Abstract
We review the literature on Tau and TDP-43 proteinopathies in aged human brains and the relevant underlying pathogenetic cascades. Complex interacting pathways are implicated in Alzheimer’s disease and related dementias (ADRD), wherein multiple proteins tend to misfold in a manner that is “reactive,” but, subsequently, each proteinopathy may contribute strongly to the clinical symptoms. Tau proteinopathy exists in brains of individuals across a broad spectrum of primary underlying conditions—e.g., developmental, traumatic, and inflammatory/infectious diseases. TDP-43 proteinopathy is also expressed in a wide range of clinical disorders. Although TDP-43 proteinopathy was first described in the central nervous system of patients with amyotrophic lateral sclerosis (ALS) and in subtypes of frontotemporal dementia (FTD/FTLD), TDP-43 proteinopathy is also present in chronic traumatic encephalopathy, cognitively impaired persons in advanced age with hippocampal sclerosis, Huntington’s disease, and other diseases. There is also evidence of cellular co-localization between Tau and TDP-43, suggesting common pathways or protein interactions facilitating misfolding in one protein by the other. Multiple pleiotropic gene variants can alter risk for Tau or TDP-43 pathologies, and certain gene variants (e.g., APOE ε4, Huntingtin triplet repeats) are associated with increases of both Tau and TDP-43 proteinopathies. Studies of genetic risk factors have provided insights into multiple nodes of the pathologic cascades involved in Tau and TDP-43 proteinopathies. Variants from a specific gene can be either a low-penetrant risk factor for a group of diseases, or alternatively, a different variant of the same gene may be a disease-driving allele that is associated with a relatively aggressive and early-onset version of a clinically and pathologically specific disease type. Overall, a complex but enlightening paradigm has emerged, wherein both Tau and TDP-43 are linked to numerous overlapping upstream influences, and both are connected with multiple downstream pathologically- and clinically-defined deleterious effects.
Introduction
A common theme that has emerged in Alzheimer’s disease (AD) and related dementia (collectively termed ADRD) research is that underlying genetic and/or environmental perturbations lead to formation, aggregation, and propagation of misfolded proteins (termed a “proteinopathy”) that can be deleterious through both toxic gain-of-function and also loss of normal function mechanisms [1–3]. Over the past several decades, clinical-pathologic correlation (CPC) studies, in concert with genetic studies, provided significant new insights into the roles of proteinopathies in ADRD. Modern CPC studies have established that two of the most important pathologic hallmarks associated with clinical cognitive impairment are proteinopathies associated with the microtubule-associated protein Tau, and the transactive response DNA 43 kDa binding protein (TDP-43).
Both Tau and TDP-43 proteinopathies are caused by pathologic mechanisms that occur in a wide range of disease paradigms. In the course of these diseases, common pathologic cascades are downstream of many different primary causes, while clinical symptoms are not necessarily specific to their biologic or pathologic antecedents. Whereas there is widespread appreciation that many different conditions are associated with Tau protein pathology (“the tauopathies”) [4, 5], the study of TDP-43 proteinopathic conditions is a younger area of research. Interestingly, some of the paradigms associated with tauopathic disease also seem to apply to TDP-43 proteinopathies, including the tendency of specific genetic modifiers [6, 7] to apparently increase the proteinopathy that was caused by different primary upstream causes. Here, we review the relevant literature on these “misfoldingopathies”, beginning with a descriptions of some of the emerging themes of pathologic cascades, then reviewing some of the pleiotropic genetic influences discovered to affect Tau and TDP-43 proteinopathies, and finally discussing a subset of the downstream effects that were discovered before the age of widespread immunohistochemistry-based neuropathologic investigations.
Pathologic cascades and synergies: protein misfolding can have multiple causes and effects
Proteinopathies can provide diagnostic histologic markers for ADRD subtypes, and also may eventually represent therapeutic targets. However, there are added levels of complexity that derive from differing upstream influences, from multi-step pathologic cascades, from overlapping downstream effects, and from biochemical interactions that may occur between the different misfolding proteins. All of these factors often occur concurrently and should be considered when studying the Tau and TDP-43 proteinopathies.
A generalizable paradigm of pathologic cascades in ADRD involving misfolding proteins (Fig. 1) can be applied more specifically in the contexts of both Tau and TDP-43 proteinopathies (Fig. 2). The red arrows in Figs 1 and 2 indicate that the presence of misfolded proteins can, by themselves, contribute via positive feedback mechanism (s) to changes in the local biochemical microenvironment, propagating further misfolding of proteins. Evidence in support of this positive-feedback hypothesis comes from multiple sources including cell culture models [8, 9] and “transmission” animal models [10–12] where injection of misfolded proteins can lead to an auto-propagating degenerative changes in which intrinsic proteins are incorporated into a pathogenetic process by exogenous proteins.
Fig. 1.
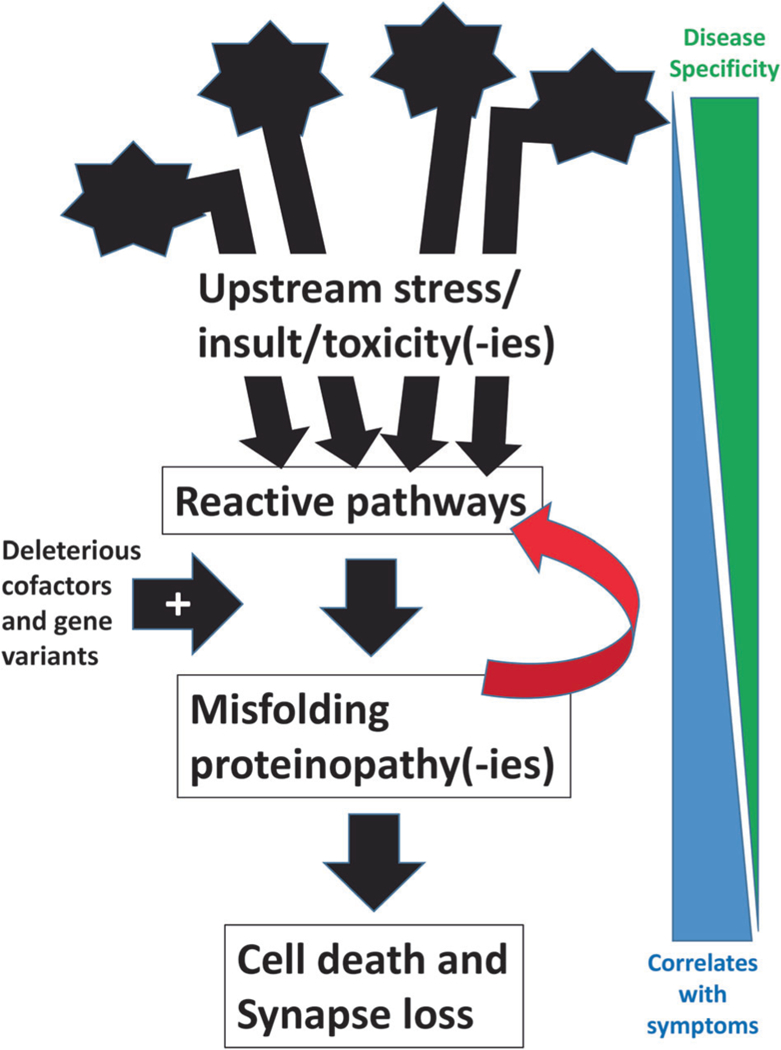
Schematic cartoon depicts a paradigm for the mechanisms underlying neurodegenerative conditions of aging. Multiple different upstream genetic and/or environmental factors have the potential to constitute a trigger for reactive changes in the brain. The reactive mechanisms and pathways may be compensatory or beneficial in some contexts. However, those same pathways may also contribute to one or multiple different proteins misfolding. The tendency to generate misfolding proteins appears to be augmented among individuals with specific genetic risk factors. Importantly, a salient feature of misfolding proteins that are impactful, in a clinical and biologic sense, is that they appear to have a propensity to create or promote a micro-environmental shift toward biochemical pathways that augment their own misfolding. This deleterious feedback mechanism (signified by the red arrow) may promote an auto-propagating cycle, greatly amplifying the impact of the primary disease mechanism(s). The net effects of the upstream trigger, reactive pathways, and misfolding proteinopathies, are cell death and synapse elimination that can culminate in clinical manifestations. Note that on the right of the figure is indicated an inverse relationship between the factors that are disease-specific (related to upstream mechanisms) and the downstream pathologic phenomena, such as synapse loss, that correlate best with antemortem clinical symptoms
Fig. 2.
