour at 37 °C with constant shaking according to manufactures instructions (Thermo Scientific, cat # C10412 and C20023). Immunoprecipitation of “Click-IT” lysates were performed under denaturing conditions. Geranylgeranylated FBXL2 was detected with streptavidin covalently conjugated to horseradish peroxidase (STREP-HRP) (Pierce#21126) followed by immunoblotting, as previously described27,29.
In vitro geranylgeranylation assay
Briefly, recombinant human FBXL2, FBXW7, K-RAS4B, CDC42, and tagged GGTase3 (MBP-PTAR1 and GST-RabGTTB, purified individually and then mixed to allow complex formation) were affinity purified from bacterial system using standard protocols. Untagged GGTase3 (a GST-PTAR1-RabGTTB complex in which the GST was cleaved following purification) was obtained as described below. Purified rat GGTase1 (GST-His-FNTA and untagged PGGT1B) was from MilliporeSigma (cat # 345852). FBXW7 (10 μM), K-RAS4B (10 μM) and various FBXL2 or CDC42 amounts (1 μM, 5 μM, 10 μM, or 25 μM) were first equilibrated to 37°C in a reaction buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 20 mM KCl, 100 μM ZnCl2, 5 mM MgCl2 buffer, 0.4 μM of [3H]-geranylgeranyl pyrophosphate (GGPP) (American Radiolabeled Chemicals no. ART 348). The geranylgeranylation reaction in a 50 μl volume was initiated with the addition of 100 ng of either GGTase3 or GGtase1. The reaction proceeded for 1.5 hours at 37°C while shaking at 900 rpm and subsequently blotted onto two Whatman Grade P81 Ion Exchange Cellulose Chromatography Paper (Lab Supply Outlaws no. 05-717-2C). The two papers were then placed into Ultrafree-MC Centrifugal Filter (Millipore no. UFC30HV00) and spun at 13000 rpm for 2 minutes. The papers were washed three times with 600uL of 95% ethanol and 75 mM K2HPO4 and the last wash was performed with PBS. The two chromatography papers were added to 5 ml of Universol™-Esliquid Scintillation Cocktail (MP Bio no. 0188248001) and the activity was measured via a scintillation counter (Scintillation Counter-Beckman 6500 Multi-Purpose Scintillation Counter). The activity was determined in counts per minute (CPM). Geranylgenranylation of the different proteins was measured by determining the transfer of [3H]-geranygeranylpyrophosphate onto purified substrates by GGTase3 or GGTase1 and plotted as μM/min using CPM counts. Michaelis-Menten kinetics was used to generate Km values using Prism Graphpad software.
Recombinant protein expression and purification
The DNAs encoding human GGTase3 heterodimer (PTAR1-RabGGTB) and FBXL2-SKP1 were subcloned into pFastBac vectors for protein co-expression in High Five (Invitrogen) monolayer insect cells. Among all four recombinant proteins, PTAR1 was expressed as a glutathione S-transferase (GST) N-terminal fusion protein and the tetrameric complex of GGTase3-FBXL2-SKP1 was isolated by glutathione affinity chromatography using buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1.0 mM DTT (dithiothreitol). The eluted tetrameric complex was further purified by anion exchange and gel filtration chromatography, after overnight TEV (tobacco etch virus) protease cleavage, in a final buffer containing 20 mM HEPES pH7.5, 150 mM NaCl and 2.0 mM DTT. The protein sample was concentrated and flash frozen for crystallization. For testing the binding activity of FBXL2-SKP1 with GGTase3 and its geranylgeranylation by GGTase3, the two individual protein complexes of FBXL2-SKP1 and GGTase3 were expressed in E. coli with the affinity tags fused on PTAR1 and FBXL2, respectively. The proteins were isolated and purified similarly as the tetrameric complex by three steps of affinity, anion exchange, and gel filtration chromatography. The affinity tag may be left on the proteins for the purposes of assays.
Crystallization, data collection and structure determination
The crystals of the GGTase3-FBXL2-SKP1 complex were obtained at room temperature by the hanging-drop vapour diffusion method, using 1.0 μl protein sample mixed with an equal volume of reservoir solution containing 0.1 M MES pH6.0, and 20%−22% (v/v) PEG 400. Crystals appeared within 1–2 days and would complete growth in approximately a week. Conducting micro-seeding substantially increased the size of crystals, and the best crystals diffracted to 2.5 Å. Crystals were cryoprotected by addition of 20% (v/v) glycerol to the reservoir solutions in harvest, and flash frozen in liquid nitrogen for data collection. All data sets were collected at 100 K with a wavelength of 1.000 Å at the BL8.2.1 and BL8.2.2 beamlines at the Advanced Light Source of the Lawrence Berkeley National Laboratory. Diffraction data were indexed, integrated, and scaled with the HKL2000 package47. The structure was determined by molecular replacement using Phaser in the CCP4 suite48 with models of human RabGGTase (PDB: 3DSS) and SKP1-SKP2 complex (PDB: 1FQV) structures. The initial models were then rebuilt manually using COOT48 and refined by Refmac5 and PHENIX49. Ramachandran plot analysis of the final model showed that 96.4% and 3.6% of the residues are in favored and allowed regions, respectively. Complete data collection and refinement statistics are summarized in Table 1. All structure figures were rendered in PyMOL.
