Abstract
Clonal hematopoiesis (CH) is a common aging-associated condition with increased risk of hematologic malignancy. Knowledge of the mechanisms driving evolution from CH to overt malignancy has been hampered by a lack of in vivo models that orthogonally activate mutant alleles. Here, we develop independently regulatable mutations in DNA methyltransferase 3A (Dnmt3a) and nucleophosmin 1 (Npm1), observed in human CH and AML, respectively. We find Dnmt3a mutation expands hematopoietic stem and multipotent progenitor cells (HSC/MPPs), modeling CH. Induction of mutant Npm1 after development of Dnmt3a-mutant CH causes progression to myeloproliferative disorder (MPD), and more aggressive MPD is observed with longer latency between mutations. MPDs uniformly progress to acute myeloid leukemia (AML) following transplant, accompanied by a decrease in HSC/MPPs and an increase in myeloid-restricted progenitors, the latter of which propagate AML in tertiary recipient mice. At a molecular level, progression of CH to MPD is accompanied by selection for mutations activating Ras/Raf/MAPK signaling. Progression to AML is characterized by additional oncogenic signaling mutations (Ptpn11, Pik3r1, Flt3) and/or mutations in epigenetic regulators (Hdac1, Idh1, Arid1a). Together, our study demonstrates that Npm1 mutation drives evolution of Dnmt3a-mutant CH to AML and rate of disease progression is accelerated with longer latency of CH.
Subject terms: Cancer models, Haematopoietic stem cells, Cancer stem cells, Acute myeloid leukaemia, Myeloproliferative disease
Introduction
Clonal hematopoiesis (CH) occurs when stem and progenitor cell clones gain one or more somatic mutations that confer a competitive advantage [1]. This can be observed in patients by an increase in the variant allele frequency (VAF) of somatic mutations in the blood or bone marrow (BM) [2]. There is a vast spectrum of outcomes in individuals with CH; in many cases, CH causes little to no clinically relevant hematopoietic phenotype. At the other extreme, CH cases represent a permissive state for development of BM failure syndromes including myelodysplastic syndrome (MDS) [3, 4], myeloproliferation including myeloproliferative disorder (MPD) [5], or leukemias including acute myeloid leukemia (AML), the last of which is accompanied by additional somatic mutations [6–8]. Pressing clinical issues include how to identify individuals with CH for which this condition should be considered a medically important event and how to develop therapies that might prevent onset of hematologic malignancies. To answer these questions, we need to understand the factors (specific somatic mutations, genetic background of the individual, environmental factors, etc.) that cause evolution of CH to hematologic malignancies. A current barrier is the lack of available animal models that undergo the evolution of CH to hematologic malignancies in a native in vivo environment.
The most common somatic mutations present in CH are in genes associated with epigenetic modifications, including the DNA methyltransferase DNMT3A, and splicing factors. Several mouse models of DNMT3A-driven CH have been generated by conditional knockout [9], mutant allele knockin [10, 11], or mutant allele overexpression [12]. Further, several studies have identified cooperating mutations that can act synergistically with Dnmt3a knockout or mutation to cause leukemia, such as Flt3ITD [13, 14], Flt3ITD and Npm1c [11], cKit [15], and KrasG12D [16]. However, existing combinatorial mutation models are limited by the use of germline alleles with conditional alleles (thereby not modeling the known somatic nature of these mutations), multiple conditional alleles that are induced simultaneously by Cre recombination, or conditional alleles combined with ex vivo transduction/overexpression of cooperating mutations. Thus, based on the design of previous mouse models, none are able to evaluate the evolution of CH to hematologic malignancies within a native in vivo environment.
To overcome these limitations, we have developed an inducible, dual-recombinase system combining flippase-FRT (Flp-FRT) and Cre-loxP recombination technologies [17, 18] to improve genetically engineered mouse models of CH and to allow study of the evolution of CH to hematopoietic malignancy. This dual-recombinase system allows for sequential induction of a Dnmt3a hotspot mutation in R878H (Dnmt3aR878H), replicating human R882H [1], and the recurrent hotspot mutation in the multifunctional nuclear protein Npm1 (Npm1c type A; Npm1cA) [19, 20]. Previously, concurrent expression of Dnmt3aR878H and Npm1cA mutations was not found to cause a lethal malignancy within a 45-week observation period [11]; however, it remains unknown whether malignancy would have occurred over a longer timeframe or with a latency period between expression of these two mutations. Therefore, we have utilized our model to examine the cellular and molecular alterations during evolution of CH to hematologic malignancy within a native in vivo environment.
Materials and methods
Genetic engineering of Dnmt3afl-R878H/+ and Npm1frt-cA/+ mice
Targeting vectors mDnmt3a_LSL_exon23_bGHpA_pBlight and mNpm1_FS12_exon11_FSF_pBlight were generated by in vitro synthesis (GenScript). The Dnmt3afl-R878H/+ construct design placed wild-type exon 23 in a loxP-flanked STOP cassette (4xSV40pA) into the intron between exons 22 and 23, and the 3′ homology arm included the 2633 G>A in exon 23 to encode the R878H mutation. The Npm1frt-cA/+ construct design placed wild-type exon 11 in an FRT-flanked STOP cassette (4xSV40pA) into the intron between exons 10 and 11, and the 3’ homology arm included a frameshift mutation to create a humanized mutant exon 11 (p.W288fs*12). Constructs were linearized and electroporated into C57BL/6 embryonic stem (ES) cells. Clones were screened by loss-of-allele PCR, confirmed by Southern blot and microinjected into C57BL/6 blastocysts. PCR genotyping of pups was used to screen for germline transmission (primer sequences in Table S1).