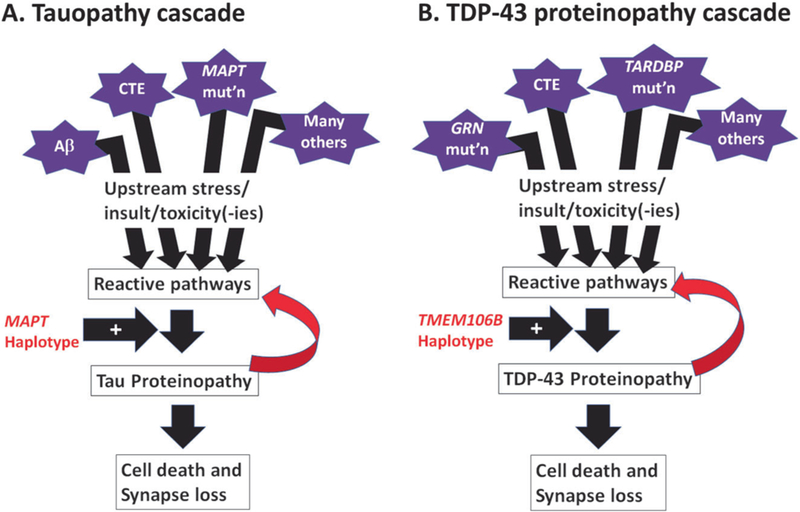
Schematic cartoons depict the overlapping elements of the pathological cascades that are seen in Tau (a) and TDP-43 (b) proteinopathies. Multiple factors can contribute to Tau or TDP-43 proteinopathies, or both (see Table 1). For Tau and TDP-43 proteins, reactive pathways—perhaps related to oxidative stress, phosphorylation, autophagy, proteolysis, inflammation, and/or other biochemical changes—appear to have an influence on the proteins’ structural properties, making the polypeptides more likely to misfold and to generate both toxicity and loss of normal function. Whereas the MAPT haplotype appears to be an important genetic risk factor in multiple different Tau proteinopathies, the TMEM106B haplotype (signaled by the rs1990662 risk variant) is associated with increased risk for multiple TDP-43 proteinopathies. Both Tau and TDP-43 proteinopathies also appear to be “transmissible” in animal models (see ref. [10]), meaning that their presence in the brain can—even without another “upstream factor”—potentiate additional “downstream” proteinopathy, contributing to cell death and synapse loss
Many questions remain regarding what, specifically, initiates Tau and TDP-43 proteinopathies. Here we focus predominantly on TDP-43 proteinopathy, as prior reviews have focused extensively on tauopathies [13–15]. Given that a majority of patients with TDP-43 proteinopathies lack strong family histories of similar illnesses (as far as we now know), it may be that exogenous factors can influence TDP-43 to form pathologic inclusions. One example of a relatively clear upstream mechanism at work is chronic traumatic encephalopathy (CTE), a progressive neurodegenerative condition in which the initiating event is recurrent brain trauma, and pathologic examination reveals both TDP-43 and Tau proteinopathies [16–19]. In one study [20], of 12 autopsied individuals with CTE, 10 had widespread TDP-43 pathology; three of these who had developed progressive motor neuron disease with weakness, atrophy, and fasciculations were found to have both Tau and TDP-43 proteinopathies. Proteins that have been found in disease-associated protein aggregates have revealed common properties—internal amino acid sequences that facilitate aggregation [21, 22]. Similar protein motifs are found in non-disease causing proteins, and their liberation and interaction with disease causing proteins allows selfpropagation and formation of cytotoxic proteins aggregations [23, 24]. In normal cells, misfolded proteins can be degraded or refolded into their homeostatic form by protein chaperones [25]. As the human brain undergoes normal aging, the chaperone proteins responsible for correct protein folding are repressed, and these changes are even more prominent in patients affected by AD, Huntington’s disease (HD), or Parkinson Disease (PD) [26], so, the lack of chaperone proteins may be contributing to these disorders.
Following or in parallel with disease initiation, specific biochemical factors, paradigms, and cellular domains have been associated with worsening (or protection from) TDP-43 proteinopathy—posttranslational modifications (phosphorylation, acetylation, proteolysis), autophagy, endosomal/lysosomal processing, mitochondrial function, nuclearcytoplasmic transport, oxidative stress, and neuroinflammation, to name a few. A comprehensive discussion of these factors is beyond the scope of this review. However, we here address several of these contributing factors.
Oxidative stress has been postulated as a triggering mechanism responsible for the formation of pathologic TDP-43 inclusions [27, 28]. Work by Iguchi et al. demonstrated that treatment of NSC34 cells and mouse primary cortical neurons with ethacrynic acid (EA) depleted their glutathione levels and causes increased oxidative stress. This caused TDP-43 phosphorylation, insolubilization, and cytoplasmic distribution [29]. The phosphorylation was reversed by N-acetylcysteine (NAC) or inhibition of CK1 and CK2, suggesting that CK1 and CK2 can mediate TDP-43 phosphorylation as a result of oxidative stress. Exploring whether other causes of oxidative stress can result in the same phenotype, the authors found that H2O2 also induced C-terminal phosphorylation, insolubilization, and cytoplasmic distribution of TDP-43 as observed under EA exposure [29]. Oxidative stress has been associated with various aging-related disorders [30, 31], and since some TDP-43 proteinopathies are aging-related diseases, this supports the hypothesis that oxidative stress can potentiate the pathologic TDP-43 modification.
TDP-43, when found in insoluble toxic inclusions, is known to be hyper-phosphorylated at C-terminal sites [29, 32, 33]. The best characterized phosphorylation sites on the TDP-43 protein are amino acid residues Ser409 and Ser410 [32, 34]. Additional phosphorylated residues that are associated with disease pathology are Ser379, Ser403, Ser404 [32, 35, 36], which have not been as well characterized as Ser409/410. Some studies demonstrate that phosphorylation of TDP-43 negatively affects its solubility [37–40]. There are multiple kinases that can phosphorylate TDP-43; however, the exact role for these molecules in each specific neurodegenerative disease has not been clearly established and more work is required in this area.
Another potential upstream contributing factor in TDP-43 pathology is believed to be defects in autophagy. Previous studies provided evidence that pathological forms of TDP-43 are cleared through autophagy [41–43]. For example, Wang et al. demonstrated that using an mTOR inhibitor rapamycin and three other autophagy activators (spermidine, carbamazepine, and tamoxifen) reduced the number of cytosolic TDP-43 inclusions in a FTLD-TDP mouse model [42]. Further strengthening the association between autophagy and pathological TDP-43 accumulation was work by Chang et al. [44], who, studying neurons deficient in progranulin (the polypeptide product of GRN), found that autophagy regulators were impaired and autophagic flux was reduced. This reduction in autophagy rendered neurons more prone to accumulate pathological TDP-43 [44]. These results suggest that, in addition to oxidative damage and phosphorylation, defects in autophagy precipitate pathologic TDP-43.
Whatever the upstream mechanisms, the accumulation of one species of misfolded protein can affect cellular processes and ultimately trigger misfolding of different proteins in the same cells [1, 3, 45–48]—a process termed “pathologic synergy” [3]. This interaction among misfolded protein species may accelerate cognitive decline in some dementing disorders [3]. Tau and TDP-43 proteinopathies appear to be have the potential for pathologic synergy. Brain conditions with Tau pathologies, including argyrophilic grain disease [49], HD [50], anti-IgLON5 tauopathy [51], corticobasal degeneration (CBD), and progressive nuclear palsy (PSP) [52–54], have been reported to also demonstrate comorbid TDP-43 pathology, often with both pathologies present in the amygdala [3]. Several studies have demonstrated co-localization of Tau and TDP-43 pathologic aggregates in the same cells [3]. For example, Higashi et al. used confocal microscopy and double-label immunostaining against TDP-43 and Tau to show that TDP-43 and Tau-positive NFT co-localize in amygdala in AD patients [55]. Smith et al., in a study of 247 subjects, found that a subset of colocalized hippocampal Tau/TDP-43 pathology was associated with advanced AD [56]. In this study, immunofluorescence experiments indicated that approximately 25% of cells with TDP-43 proteinopathy showed detectable colocalized phospho-Tau immunoreactivity [56]. Even in brains that lack appreciable AD-type amyloid plaques, there can be both Tau and TDP-43 pathologies. Note that in the representative case shown in Fig. 3, Tau and TDP-43 pathologies are demonstrated in multiple regions, including, intriguingly, near the pial surface of the brain. We conclude that these common pathologies may occur secondarily or in parallel with each other.
Fig. 3.