Octet BioLayer interferometry measurement
Binding affinity of His-tagged FBXL2-SKP1 with GGTase3 was measured using the Octet Red 96 (ForteBio, Pall Life Sciences) following the manufacturer’s procedures in triplicates. The optical probes were coated with Anti-Penta-His (HIS1K), loaded with 500 nM His-tagged FBXL2-SKP1 purified protein as ligand and quenched with 2.5 μM His-GST protein prior to kinetic binding analysis. The reactions were carried out in black 96-well plates maintained at 30°C with volume of 200 μl in each well. Binding buffer contained 25 mM HEPES pH 7.7, 150 mM NaCl, 2.0 mM DTT, and 0.1% (w/v) BSA. Purified GGTase3 as the analyte were serially diluted in the binding buffer to acquire complete measurements. No binding of the analyte to the unloaded probes was observed. Binding kinetics of the analyte at all concentrations were obtained simultaneously using instrumental defaults. All data were analyzed with the Octet data analysis software. The association and dissociation curves were globally fit with a 1:1 ligand model. The kon and kdis values were used to calculate the dissociation constant KD with direct binding analysis.
Cell lines
All cell lines were purchased from ATCC and routinely tested for mycoplasma.
Live cell Imaging
HeLa cells incubated with various siRNA oligos were plated in dishes with glass base (Thermo Scientific #150682) in DMEM medium supplemented with 10% FBS overnight before transient transfection of GFP-tagged cDNAs of indicated genes with Lipofactemine®3000 reagent. One-hour post-transfection protocols the cells were incubated in fresh medium and incubated for sixteen hours at 37°C supplemented with 5% CO2. Live cell imaging of cells was carried out with Zeiss LSM-510 META confocal microscopes using 63× oil-based objective in incubation chamber at 37°C supplemented with 5% CO2. In each experiment 10 independent frames were captured. Images were captured and processed with ZEN/ZEN lite imaging software from Zeiss.
Antibodies, Plasmids, and Chemicals
A rabbit polyclonal antibody against the peptide corresponding to amino acids 339 to 359 (CDGLNDSSKQGYSQETKRLKRT) of human PTAR1 primary sequence was custom made by YenZyme antibodies, LLC, CA 94080. The specificity of the antibody was validated by silencing PTAR1 in primary fibroblasts NHFs and HEK293T cell lines (Supplementary Fig. 1A). Antibodies were from Abcam (FNTA, FNTB), Abnova (PGGT1B), Bethyl (RabGGTB), Aviva Biosystems (RabGGTA), Sigma (anti-FLAG M2, anti-FLAG, β-actin), Santa Cruz Biotechnology (SKP1), and Covance (anti-HA). A polyclonal rabbit antibody to FBXL2 was generated using a peptide against FBXL2 as previously described27. Human cDNA ORFs for PTAR1, FNTA, and RabGGTA were purchased from Origene. GFP-tagged cDNA of RAB1a, RAB7a, RAB11, RAB34, and RAB35 were obtained from Addgene. The variously tagged-genes and mutants were generated in pcDNA3 and pEGFPC1 vectors either by sub-cloning or site-directed mutagenesis methodologies, as described.
Immunoprecipitation and Immunoblotting
RPE1-HTERT, HeLa, and HEK-293T cells were transiently transfected using Lipofectamine® 3000 reagent. Sixteen hours after transfection HEK-293T cells were incubated with MG132 for three hours before harvesting. Lysis of cell pellets was carried out with lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, glycerol 10%, 1 mM EDTA, 50 mM NaF, NP-40 0.1%) supplemented with protease and phosphatase inhibitors. Protein estimation in soluble lysates was carried out with Bio-Rad DC™ protein assay according to manufactures protocol (cat# 500–113/4). Immunoprecipitation and western blotting were performed as previously described27,28. For trimeric complex formation, sixteen hours after transfection with indicated plasmids, cells were harvested and WCLs were used for a first immunoprecipitation with an anti-FLAG antibody. Immunoprecipitated proteins were eluted with an excess of FLAG peptide. 10% of the eluate was set aside, and the remaining 90% was subjected to a second immunoprecipitation using an anti-GFP antibody and elution with 1% SDS. The two eluates were then immunoblotted.