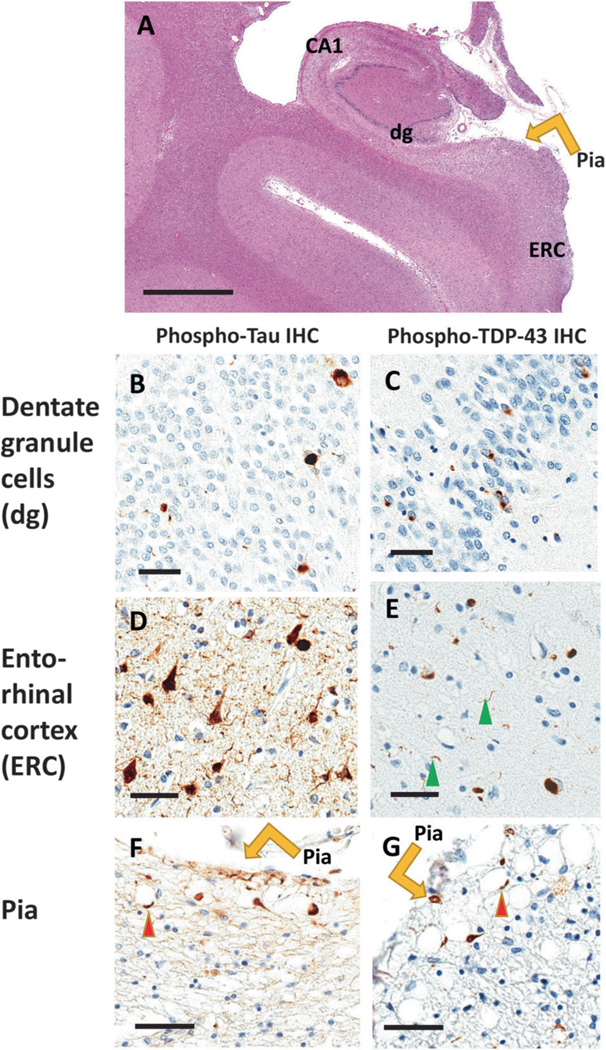
Comorbid Tau and TDP-43 pathologies are relatively common pathologic phenomena. Here are shown photomicrographs depicting stained sections from the brain of a 102-year-old woman who died with a history of dementia. Autopsy showed minimal Alzheimer’s disease-type changes (no neuritic Aβ plaques and Braak NFT stage II). Portions of the brain were stained using hematoxylin and eosin (H&E; a), and near-adjacent sections were stained for phospho-Tau immunohistochemistry (IHC; b, d, f), and phospho-TDP-43 IHC (c, e, g). The hippocampus is shown in the coronal plane (a) with anatomic regions labeled. Panels b and c show dentate granule (dg) cells, d and e show entorhinal cortex (ERC), and f and g demonstrate IHC staining near the pia lining (orange arrows). Sections are counterstained using hematoxylin (blue nuclei) and IHC reaction product is brown. Note that both Tau and TDP-43 proteinopathy are seen in dentate granule cells, entorhinal cortex, and within twiglike processes around corpora amylacea (red arrowheads) near the pia layer at the surface of the medial temporal lobes. Scale bars = 4 mm (a), 70 μm (b–e), and 100 μm (f, g)
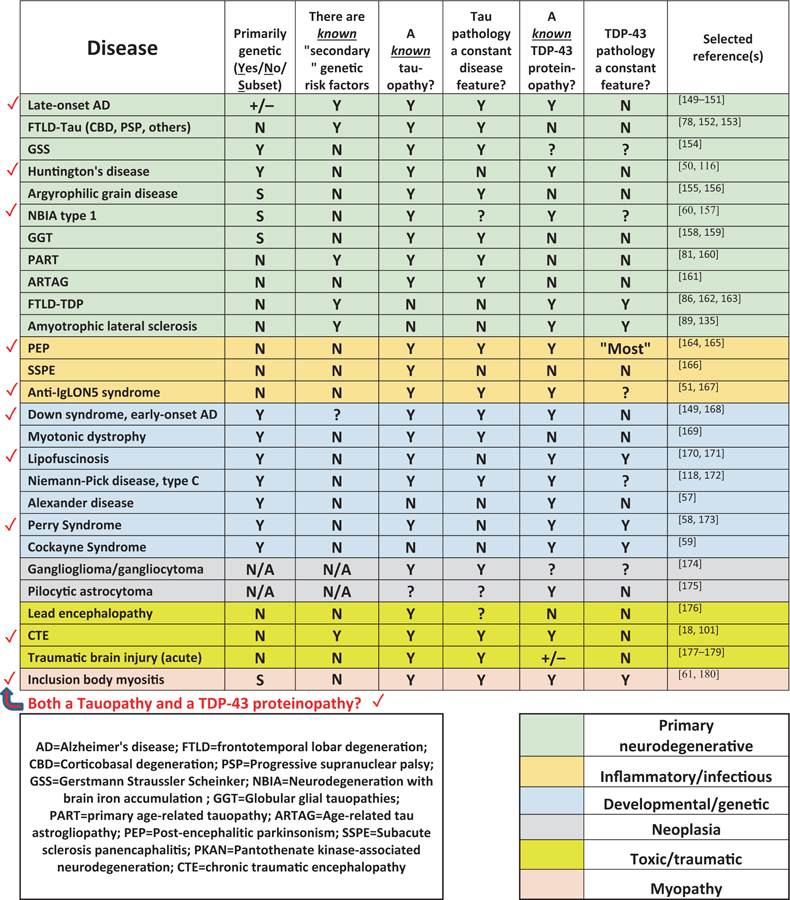
It is widely accepted that tauopathies span diverse disease paradigms such as developmental/genetic diseases, viral infection, triplet repeat disorders, head trauma, myositis, and many others (Table 1). Excellent prior reviews have been written on tauopathies [13–15]. It is less well-known that pathologic promiscuity also characterizes TDP-43 proteinopathies (Table 1). For example, to the relatively well-known diseases with TDP-43 proteinopathy (ALS and FTLD), one can add Alexander disease [57], Perry syndrome [58], Cockayne syndrome [59], neurodegeneration with brain iron accumulation [60], inclusion body myositis [61], HD [50], and other conditions (Table 1). Brain diseases that have been characterized to include both Tau and TDP-43 proteinopathies are indicated by red checkmarks in Table 1. The full implications of these observations are still not well understood but some inferences can be made: (1) Both Tau and TDP-43 proteinopathies can be driven by a diverse set of upstream factors; (2) Most of the upstream factors associated with Tau and TDP-43 proteinopathies involve some sort of chronic stress or insult; (3) Both Tau and TDP-43 proteinopathies are not disease-specific, but they still may play a role in influencing—perhaps dramatically exacerbating—multiple different upstream diseasedriving mechanisms.
Table 1.
A selection of Tau and/or TDP-43 proteinopathies
 |
Genetic pleiotropy in neurodegeneration and Tau/TDP-43 proteinopathies
Genetics is (for good reasons) considered to exert upstream influences on disease phenotypes. However, genetic factors may influence diseases at different nodes of a pathologic cascade—perhaps causing protein misfolding to be worsened only after it has begun to develop. As such, there is abundant evidence of pleiotropy, wherein a given gene, or even a specific gene mutation, may be associated with more than one different phenotypes.
Pleiotropic influences have been shown to be at work in both Tau and TDP-43 proteinopathies, which should come as no surprise. Since the first genomewide association study (GWAS) was conducted, there have been over 80,000 unique single nucleotide polymorphism (SNP)-trait associations discovered and published in over 3000 manuscripts [62]. As more replicable associations were found, the magnitude of pleiotropy in human disease has been increasingly appreciated. In 2011, a study of curated results from the GWAS Catalog indicated pleiotropy in nearly 17% of genes and 4.6% of SNPs [62, 63]. Notably, the conservative criteria used in this study undoubtedly induced underestimation and the distinction between a cross-phenotype association, and, true biological pleiotropy cannot be guaranteed from these database studies [64]. In a more recent survey of GWAS results, 44% of trait-associated SNPs were found to associate with more than one phenotype [65]. The increased availability of GWAS summary statistics in concert with methodological developments for estimation of the genetic covariance shared between distinct traits—including extensions for annotationbased partitioning—has enabled accelerated progress in the investigation of genetic mechanisms shared between multiple traits [66, 67]. These exciting developments will continue to help resolve the etiologies of complex diseases that affect the aged human brain.
Before discussing pleiotropy in ADRD proteinopathies, it is important to note that there is some conceptual ambiguity in the term “pleiotropy”. The detectable crossphenotype genetic associations can be classified, and only a subset deserve the designation of true biologic pleiotropy. Hence, there are subtypes of cross-phenotype associations that could be delineated when a genetic variant or gene is correlated with more than one trait: biological pleiotropy, mediated pleiotropy and spurious pleiotropy [64]. These distinctions are important but it is notable that even when some degree of reported results are “spurious” (the study design, genotype, and/or phenotype are biasing the result), there could still be important biological commonalities between two conditions as highlighted by the genetic association; such is apparently the case for schizophrenia and bipolar disorder [64]. For an excellent discussion of this topic, please see ref. [64]. Here, we focus on four genes relevant to Tau and/or TDP-43 proteinopathies: MAPT, TMEM106B, GRN, and APOE (Table 2).
Table 2.
Pleiotropy in neurodegenerative conditions: select genes
| Gene | # Unique traits with GWAS hitsa | # Total SNP-trait GWAS associationsa | Associated conditions related to neurologic/ neurodegenerative diseases | Selected reference (s) |
|---|---|---|---|---|
| MAPT | 26 | 69 | FTLD-Tau (including CBD, PSP, others) | [78, 162, 181] |
| Alzheimer’s disease | [78, 79] | |||
| Parkinson’s disease | [78, 182, 183] | |||
| Primary age-related tauopathy | [81] | |||
| Globular glial tauopathy | [159, 184] | |||
| TMEM106B | 9 | 18 | FTLD-TDP and ALS | [86, 89] |
| TDP-43/hippocampal sclerosis | [92, 108, 185] | |||
| Chronic traumatic encephalopathy | [101] | |||
| Healthy brain aging | [186, 187] | |||
| GRN | 5 | 3 | FTLD-TDP | [103, 104, 188, |
| 189] | ||||
| TDP-43/hippocampal sclerosis | [93, 94, 106] | |||
| Neuronal ceroid lipofuscinosisb | [171, 190, 191] | |||
| APOE | 77 | 271 | Alzheimer’s disease | [111, 192, 193] |
| TDP-43/hippocampal sclerosis | [112, 114, 115] | |||
| Lewy body disease | [193–195] | |||
| ABCC9 | 2 | 2 | TDP-43/hippocampal sclerosis | [94, 108, 196] |
| Cantu syndrome | [144, 197] |
Association counts were based on the database from the NHGRI-EBI GWAS Catalog [62]: https://www.ebi.ac.uk/gwas/ as of 26 November 2018 and filtered to include only results with p < 5e−8
Due to homozygous GRN mutations
Specific mutations in MAPT produce different pathologic features and correspondingly different clinical symptoms. The MAPT gene codes for the Tau protein. Alternative splicing of MAPT is able to generate six Tau isoforms ranging from 352 to 441 amino acids in adult human brain [68]. Direct associations between MAPT mutation and neurodegenerative diseases were established when dominantly inherited forms of frontotemporal dementia and parkinsonism (FTDP) were linked to chromosome 17q21–22 (FTDP-17) [69–71]. There are now dozens of known pathogenic mutations identified in MAPT [72]. The MAPT mutations exert different effects—the mutations in exons 9–12 typically impair the function of Tau microtubulebinding repeats, whereas other mutations affect alternative splicing of Tau pre-mRNA [73].
Underscoring the pleiotropic effects of MAPT gene variants, two haplotypes of MAPT exist and they are associated with a variety of different ADRD phenotypes. The haplotypes themselves are characterized by a 900-kilobase inversion (H1) or noninversion (H2) polymorphism [74]. Prior to discovery of H1 haplotype in MAPT, it was demonstrated that TG repeats (A0/A0 genotype, which is present in > 50% of Caucasians [75, 76]) in exon 9 were linked with increased risk of development of PSP [77]. Further work demonstrated that the H1 allele is also associated with increased risk of developing other tauopathies including FTLD-Tau, AD, and primary age-related tauopathy (PART) (see refs [78–81]). Perhaps surprisingly, Pastor et al. genotyped 152 PD and 52 AD patients and demonstrated that the tau A0/A0 allelic frequency was increased in PD (not considered a tauopathy) as well as AD [82]. Other groups confirmed that the H1 polymorphism was associated with PD risk [79, 83]. Collectively, these studies indicate that single nucleotide mutations, nucleotide duplications, and inversions in MAPT produce pleiotropic phenotypes, linking one gene to numerous downstream diseases. More specifically, MAPT demonstrates that a particular gene can harbor rare, high-penetrance, diseasedriving mutations that cause one set of diseases (PSP, CBD, argyrophilic grain disease, or globular glial tauopathy), or, alternatively, a separate variant in the same gene can contain relatively common, low-penetrance alleles (MAPT haplotypes) that are associated with altered risk for separate complex diseases (AD, PD, and PART). We recognize that it is debatable how one fits these observations into the concept of pleiotropy, however, we emphasize that these diseases differ importantly in terms of clinical and pathological features, and the genetic associations provide important clues of pathogenetic overlap.
As is the case for MAPT/Tau in neurodegenerative disease, evidence has emerged of genetic modifiers in TDP-43 proteinopathies [6, 7]. TDP-43 is the polypeptide product of the TARDBP gene, which is mutated in some cases of amyotrophic lateral sclerosis (ALS) [84, 85]. However, the far greater number of “sporadic” TDP-43 proteinopathy cases remain largely unexplained. In an attempt to find genetic risk factors for FTLD-TDP, Van Deerlin at al. studied patients with FTLD-TDP pathology and conducted a GWAS [86]. This led to the discovery of SNPs on chromosome 7p21.3 that were associated with FTLD-TDP risk. Nine SNPs in the original Van Deerlin study were found to span the transmembrane protein 106B (TMEM106B) gene locus. Of these gene variants, only one SNP (rs3173615) is located within the coding region of TMEM106B and results in a threonine to serine substitution at amino acid 185 (T185S). The missense variant affecting codon 185 is only two amino acids downstream from a critical site of N-glycosylation, residue 183 that is part of the N-X-T/S glycosylation consensus sequence [87]. The T185 isoform of TMEM106B produces ~2-fold increase in TMEM106B protein levels compared to the S185 isoform [88].
TMEM106B gene variants now have been associated with risk for numerous TDP-43 proteinopathies [86, 89–96]. However, the function of TMEM106B protein in the context of brain physiology has only partly been explored. TMEM106B is a 274 amino acid single pass, type-II transmembrane protein that localizes to cellular lysosomes [87]. It contains a highly glycosylated luminal domain and this posttranslational modification is required to transport TMEM106B from the endoplasmic reticulum to late endosomes and lysosomes. Interestingly when vacuolar H+-ATPases are inhibited, the levels of TMEM106B and GRN/progranulin (see below) significantly increase, suggesting a link between these two proteins [87]. Further, it has been demonstrated that TMEM106B has a role in regulation of lysosome synthesis, size, trafficking, and localization [87, 97–100]. Schwenk et al. [100] found that TMEM106B knockdown in primary neurons alters neuronal trafficking and blunts dendritic arborization, increasing retrograde transport of lysosomes in dendrites. This supports the finding by Stagi et al. [99] that demonstrated TMEM106B knockdown resulted in distribution of lysosomes in cell soma. Collectively, these studies provide strong evidence that TMEM106B participates in lysosomal function, and suggest that alterations in lysosomal cell physiology can be linked to neurodegeneration. Beyond FTDP-TDP, TMEM106B variants have been associated with hippocampal sclerosis (HS) pathology [92] (see below) and, in a recent study by Cherry and colleagues, TMEM106B allele rs3173615 SNP was linked to alteration of the CTE phenotype [101].
In addition to TMEM106B, another gene implicated in TDP-43 proteinopathy was granulin (GRN), as first became clear when germline mutations in GRN were discovered to cause FTLD-TDP [102–104]. Further, a common SNP, rs5848 (~40% of most human populations harbor one copy of this allele), located in the 3′-untranslated region (UTR) of GRN, was demonstrated to be associated with a ~3-fold increased risk of developing FTLD-TDP among individuals homozygous for the T-allele of rs5848 compared to C-allele [105]. Rademakers and colleagues demonstrated that the expression of GRN is partly regulated by a microRNA (miR-659) that binds to the mRNA’s 3′-UTR [105]. Moreover, GRN rs5848 allele is also associated with HS pathology and TDP-43 proteinopathy in non-FTLD aged persons [106–108]. Recent work suggests that the impact of GRN on multiple different neurodegenerative conditions may be more extensive than was previously thought [109]. Additional discussion of the genetic factors involved in TDP-43 proteinopathy in aged individuals is provided below.
As with MAPT, TMEM106B, and GRN, the APOE locus can influence more than one disease, again suggesting shared pathophysiology among neurodegenerative diseases. The APOE ε4 allele, which is strongly associated with deposition of Aβ peptide and AD pathogenesis [110, 111], has also been associated with Lewy body disease [112, 113], TDP-43 proteinopathy [112, 114], and HS pathology [115]. We note that there is an entirely different list of genes (e.g., Huntingtin, NPC1) in which mutations can lead to both Tau and TDP-43 pathologies [116–118]. Collectively, these results underscore that—reflecting differing upstream causes and downstream pathologies—individual gene changes can exert pleiotropic effects that influence misfolded proteins and help drive resulting degenerative brain changes. The mechanistic influences of each individual gene change are still being debated although the importance of genetics is beyond doubt.
Downstream effects: insights and controversies in an evolving research field
The field of ADRD research is characterized by both a rapid pace of discovery and also areas of controversy, some of which directly pertain to current ideas on Tau and TDP-43 proteinopathies. Although each novel scientific discovery builds on the prior corpus of knowledge, the resulting new paradigms can be challenging to reconcile with prior assumptions and hypotheses. One contributing factor that generates both insights and confusion is the constantly changing methods used for making pathologic observations—notably, the immunohistochemical detection of proteinopathies. Here we highlight several important perceptions that have affected the field of ADRD research: first, since AD itself (Tau tangles and amyloid plaques) was discovered relatively early as a driver of cognitive impairment in the elderly, this also led to an assumption that most or all of non-vascular dementia was secondary to AD; and, second, relatively nonspecific neuropathologic features including synapse loss and HS pathology were attributed to specific causes, and therefore considered to define specific diseases. New data contradict these assumptions.
The neuropathologic hallmarks of AD (AD neuropathologic changes, or ADNC) were discovered at the beginning of the 20th century using silver-based histologic stains [119]. Since then, the strong correlative impact of Tau tangles on cognition has been firmly established [4, 5, 13]. However, many CPC studies have revealed that the neuropathologic substrate(s) of amnestic cognitive impairment are not usually “pure” ADNC [2, 93, 120–122], and there is now ample evidence of common non-ADNC pathways to dementia [123, 124]. The reasons that this is important are several: (1) there are multiple common, clinically impactful diseases that affect aged persons’ brains and which need to be addressed in clinical and research contexts; (2) different proteinopathic changes (e.g., Tau, TDP-43, and α-synuclein proteinopathies) often co-occur in the same brain; (3) Tau and TDP-43 proteinopathies are both evidently the result of pathologic cascades that provide possible overlap in therapeutic targets for conditions that have different upstream causes. Some of the hypothesized pathogenetic mechanisms are described above; it is important to note the downstream effects were discovered during an era when the underlying mechanistic explanations were much less well understood.
We note two phenomena—synapse loss and HS pathology—that have been associated with dementia, and which relate to our central foci of Tau and TDP-43 proteinopathies. Synapse loss was considered for a time to be a relatively specific feature of AD, but this has been shown to be an incorrect hypothesis, for the simple reason that synapse loss characterizes numerous different neurodegenerative conditions. Multiple studies have reported ADassociated changes in specific synaptic proteins in different areas of the brain, and, these changes include both presynaptic (synaptophysin, synaptobrevin) and postsynaptic proteins (PSD-95, drebrin) [125]. The overall picture from these studies is that the loss of synapses and/or synaptic proteins is widespread in AD [125]. However, synapse loss is a nonspecific proxy for multiple diseases, a good indicator for neurodegeneration without specifically indicating AD or indeed any underlying ADRD subtype [125]. Recent CPC studies helped explain why there is an imperfect correlation between AD-type pathology per se (plaques and tangles) and cognitive status [124]. Thus, particularly in the aged brain where coexisting/mixed pathologies are common, non-specific markers of neurodegeneration (synapse loss, or neuronal loss) have stronger correlations with cognitive status than specific markers related to any one disease entity.
Another nonspecific histopathologic hallmark that has been associated with dementia, and that is directly related to TDP-43 proteinopathy, is HS pathology. This ill-defined pathologic endpoint has been associated with various underlying disease categories including epilepsy, hypoxia/ anoxia, infectious diseases, and various neurodegenerative conditions [52, 126–128]. The term HS is applied in clinical radiology, usually in relation to seizure disorders, and ~90% of PubMed hits for the search term “hippocampal sclerosis” are papers related to seizure disorders. From the neuropathologic standpoint, there is no specific terminology or classification system to characterize HS pathology in older individuals. In 1993, Dickson et al. [129] identified 13 elderly subjects with documented antemortem dementia and HS pathology, yet who lacked substantial ADNC. Larger case series of persons with HS and dementia were subsequently reported [130–134]. Following the 2006 discovery of TDP-43 in subjects with rare diseases (ALS and FTLD) [135], TDP-43 proteinopathy was also discovered to be a common pathologic features of brains of elderly persons lacking ALS or FTLD, including in individuals with comorbid HS pathology [136, 137]. These persons tended to be diagnosed clinically as AD [107, 138]. For the brain disease of which HS pathology was the initially discovered histopathologic indicator, it is now clear that TDP-43 proteinopathy is a more sensitive and specific feature than HS itself [48, 139–143]. A hypothetical pathologic cascade that may underlie the age-related TDP-43 proteinopathy is shown in Fig. 4, integrating findings from multiple prior studies [48, 92, 94, 144–148]. This proposed sequence illustrates how the upstream influences (ABCC9 gene variant and arteriolosclerosis) may occupy different pathogenetic nodes in comparison to the misfolding of TDP-43 that may be affected more directly by the genetic modifiers— TMEM106B and GRN genotypes. It is hoped that the various causes of the common TDP-43 proteinopathy/ies of aging will be better characterized in future studies.