Gene silencing
Cells were grown in live cell imaging compatible dish (Thermo Scientific #150682), and 10 cm dish formats. ON TARGET siRNA oligos from Dharmacon, Inc.2d for various genes (PTAR1, PGGT1B, FNTA, and RabGGTA) and controls were used at 5 nM final concentration for 24–72 hours using Lipofectamine® RNAimax reagent, according to the manufacturer’s instruction (invitrogen). ON-TARGETplus human PTAR1 siRNA (L-010311–01, LU-010311-01, J-010311-09-0005, J-010311-10-0002): GAAGCUAGGUAUAACCGGA, GAACAGGAGAUGAAUUGAUA, CCAUAGUCCUGGUUGAAAA, GGAGGAACCCACACAUAGA. ON-TARGETplus human FNTA siRNA (L-008807, LU-008807), ON-TARGETplus human RABGGTA siRNA (L-005097), ON-TARGETplus human PGGT1B (L-008703-00-0005) and ON-TARGET Non-targeting siRNA #1 (D-001810-01-05).
Statistics and Reproducibility
Dataset were analyzed by GraphPad Prism® software with student’s unpaired T-test and ANOVA. Error bars represent either S.D. or S.E.M as indicated in the text. All immunoblot images and immunofluorescence images are representative of at least three independent experiments, except where specified in the figure legends.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Structural coordinates and structural factors have been deposited to the Protein Data Bank (PDB) under accession number 6O60. Source data for Figure 2a, b and Supplementary Figure 2b are available with the paper online. All other data are available from the authors upon request.
All experiments were repeated at least three times. Uncropped blot/gel images are shown in Supplementary Data Set 1.
Supplementary Material
Acknowledgments:
The authors thank G. Rona for his contribution, C. Fierke, J. Ramalho, and M. Seabra for reagents, T.R. Hinds for Octet BLI analysis, and M. Bergo for critically reading the manuscript. M. Pagano is grateful to T.M. Thor for continuous support. This work was funded by grants from the NIH (R01-GM057587 and R01-CA076584) to M. Pagano. M. Pagano and N. Zheng are Investigators with the Howard Hughes Medical Institute.
Footnotes
Competing Interests:
The authors declare no competing financial interests. M.P. is a consultant for BeyondSpring Pharmaceuticals and a member of the scientific advisory boards of CullGen, Inc. and Kymera Therapeutics. N.Z. is a member of the scientific advisory board of Kymera Therapeutics.
References
- 1.Zhang FL & Casey PJ Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65, 241–269, doi: 10.1146/annurev.bi.65.070196.001325 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Wang M & Casey PJ Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol 17, 110–122, doi: 10.1038/nrm.2015.11 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Wright LP & Philips MR Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res 47, 883–891, doi: 10.1194/jlr.R600004-JLR200 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Ahearn IM, Haigis K, Bar-Sagi D & Philips MR Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 13, 39–51, doi: 10.1038/nrm3255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hougland JL & Fierke CA Getting a handle on protein prenylation. Nat Chem Biol 5, 197–198, doi: 10.1038/nchembio0409-197 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Berndt N, Hamilton AD & Sebti SM Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11, 775–791, doi: 10.1038/nrc3151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen UT, Goody RS & Alexandrov K Understanding and exploiting protein prenyltransferases. Chembiochem 11, 1194–1201, doi: 10.1002/cbic.200900727 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Cox AD, Der CJ & Philips MR Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res 21, 1819–1827, doi: 10.1158/1078-0432.CCR-14-3214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casey PJ & Seabra MC Protein prenyltransferases. J Biol Chem 271, 5289–5292 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Maurer-Stroh S, Washietl S & Eisenhaber F Protein prenyltransferases. Genome Biol 4, 212(2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane KT & Beese LS Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res 47, 681–699, doi: 10.1194/jlr.R600002-JLR200 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Benetka W, Koranda M, Maurer-Stroh S, Pittner F & Eisenhaber F Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem 7, 6, doi: 10.1186/1471-2091-7-6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Z et al. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J 27, 2444–2456, doi: 10.1038/emboj.2008.164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rak A, Pylypenko O, Niculae A, Goody RS & Alexandrov K Crystallization and preliminary X-ray diffraction analysis of monoprenylated Rab7 GTPase in complex with Rab escort protein 1. J Struct Biol 141, 93–95 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Pylypenko O et al. Structure of Rab escort protein-1 in complex with Rab geranylgeranyltransferase. Mol Cell 11, 483–494 (2003). [DOI] [PubMed] [Google Scholar]
- 16.James GL, Goldstein JL & Brown MS Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J Biol Chem 270, 6221–6226 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Baron R et al. RhoB prenylation is driven by the three carboxyl-terminal amino acids of the protein: evidenced in vivo by an anti-farnesyl cysteine antibody. Proc Natl Acad Sci U S A 97, 11626–11631, doi: 10.1073/pnas.97.21.11626 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carboni JM et al. Farnesyltransferase inhibitors are inhibitors of Ras but not R-Ras2/TC21, transformation. Oncogene 10, 1905–1913 (1995). [PubMed] [Google Scholar]