Fig. 4.
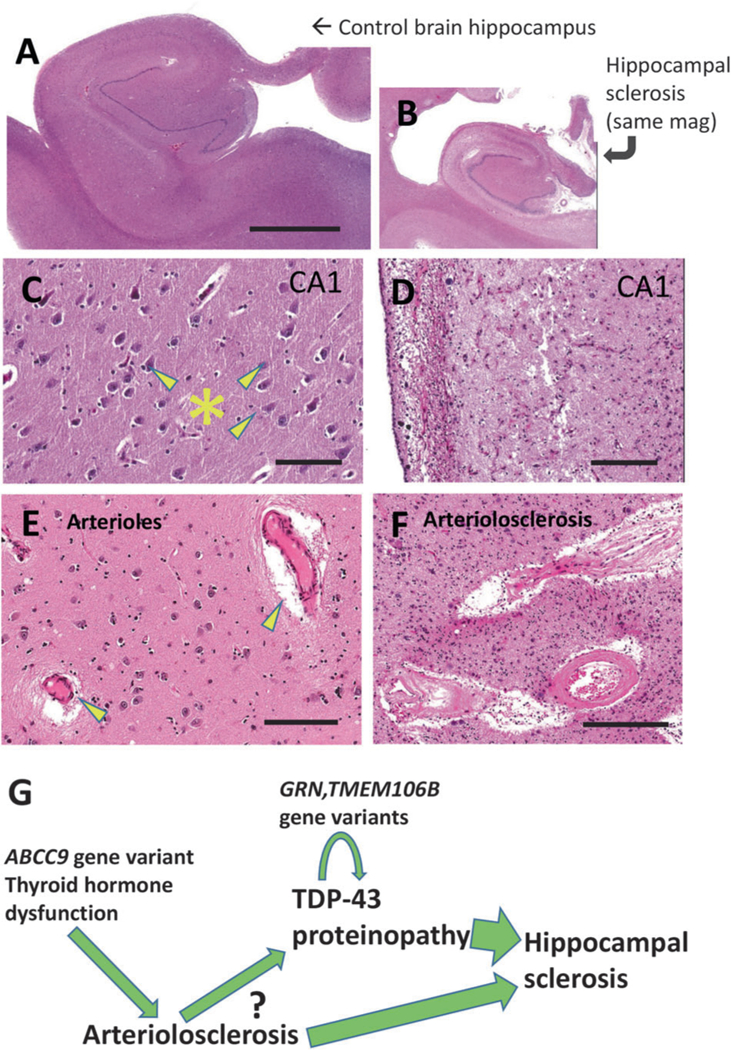
TDP-43 proteinopathy with hippocampal sclerosis (HS) pathology and arteriolosclerosis pathology provide the basis for a hypothetical pathologic cascade in aged human brains. Shown are aged control brain (a, c, e) and HS brain (b, d, f) for comparison’s sake, stained using H&E. This HS brain is the same one that is depicted in Fig. 3. Note that the photomicrographs in (a) and (b) are at the same magnification, indicating the amount of atrophy in the hippocampus shown in (b). Portions of the hippocampal CA1 sector are shown for comparison in panels c, d. Note that the control brain has large pyramidal neurons (arrowheads) and dense, intact eosinophilic neuropil (*). By contrast, the CA1 sector in the brain with HS pathology shows astrocytosis, dropout of neurons, and neuropil that is looser and rarefied. Brain arterioles that have histopathologic features within normal limits in aged brains (arrowheads in panel e) can be contrasted with arteriolosclerosis pathology (f) where the arteriolar walls are thickened and dysmorphic with eosinophilic material in the vessel wall that may impair cerebral blood flow. A hypothetical sequence, influenced by various factors, is shown (g) that incorporates findings from multiple prior studies [48, 92, 94, 144–148]. Upstream genetic risk factors may contribute to brain arteriolosclerosis in a manner that induces chronic stress in the brain, potentiating TDP-43 proteinopathy that also is influenced by additional genetic risk factors (e.g., TMEM106B, GRN). The question-mark conveys that the detailed mechanisms are as yet mostly unknown. The combination of ‘upstream’ stresses and TDP-43 proteinopathy may contribute to the cell loss and gliosis that manifests as HS pathology and contributes to the dementia syndrome. Scale bars = 6 mm (a, b), 120 μm (c, d), and 200 μm (f, g)
Summary
The aged brain is an extremely complex milieu and the pathologies of old age are correspondingly challenging to understand for researchers and clinicians alike. Neurodegenerative diseases evolve in a multi-step and multi-factorial manner, yet coherent paradigms are emerging (Fig. 5). Both Tau and TDP-43 proteinopathies are linked to multiple upstream influences, and both are connected with numerous deleterious downstream endpoints. Gene variants can be either disease-specific, or, they appear to be able to exert influence on the misfolding pathology itself rather than the upstream cause. Therefore, the gene variants may be associated with pleiotropic effects in multiple disease conditions. We conclude that the intriguing similarities between the pathogenetic cascades involved in Tau and TDP-43 proteinopathies may—when considered together—shed light on both, and help to guide researchers toward much-needed diagnostic and therapeutic strategies.
Fig. 5.
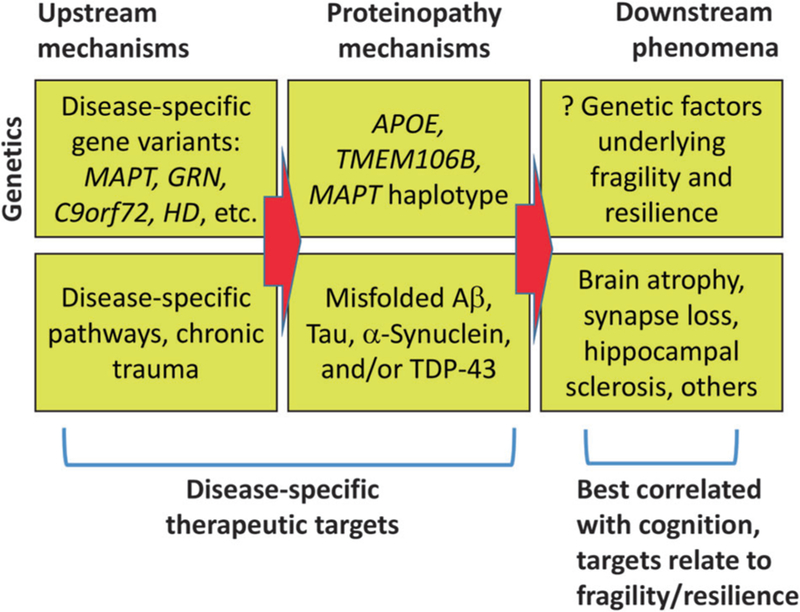
Schematic depiction of the genetic (top) and non-genetic (bottom) factors that are contributory to the complex, multi-stage pathologic cascades of neurodegenerative diseases. Different genetic factors may either contribute to “primary” condition-specific aspects of disease progression, or may induce pleiotropic effects that can influence (protect from, or exacerbate) misfolded proteins in multiple diseases, resulting in phenotypes that overlap in both clinical and pathological contexts. The aspects that may be most amenable to disease-specific therapeutic strategies are “upstream” influences, whereas the “downstream” pathologic manifestations correlate best with clinical symptoms
Acknowledgements
We are very grateful to the research volunteers, clinicians, and staff for their hard work. Grant support was received via National Institutes of Health grants P30 AG028303, R01 AG042475, R01 AG057187, and R21 AG050146.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Trojanowski JQ, Lee VM. “Fatal attractions” of proteins. A comprehensive hypothetical mechanism underlying Alzheimer’s disease and other neurodegenerative disorders. Ann N Y Acad Sci 2000;924:62–67. [DOI] [PubMed] [Google Scholar]
- 2.Rahimi J, Kovacs GG. Prevalence of mixed pathologies in the aging brain. Alzheimers Res Ther 2014;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson PT, Abner EL, Patel E, et al. The amygdala as a locus of pathologic misfolding in neurodegenerative diseases. J Neuropathol Exp Neurol 2018;77:2–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delacourte A Tau pathology and neurodegeneration: an obvious but misunderstood link. J Alzheimers Dis 2008;14:437–40. [DOI] [PubMed] [Google Scholar]
- 5.Abisambra JF, Scheff S. Brain injury in the context of tauopathies. J Alzheimers Dis 2014;40:495–518. [DOI] [PubMed] [Google Scholar]
- 6.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am J Neurodegener Dis 2013;2:145–75. [PMC free article] [PubMed] [Google Scholar]
- 7.Jain N, Chen-Plotkin AS. Genetic modifiers in neurodegeneration. Curr Genet Med Rep 2018;6:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost B, Ollesch J, Wille H, et al. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem 2009;284:3546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JL, Covell DJ, Daniels JP, et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell 2013;154:103–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porta S, Xu Y, Restrepo CR, et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat Commun 2018;9:4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furman JL, Vaquer-Alicea J, White CL 3rd, et al. Widespread tau seeding activity at early Braak stages. Acta Neuropathol 2017;133:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 2009;284:12845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams DR. Tauopathies: classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Intern Med J 2006;36:652–60. [DOI] [PubMed] [Google Scholar]
- 14.Takashima A Hyperphosphorylated tau is a cause of neuronal dysfunction in tauopathy. J Alzheimers Dis 2008;14:371–5. [DOI] [PubMed] [Google Scholar]
- 15.Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol 2013;12:609–22. [DOI] [PubMed] [Google Scholar]
- 16.Bieniek KF, Ross OA, Cormier KA, et al. Chronic traumatic encephalopathy pathology in a neurodegenerative disorders brain bank. Acta Neuropathol 2015. [DOI] [PMC free article] [PubMed]
- 17.Grinberg LT, Anghinah R, Nascimento CF, et al. Chronic traumatic encephalopathy presenting as Alzheimer’s disease in a retired soccer player. J Alzheimers Dis 2016;54:169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKee AC, Cairns NJ, Dickson DW, et al. The first NINDS/ NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol 2016;131:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mez J, Daneshvar DH, Kiernan PT, et al. Clinicopathological evaluation of chronic traumatic encephalopathy in players of american football. JAMA 2017;318:360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee AC, Gavett BE, Stern RA, et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J Neuropathol Exp Neurol 2010;69:918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol 2014;15:384–96. [DOI] [PubMed] [Google Scholar]
- 22.Tzotzos S, Doig AJ. Amyloidogenic sequences in native protein structures. Protein Sci 2010;19:327–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guijarro JI, Sunde M, Jones JA, et al. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci USA 1998;95:4224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirschner DA, Inouye H, Duffy LK, et al. Synthetic peptide homologous to beta protein from Alzheimer disease forms amyloid-like fibrils in vitro. Proc Natl Acad Sci USA 1987;84:6953–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 2009;16:574–81. [DOI] [PubMed] [Google Scholar]
- 26.Brehme M, Voisine C, Rolland T, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep 2014;9:1135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jellinger KA. Recent advances in our understanding of neuro-degeneration. J Neural Transm 2009;116:1111–62. [DOI] [PubMed] [Google Scholar]
- 28.Joyce PI, Fratta P, Fisher EM, et al. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mamm Genome 2011;22:420–48. [DOI] [PubMed] [Google Scholar]
- 29.Iguchi Y, Katsuno M, Takagi S, et al. Oxidative stress induced by glutathione depletion reproduces pathological modifications of TDP-43 linked to TDP-43 proteinopathies. Neurobiol Dis 2012;45:862–70. [DOI] [PubMed] [Google Scholar]
- 30.Butterfield DA, Perluigi M, Sultana R. Oxidative stress in Alzheimer’s disease brain: new insights from redox proteomics. Eur J Pharmacol 2006;545:39–50. [DOI] [PubMed] [Google Scholar]
- 31.Calabrese V, Guagliano E, Sapienza M, et al. Redox regulation of cellular stress response in neurodegenerative disorders. Ital J Biochem 2006;55:263–82. [PubMed] [Google Scholar]
- 32.Hasegawa M, Arai T, Nonaka T, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol 2008;64:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampathu DM, Neumann M, Kwong LK, et al. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol 2006;169:1343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inukai Y, Nonaka T, Arai T, et al. Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett 2008;582:2899–904. [DOI] [PubMed] [Google Scholar]
- 35.Kametani F, Obi T, Shishido T, et al. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci Rep 2016;6:23281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann M, Kwong LK, Lee EB, et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol 2009;117:137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moujalled D, James JL, Parker SJ, et al. Kinase inhibitor screening identifies cyclin-dependent kinases and glycogen synthase kinase 3 as potential modulators of TDP-43 cytosolic accumulation during cell stress. PLoS ONE 2013;8:e67433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furukawa Y, Kaneko K, Nukina N. Molecular properties of TAR DNA binding protein-43 fragments are dependent upon its cleavage site. Biochim Biophys Acta 2011;1812:1577–83. [DOI] [PubMed] [Google Scholar]
- 39.Zhang YJ, Gendron TF, Xu YF, et al. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener 2010;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Igaz LM, Kwong LK, Chen-Plotkin A, et al. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J Biol Chem 2009;284:8516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barmada SJ, Serio A, Arjun A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 2014;10:677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang IF, Guo BS, Liu YC, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinpathies of the TAR DNA binding protein 43. Proc Natl Acad Sci USA 2012;109:15024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghavami S, Shojaei S, Yeganeh B, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol 2014;112:24–49. [DOI] [PubMed] [Google Scholar]
- 44.Chang MC, Srinivasan K, Friedman BA, et al. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med 2017;214:2611–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spires-Jones TL, Attems J, Thal DR. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol 2017;134:187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy J Alzheimer’s disease: the amyloid cascade hypothesis: an update and reappraisal. J Alzheimers Dis 2006;9:151–3. (3 Suppl) [DOI] [PubMed] [Google Scholar]
- 47.Irwin DJ, Lee VM, Trojanowski JQ. Parkinson’s disease dementia: convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat Rev Neurosci 2013;14:626–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson PT, Trojanowski JQ, Abner EL, et al. “New old pathologies”: AD, PART, and cerebral age-related TDP-43 with sclerosis (CARTS). J Neuropathol Exp Neurol 2016;75:482–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold SJ, Dugger BN, Beach TG. TDP-43 deposition in prospectively followed, cognitively normal elderly individuals: correlation with argyrophilic grains but not other concomitant pathologies. Acta Neuropathol 2013;126:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davidson Y, Amin H, Kelley T, et al. TDP-43 in ubiquitinated inclusions in the inferior olives in frontotemporal lobar degeneration and in other neurodegenerative diseases: a degenerative process distinct from normal ageing. Acta Neuropathol 2009;118:359–69. [DOI] [PubMed] [Google Scholar]
- 51.Cagnin A, Mariotto S, Fiorini M, et al. Microglial and neuronal TDP-43 pathology in anti-IgLON5-related tauopathy. J Alzheimers Dis 2017;59:13–20. [DOI] [PubMed] [Google Scholar]
- 52.Yokota O, Davidson Y, Bigio EH, et al. Phosphorylated TDP-43 pathology and hippocampal sclerosis in progressive supranuclear palsy. Acta Neuropathol 2010;120:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kertesz A, Finger E, Murrell J, et al. Progressive supranuclear palsy in a family with TDP-43 pathology. Neurocase 2015;21:178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kouri N, Oshima K, Takahashi M, et al. Corticobasal degeneration with olivopontocerebellar atrophy and TDP-43 pathology: an unusual clinicopathologic variant of CBD. Acta Neuropathol 2013;125:741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Higashi S, Iseki E, Yamamoto R, et al. Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res 2007;1184:284–94. [DOI] [PubMed] [Google Scholar]
- 56.Smith VD, Bachstetter AD, Ighodaro E, et al. Overlapping but distinct TDP-43 and tau pathologic patterns in aged hippocampi. Brain Pathol 2017;28:264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker AK, Daniels CM, Goldman JE, et al. Astrocytic TDP-43 pathology in Alexander disease. J Neurosci 2014;34:6448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishima T, Koga S, Lin WL, et al. Perry syndrome: a distinctive type of TDP-43 proteinopathy. J Neuropathol Exp Neurol 2017;76:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakurai A, Makioka K, Fukuda T, et al. Accumulation of phosphorylated TDP-43 in the CNS of a patient with Cockayne syndrome. Neuropathology 2013;33:673–7. [DOI] [PubMed] [Google Scholar]
- 60.Haraguchi T, Terada S, Ishizu H, et al. Coexistence of TDP-43 and tau pathology in neurodegeneration with brain iron accumulation type 1 (NBIA-1, formerly Hallervorden-Spatz syndrome). Neuropathology 2011;31:531–9. [DOI] [PubMed] [Google Scholar]
- 61.Weihl CC, Temiz P, Miller SE, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry 2008;79:1186–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017;45:D896–D901. (D1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 2011;89:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solovieff N, Cotsapas C, Lee PH, et al. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 2013;14:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chesmore K, Bartlett J, Williams SM. The ubiquity of pleiotropy in human disease. Hum Genet 2018;137:39–44. [DOI] [PubMed] [Google Scholar]
- 66.Lu Q, Li B, Ou D, et al. A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am J Hum Genet 2017;101:939–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goedert M Tau protein and neurodegeneration. Semin Cell Dev Biol 2004;15:45–49. [DOI] [PubMed] [Google Scholar]
- 69.Spillantini MG, Murrell JR, Goedert M, et al. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA 1998;95:7737–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poorkaj P, Bird TD, Wijsman E, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 1998;43:815–25. [DOI] [PubMed] [Google Scholar]
- 71.Baker M, Kwok JB, Kucera S, et al. Localization of frontotemporal dementia with parkinsonism in an Australian kindred to chromosome 17q21–22. Ann Neurol 1997;42:794–8. [DOI] [PubMed] [Google Scholar]
- 72.Ingram EM, Spillantini MG. Tau gene mutations: dissecting the pathogenesis of FTDP-17. Trends Mol Med 2002;8:555–62. [DOI] [PubMed] [Google Scholar]
- 73.Goedert M, Spillantini MG. Tau mutations in frontotemporal dementia FTDP-17 and their relevance for Alzheimer’s disease. Biochim Biophys Acta 2000;1502:110–21. [DOI] [PubMed] [Google Scholar]
- 74.Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet 2005;37:129–37. [DOI] [PubMed] [Google Scholar]
- 75.Conrad C, Amano N, Andreadis A, et al. Differences in a dinucleotide repeat polymorphism in the tau gene between Caucasian and Japanese populations: implication for progressive supranuclear palsy. Neurosci Lett 1998;250:135–7. [DOI] [PubMed] [Google Scholar]
- 76.Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 1999;8:711–5. [DOI] [PubMed] [Google Scholar]
- 77.Conrad C, Andreadis A, Trojanowski JQ, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol 1997;41:277–81. [DOI] [PubMed] [Google Scholar]
- 78.Zhang CC, Zhu JX, Wan Y, et al. Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget 2017;8:44994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou F, Wang D. The associations between the MAPT polymorphisms and Alzheimer’s disease risk: a meta-analysis. Oncotarget 2017;8:43506–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jun G, Ibrahim-Verbaas CA, Vronskaya M, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 2015. [DOI] [PMC free article] [PubMed]
- 81.Santa-Maria I, Haggiagi A, Liu X, et al. The MAPT H1 haplotype is associated with tangle-predominant dementia. Acta Neuropathol 2012;124:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pastor P, Ezquerra M, Munoz E, et al. Significant association between the tau gene A0/A0 genotype and Parkinson’s disease. Ann Neurol 2000;47:242–5. [PubMed] [Google Scholar]
- 83.Zabetian CP, Hutter CM, Factor SA, et al. Association analysis of MAPT H1 haplotype and subhaplotypes in Parkinson’s disease. Ann Neurol 2007;62:137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008;319:1668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol 2008;7:409–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 2010;42:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lang CM, Fellerer K, Schwenk BM, et al. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 2012;287:19355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nicholson AM, Finch NA, Wojtas A, et al. TMEM106B p. T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem 2013;126:781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vass R, Ashbridge E, Geser F, et al. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol 2010. [DOI] [PMC free article] [PubMed]