- 19.Rowell CA, Kowalczyk JJ, Lewis MD & Garcia AM Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem 272, 14093–14097 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Whyte DB et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272, 14459–14464 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Skaar JR, Pagan JK & Pagano M Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14, 369–381, doi: 10.1038/nrm3582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaar JR, Pagan JK & Pagano M SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov 13, 889–903, doi: 10.1038/nrd4432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao I et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell 130, 943–957, doi: 10.1016/j.cell.2007.06.052 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koga K, Yao I, Setou M & Zhuo M SCRAPPER Selectively Contributes to Spontaneous Release and Presynaptic Long-Term Potentiation in the Anterior Cingulate Cortex. J Neurosci 37, 3887–3895, doi: 10.1523/JNEUROSCI.0023-16.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen BB, Coon TA, Glasser JR & Mallampalli RK Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol 31, 1905–1920, doi: 10.1128/MCB.00723-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell 18, 425–434 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Kuchay S et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol 15, 472–480, doi: 10.1038/ncb2731 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuchay S et al. PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546, 554–558, doi: 10.1038/nature22965(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchay S et al. NS5A Promotes Constitutive Degradation of IP3R3 to Counteract Apoptosis Induced by Hepatitis C Virus. Cell Rep 25, 833–840 e833, doi: 10.1016/j.celrep.2018.09.088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin TB et al. Fbxo3-Dependent Fbxl2 Ubiquitination Mediates Neuropathic Allodynia through the TRAF2/TNIK/GluR1 Cascade. J Neurosci 35, 16545–16560, doi: 10.1523/JNEUROSCI.2301-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai CY et al. Spinal Fbxo3-Dependent Fbxl2 Ubiquitination of Active Zone Protein RIM1alpha Mediates Neuropathic Allodynia through CaV2.2 Activation. J Neurosci 36, 9722–9738, doi: 10.1523/JNEUROSCI.1732-16.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S et al. Lipopolysaccharide Primes the NALP3 Inflammasome by Inhibiting Its Ubiquitination and Degradation Mediated by the SCFFBXL2 E3 Ligase. J Biol Chem 290, 18124–18133, doi: 10.1074/jbc.M115.645549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan MK, Lim HJ, Bennett EJ, Shi Y & Harper JW Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol Cell 52, 9–24, doi: 10.1016/j.molcel.2013.08.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huttlin EL et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440, doi: 10.1016/j.cell.2015.06.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellacheruvu D et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736, doi: 10.1038/nmeth.2557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoma NH, Iakovenko A, Goody RS & Alexandrov K Phosphoisoprenoids modulate association of Rab geranylgeranyltransferase with REP-1. J Biol Chem 276, 48637–48643, doi: 10.1074/jbc.M108241200 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Taylor JS, Reid TS, Terry KL, Casey PJ & Beese LS Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J 22, 5963–5974, doi: 10.1093/emboj/cdg571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furfine ES, Leban JJ, Landavazo A, Moomaw JF & Casey PJ Protein farnesyltransferase: kinetics of farnesyl pyrophosphate binding and product release. Biochemistry 34, 6857–6862 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Park HW, Boduluri SR, Moomaw JF, Casey PJ & Beese LS Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science 275, 1800–1804 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Schulman BA et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386, doi: 10.1038/35042620 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Zheng N et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709, doi: 10.1038/416703a (2002). [DOI] [PubMed] [Google Scholar]
- 42.Xing W et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68, doi: 10.1038/nature11964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan X et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645, doi: 10.1038/nature05731 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Long SB, Casey PJ & Beese LS The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures. Structure 8, 209–222 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Long SB, Casey PJ & Beese LS Reaction path of protein farnesyltransferase at atomic resolution. Nature 419, 645–650, doi: 10.1038/nature00986 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Blomen VA et al. Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096, doi: 10.1126/science.aac7557 (2015). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 47.Otwinowski Z & Minor W in Methods in Enzymology Vol. 276 (eds Carter CW & Sweet RM) 307–326 (Academic Press, New York, 1997). [DOI] [PubMed] [Google Scholar]
- 48.CCP4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr D50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–1954, doi:S0907444902016657 [pii] (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural coordinates and structural factors have been deposited to the Protein Data Bank (PDB) under accession number 6O60. Source data for Figure 2a, b and Supplementary Figure 2b are available with the paper online. All other data are available from the authors upon request.
All experiments were repeated at least three times. Uncropped blot/gel images are shown in Supplementary Data Set 1.