- 90.Cruchaga C, Graff C, Chiang HH, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol 2011;68:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen-Plotkin AS, Unger TL, Gallagher MD, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci 2012;32:11213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rutherford NJ, Carrasquillo MM, Li M, et al. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology 2012;79:717–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murray ME, Cannon A, Graff-Radford NR, et al. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol 2014;128:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nelson PT, Estus S, Abner EL, et al. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol 2014;127:825–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu L, De Jager PL, Yang J, et al. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology 2015;84:927–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pottier C, Zhou X, Perkerson RB 3rd, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genomewide association study. Lancet Neurol 2018;17:548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein ZA, Takahashi H, Ma M, et al. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 2017;95:281–96 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou X, Sun L, Brady OA, et al. Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol Commun 2017;5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stagi M, Klein ZA, Gould TJ, et al. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci 2014;61:226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwenk BM, Lang CM, Hogl S, et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J 2014;33:450–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cherry JD, Mez J, Crary JF, et al. Variation in TMEM106B in chronic traumatic encephalopathy. Acta Neuropathol Commun 2018;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu CE, Bird TD, Bekris LM, et al. The spectrum of mutations in progranulin: a collaborative study screening 545 cases of neurodegeneration. Arch Neurol 2010;67:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boeve BF, Baker M, Dickson DW, et al. Frontotemporal dementia and parkinsonism associated with the IVS1 + 1G- > A mutation in progranulin: a clinicopathologic study. Brain 2006;129:3103–14. (Pt 11) [DOI] [PubMed] [Google Scholar]
- 104.Gass J, Cannon A, Mackenzie IR, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 2006;15:2988–3001. [DOI] [PubMed] [Google Scholar]
- 105.Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet 2008;17:3631–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dickson DW, Baker M, Rademakers R. Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener Dis 2010;7:170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pao WC, Dickson DW, Crook JE, et al. Hippocampal sclerosis in the elderly: genetic and pathologic findings, some mimicking Alzheimer disease clinically. Alzheimer Dis Assoc Disord 2011;25:364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nelson PT, Wang WX, Partch AB, et al. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J Neuropathol Exp Neurol 2015;74:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swarup V, Hinz FI, Rexach JE, et al. Identification of evolutionarily conserved gene networks mediating neurodegenerative dementia. Nat Med 2018. [DOI] [PMC free article] [PubMed]
- 110.Schmechel DE, Saunders AM, Strittmatter WJ, et al. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA 1993;90:9649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 1993;90:1977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang HS, Yu L, White CC, et al. Evaluation of TDP-43 proteinopathy and hippocampal sclerosis in relation to APOE epsilon4 haplotype status: a community-based cohort study. Lancet Neurol 2018;17:773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinson JL, Lee EB, Xie SX, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 2018;141:2181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wennberg AM, Tosakulwong N, Lesnick TG, et al. Association of apolipoprotein E epsilon4 with transactive response DNA-binding protein 43. JAMA Neurol 2018;75:1347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farfel JM, Yu L, Buchman AS, et al. Relation of genomic variants for Alzheimer disease dementia to common neuropathologies. Neurology 2016;87:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vuono R, Winder-Rhodes S, de Silva R, et al. The role of tau in the pathological process and clinical expression of Huntington’s disease. Brain 2015;138:1907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.St-Amour I, Turgeon A, Goupil C, et al. Co-occurrence of mixed proteinopathies in late-stage Huntington’s disease. Acta Neuropathol 2018;135:249–65. [DOI] [PubMed] [Google Scholar]
- 118.Dardis A, Zampieri S, Canterini S, et al. Altered localization and functionality of TAR DNA binding protein 43 (TDP-43) in niemann-pick disease type C. Acta Neuropathol Commun 2016;4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alzheimer A Uber eigenartige Krankheitsfälle des spateren Alters. Z Ges Neurol 1911;4:356–85. [Google Scholar]
- 120.Schneider JA, Arvanitakis Z, Bang W, et al. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology 2007;69:2197–204. [DOI] [PubMed] [Google Scholar]
- 121.Korczyn AD. Mixed dementia—the most common cause of dementia. Ann N Y Acad Sci 2002;977:129–34. [DOI] [PubMed] [Google Scholar]
- 122.Jellinger KA, Attems J. Neuropathological evaluation of mixed dementia. J Neurol Sci 2007;257:80–87. [DOI] [PubMed] [Google Scholar]
- 123.Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol 2009;68:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nelson PT, Alafuzoff I, Bigio EH, et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol 2012;71:362–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scheff SW, Neltner JH, Nelson PT. Is synaptic loss a unique hallmark of Alzheimer’s disease? Biochem Pharmacol 2014;88:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thom M, Eriksson S, Martinian L, et al. Temporal lobe sclerosis associated with hippocampal sclerosis in temporal lobe epilepsy: neuropathological features. J Neuropathol Exp Neurol 2009;68:928–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sen A, Dugan P, Perucca P, et al. The phenotype of bilateral hippocampal sclerosis and its management in “real life” clinical settings. Epilepsia 2018;59:1410–20. [DOI] [PubMed] [Google Scholar]
- 128.Malek-Ahmadi M, Kahlon V, Adler CH, et al. Prevalence of Hippocampal Sclerosis in a Clinicopathologically Characterized Cohort. Clin Exp Med Sci 2013;1:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dickson DW, Davies P, Bevona C, et al. Hippocampal sclerosis: a common pathological feature of dementia in very old (> or = 80 years of age) humans. Acta Neuropathol 1994;88:212–21. [DOI] [PubMed] [Google Scholar]
- 130.Leverenz JB, Agustin CM, Tsuang D, et al. Clinical and neuropathological characteristics of hippocampal sclerosis: a community-based study. Arch Neurol 2002;59:1099–106. [DOI] [PubMed] [Google Scholar]
- 131.Attems J, Jellinger KA. Hippocampal sclerosis in Alzheimer disease and other dementias. Neurology 2006;66:775. [DOI] [PubMed] [Google Scholar]
- 132.Zarow C, Vinters HV, Ellis WG, et al. Correlates of hippocampal neuron number in Alzheimer’s disease and ischemic vascular dementia. Ann Neurol 2005;57:896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brayne C, Richardson K, Matthews FE, et al. Neuropathological correlates of dementia in over-80-year-old brain donors from the population-based Cambridge city over-75s cohort (CC75C) study. J Alzheimers Dis 2009;18:645–58. [DOI] [PubMed] [Google Scholar]
- 134.White L, Petrovitch H, Hardman J, et al. Cerebrovascular pathology and dementia in autopsied Honolulu-Asia Aging Study participants. Ann N Y Acad Sci 2002;977:9–23. [DOI] [PubMed] [Google Scholar]
- 135.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006;314:130–3. [DOI] [PubMed] [Google Scholar]
- 136.Amador-Ortiz C, Lin WL, Ahmed Z, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol 2007;61:435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Amador-Ortiz C, Ahmed Z, Zehr C, et al. Hippocampal sclerosis dementia differs from hippocampal sclerosis in frontal lobe degeneration. Acta Neuropathol (Berl) 2007;113:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brenowitz WD, Monsell SE, Schmitt FA, et al. Hippocampal sclerosis of aging is a key Alzheimer’s disease mimic: clinical-pathologic correlations and comparisons with both alzheimer’s disease and non-tauopathic frontotemporal lobar degeneration. J Alzheimers Dis 2014;39:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nelson PT, Schmitt FA, Lin Y, et al. Hippocampal sclerosis in advanced age: clinical and pathological features. Brain 2011;134 (Pt 5):1506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nelson PT, Smith CD, Abner EL, et al. Hippocampal sclerosis of aging, a prevalent and high-morbidity brain disease. Acta Neuropathol 2013;126:161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cykowski MD, Takei H, Van Eldik LJ, et al. Hippocampal sclerosis but not normal aging or Alzheimer disease is associated with TDP-43 pathology in the basal forebrain of aged persons. J Neuropathol Exp Neurol 2016;75:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cykowski MD, Powell SZ, Schulz PE, et al. Hippocampal sclerosis in older patients: practical examples and guidance with a focus on cerebral age-related TDP-43 with sclerosis. Arch Pathol Lab Med 2017;141:1113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nag S, Yu L, Boyle PA, et al. TDP-43 pathology in anterior temporal pole cortex in aging and Alzheimer’s disease. Acta Neuropathol Commun 2018;6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nelson PT, Jicha GA, Wang WX, et al. ABCC9/SUR2 in the brain: implications for hippocampal sclerosis of aging and a potential therapeutic target. Ageing Res Rev 2015;24(Pt B):111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ighodaro ET, Abner EL, Fardo DW, et al. Risk factors and global cognitive status related to brain arteriolosclerosis in elderly individuals. J Cereb Blood Flow Metab 2016. [DOI] [PMC free article] [PubMed]
- 146.Nelson PT, Katsumata Y, Nho K, et al. Genomics and CSF analyses implicate thyroid hormone in hippocampal sclerosis of aging. Acta Neuropathol 2016;132:841–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Trieu T, Sajjadi SA, Kawas CH, et al. Risk factors of hippocampal sclerosis in the oldest old: The 90 + Study. Neurology 2018;91:e1788–e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Katsumata Y, Nelson PT, Ellingson SR, et al. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol Aging 2017;53:193.e17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Davidson YS, Raby S, Foulds PG, et al. TDP-43 pathological changes in early onset familial and sporadic Alzheimer’s disease, late onset Alzheimer’s disease and Down’s syndrome: association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol 2011;122:703–13. [DOI] [PubMed] [Google Scholar]
- 150.Josephs KA, Whitwell JL, Knopman DS, et al. Abnormal TDP-43 immunoreactivity in AD modifies clinicopathologic and radiologic phenotype. Neurology 2008;70(19 pt 2):1850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilson AC, Dugger BN, Dickson DW, et al. TDP-43 in aging and Alzheimer’s disease—a review. Int J Clin Exp Pathol 2011;4:147–55. [PMC free article] [PubMed] [Google Scholar]
- 152.Kaivorinne AL, Kruger J, Kuivaniemi K, et al. Role of MAPT mutations and haplotype in frontotemporal lobar degeneration in Northern Finland. BMC Neurol 2008;8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clark LN, Liu X, Parmalee NL, et al. The microtubule associated protein tau H1 haplotype and risk of essential tremor. Eur J Neurol 2014;21:1044–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tagliavini F, Giaccone G, Prelli F, et al. A68 is a component of paired helical filaments of Gerstmann-Straussler-Scheinker disease, Indiana kindred. Brain Res 1993;616:325–9. [DOI] [PubMed] [Google Scholar]
- 155.Kovacs GG, Pittman A, Revesz T, et al. MAPT S305I mutation: implications for argyrophilic grain disease. Acta Neuropathol 2008;116:103–18. [DOI] [PubMed] [Google Scholar]
- 156.Togo T, Sahara N, Yen SH, et al. Argyrophilic grain disease is a sporadic 4-repeat tauopathy. J Neuropathol Exp Neurol 2002;61:547–56. [DOI] [PubMed] [Google Scholar]
- 157.Wakabayashi K, Fukushima T, Koide R, et al. Juvenile-onset generalized neuroaxonal dystrophy (Hallervorden-Spatz disease) with diffuse neurofibrillary and lewy body pathology. Acta Neuropathol 2000;99:331–6. [DOI] [PubMed] [Google Scholar]
- 158.Ahmed Z, Bigio EH, Budka H, et al. Globular glial tauopathies (GGT): consensus recommendations. Acta Neuropathol 2013;126:537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tacik P, DeTure M, Lin WL, et al. A novel tau mutation, p. K317N, causes globular glial tauopathy. Acta Neuropathol 2015;130:199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Crary JF, Trojanowski JQ, Schneider JA, et al. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol 2014;128:755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kovacs GG, Ferrer I, Grinberg LT, et al. Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol 2016;131:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cairns NJ, Bigio EH, Mackenzie IR, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol (Berl) 2007;114:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bieniek KF, Murray ME, Rutherford NJ, et al. Tau pathology in frontotemporal lobar degeneration with C9ORF72 hexanucleotide repeat expansion. Acta Neuropathol 2013;125:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wong KT, Allen IV, McQuaid S, et al. An immunohistochemical study of neurofibrillary tangle formation in post-encephalitic Parkinsonism. Clin Neuropathol 1996;15:22–25. [PubMed] [Google Scholar]
- 165.Ling H, Holton JL, Lees AJ, et al. TDP-43 pathology is present in most post-encephalitic parkinsonism brains. Neuropathol Appl Neurobiol 2014;40:654–7. [DOI] [PubMed] [Google Scholar]
- 166.Paula-Barbosa MM, Brito R, Silva CA, et al. Neurofibrillary changes in the cerebral cortex of a patient with subacute sclerosing panencephalitis (SSPE). Acta Neuropathol 1979;48:157–60. [DOI] [PubMed] [Google Scholar]
- 167.Gelpi E, Hoftberger R, Graus F, et al. Neuropathological criteria of anti-IgLON5-related tauopathy. Acta Neuropathol 2016;132:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lippa CF, Rosso AL, Stutzbach LD, et al. Transactive response DNA-binding protein 43 burden in familial Alzheimer disease and Down syndrome. Arch Neurol 2009;66:1483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vermersch P, Sergeant N, Ruchoux MM, et al. Specific tau variants in the brains of patients with myotonic dystrophy. Neurology 1996;47:711–7. [DOI] [PubMed] [Google Scholar]
- 170.Wisniewski K, Jervis GA, Moretz RC, et al. Alzheimer neurofibrillary tangles in diseases other than senile and presenile dementia. Ann Neurol 1979;5:288–94. [DOI] [PubMed] [Google Scholar]
- 171.Gotzl JK, Mori K, Damme M, et al. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 2014;127:845–60. [DOI] [PubMed] [Google Scholar]
- 172.Auer IA, Schmidt ML, Lee VM, et al. Paired helical filament tau (PHFtau) in Niemann-Pick type C disease is similar to PHFtau in Alzheimer’s disease. Acta Neuropathol 1995;90:547–51. [DOI] [PubMed] [Google Scholar]
- 173.Honda H, Sasagasako N, Shen C, et al. DCTN1 F52L mutation case of Perry syndrome with progressive supranuclear palsy-like tauopathy. Park Relat Disord 2018;51:105–10. [DOI] [PubMed] [Google Scholar]
- 174.Brat DJ, Gearing M, Goldthwaite PT, et al. Tau-associated neuropathology in ganglion cell tumours increases with patient age but appears unrelated to ApoE genotype. Neuropathol Appl Neurobiol 2001;27:197–205. [DOI] [PubMed] [Google Scholar]
- 175.Lee EB, Lee VM, Trojanowski JQ, et al. TDP-43 immunoreactivity in anoxic, ischemic and neoplastic lesions of the central nervous system. Acta Neuropathol 2008;115:305–11. [DOI] [PubMed] [Google Scholar]
- 176.Niklowitz WJ, Mandybur TI. Neurofibrillary changes following childhood lead encephalopathy. J Neuropathol Exp Neurol 1975;34:445–55. [DOI] [PubMed] [Google Scholar]
- 177.Smith DH, Johnson VE, Stewart W. Chronic neuropathologies of single and repetitive TBI: substrates of dementia? Nat Rev Neurol 2013;9:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Johnson VE, Stewart W, Trojanowski JQ, et al. Acute and chronically increased immunoreactivity to phosphorylation-independent but not pathological TDP-43 after a single traumatic brain injury in humans. Acta Neuropathol 2011;122:715–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Daneshvar DH, Goldstein LE, Kiernan PT, et al. Post-traumatic neurodegeneration and chronic traumatic encephalopathy. Mol Cell Neurosci 2015;66(Pt B):81–90. [DOI] [PubMed] [Google Scholar]
- 180.Askanas V, Engel WK. Sporadic inclusion-body myositis: conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-beta42 oligomers and phosphorylated tau. Presse Med 2011;40 (4 Pt 2):e219–235. [DOI] [PubMed] [Google Scholar]
- 181.Dickson DW, Kouri N, Murray ME, et al. Neuropathology of frontotemporal lobar degeneration-tau (FTLD-tau). J Mol Neurosci 2011;45:384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kwok JB, Teber ET, Loy C, et al. Tau haplotypes regulate transcription and are associated with Parkinson’s disease. Ann Neurol 2004;55:329–34. [DOI] [PubMed] [Google Scholar]
- 183.Elbaz A, Ross OA, Ioannidis JP, et al. Independent and joint effects of the MAPT and SNCA genes in Parkinson disease. Ann Neurol 2011;69:778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tacik P, Sanchez-Contreras M, DeTure M, et al. Clinicopathologic heterogeneity in frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) due to microtubule-associated protein tau (MAPT) p.P301L mutation, including a patient with globular glial tauopathy. Neuropathol Appl Neurobiol 2017;43:200–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dickson DW, Rademakers R, Nicholson AM, et al. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology 2015;85:1354–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rhinn H, Abeliovich A. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst 2017;4:404–15. e405 [DOI] [PubMed] [Google Scholar]
- 187.Ren Y, van Blitterswijk M, Allen M, et al. TMEM106B haplotypes have distinct gene expression patterns in aged brain. Mol Neurodegener 2018;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Masellis M, Momeni P, Meschino W, et al. Novel splicing mutation in the progranulin gene causing familial corticobasal syndrome. Brain 2006;129(Pt 11):3115–23. [DOI] [PubMed] [Google Scholar]
- 189.Snowden JS, Pickering-Brown SM, Mackenzie IR, et al. Progranulin gene mutations associated with frontotemporal dementia and progressive non-fluent aphasia. Brain 2006;129 (Pt 11):3091–102. [DOI] [PubMed] [Google Scholar]
- 190.Geier EG, Bourdenx M, Storm NJ, et al. Rare variants in the neuronal ceroid lipofuscinosis gene MFSD8 are candidate risk factors for frontotemporal dementia. Acta Neuropathol 2018. [DOI] [PMC free article] [PubMed]
- 191.Smith KR, Damiano J, Franceschetti S, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 2012;90:1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993;261:921–3. [DOI] [PubMed] [Google Scholar]
- 193.Galasko D, Saitoh T, Xia Y, et al. The apolipoprotein E allele epsilon 4 is overrepresented in patients with the Lewy body variant of Alzheimer’s disease. Neurology 1994;44:1950–1. [DOI] [PubMed] [Google Scholar]
- 194.Tsuang DW, Wilson RK, Lopez OL, et al. Genetic association between the APOE*4 allele and Lewy bodies in Alzheimer disease. Neurology 2005;64:509–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jicha GA, Parisi JE, Dickson DW, et al. Age and apoE associations with complex pathologic features in Alzheimer’s disease. J Neurol Sci 2008;273:34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nho K, Saykin AJ, Disease Neuroimaging Initiative As, et al. Hippocampal sclerosis of aging, a common Alzheimer’s disease ‘Mimic’: risk genotypes are associated with brain atrophy outside the temporal lobe. J Alzheimers Dis 2016;52:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Grange DK, Nichols CG, Singh GK. Cantu syndrome and related disorders. In: Pagon RA, Adam MP, Ardinger HH, et al. , editors. Seattle: GeneReviews(R); 1993. [Google Scholar]