Experimental animals
C57BL/6J and B6.SJL-PtprcaPepcb/BoyJ (referred to as “B6.CD45.1”) mice were obtained from, and aged within, The Jackson Laboratory. Dnmt3afl-R878H/+ mice were crossed to B6.Cg-Tg(Mx1-cre)1Cgn/J mice (referred to as Mx1-Cre) [21]. Npm1frt-cA/+ mice were crossed to B6N.129S6(Cg)-Gt(ROSA)26Sortm3(CAG-flpo/ERT2)Alj/J (referred to as R26FlpoER) [22]. Unless otherwise noted, all mice were female and experiments initiated at 8–16 weeks of age. Mice were injected five times (once every other day) via intraperitoneal (IP) injection with 15 mg/kg high molecular weight polyinosinic-polycytidylic acid (pIpC) (InvivoGen) to induce Mx1-Cre recombinase expression. Mice were administered 125 mg/kg tamoxifen three times (on consecutive days) via oral gavage to induce R26FlpoER recombinase expression. Before and after pIpC or tamoxifen administration, genomic DNA was extracted from PB cells for recombination PCR using primers specified above (Table S1). In addition, RNA was extracted and cDNA synthesized from whole BM cells for Sanger sequencing to verify mutant allele expression. Real-time PCR was utilized to examine relative expression of Dnmt3a in Dnmt3afl-R878H/+ Mx1-cre+ animals prior to pIpC injection (primer sequences in Table S1). To verify cellular localization of Npm1cA, spleens from Npm1cA/+ R26FlpoER or control Npm1+/+ R26FlpoER mice were fixed, paraffin embedded, and sectioned for immunohistochemistry using a polyclonal antibody raised against human NPM1 (Cell Signaling Technology). The Jackson Laboratory’s Institutional Animal Care and Use Committee (IACUC) approved all experiments.
Primary cell isolation and hematopoietic stem/progenitor cell phenotyping
Single-cell suspensions of BM were prepared by filtering crushed, pooled femurs, tibiae, and iliac crests from each mouse. BM mononuclear cells (MNCs) were isolated by Ficoll-Paque (GE Healthcare Life Sciences) density centrifugation and stained with a combination of fluorochrome-conjugated antibodies from eBioscience, BD Biosciences, or BioLegend: CD45.2 (clone 104), c-Kit (clone 2B8), Sca-1 (clone 108129), CD150 (clone TC15-12F12.2), CD48 (clone HM48-1), FLT3 (Clone A2F10), CD34 (clone RAM34), FcγR (clone 2.4G2), mature lineage (Lin) marker mix (B220 (clone RA3-6B2), CD11b (clone M1/70), CD4 (clone RM4-5), CD8a (clone 53-6.7), Ter-119 (clone TER-119), Gr-1 (clone RB6-8C5)), and the viability stain propidium iodide (PI). Stained cells were either analyzed or sorted using a FACSAria with Diva software (BD Biosciences) based on the following surface marker profiles: LSK (Lin− Sca-1+ c-Kit+), LT-HSC (Lin− Sca-1+ c-Kit+ Flt3− CD150+ CD48−), ST-HSC (Lin− Sca-1+ c-Kit+ Flt3− CD150− CD48−), MPP2 (Lin− Sca-1+ c-Kit+ Flt3− CD150+ CD48+), MPP3 (Lin− Sca-1+ c-Kit+ Flt3− CD150− CD48+), MPP4 (Lin− Sca-1+ c-Kit+ Flt3+), myeloid-restricted progenitors (MyPro; Lin− Sca-1− c-Kit+), GMP (Lin− Sca-1- c-Kit+ CD150− CD34+ FcγR+), CMP (Lin− Sca-1− c-Kit+ CD150− CD34+ FcγRlo) and MEP (Lin− Sca-1− c-Kit+ CD150− CD34− FcγR−). All flow cytometry data was analyzed using FlowJo software.
Colony formation unit (CFU) assay
De novo isolated cells were plated in MethoCult GF M3434 (StemCell Technologies) at the indicated numbers and cultured at 37 °C and 5% CO2. Colonies were scored between 6 and 14 days post-plating using a Nikon Eclipse TS100 inverted microscope. For CFU replating assay, colonies were harvested, total viable cell counts obtained, and then 5 × 104 cells were replated in MethoCult GF M3434. To examine allele recombination at the clonal level, whole BM was plated from Dnmt3aR878H/+ Npm1cA/+ mice with MDS/MPD or MPD phenotypes. Individual colonies were picked under a Nikon Eclipse microscope (4× magnification) and used for colony PCR to examine recombination of Dnmt3aR878H and Npm1cA alleles (primers listed in Table S1).
In vivo transplantation
For competitive transplantation, 2 × 106 BM MNCs from Dnmt3afl-R878H/+ Mx1-Cre or Dnmt3a+/+ Mx1-Cre mice were combined at a 1:1 ratio with 2 × 106 BM MNCs from B6.CD45.1 mice and intravenously injected into female recipient B6.CD45.1 mice that received a lethal dose of gamma irradiation (1200 rads, split dose). Recipient mice received pIpC injections as described at 5 weeks post-transplant. For non-competitive transplantation, 106 BM MNCs from Dnmt3aR878H/+ Npm1cA/+ Mx1-Cre R26FlpoER or Dnmt3a+/+ Npm1+/+ Mx1-Cre R26FlpoER mice were intravenously injected into female recipient B6.CD45.1 mice that received a lethal dose of gamma irradiation (1200 rads, split dose). Recipient mice received pIpC injections as described at 4–5 weeks post-transplant, and tamoxifen administration at various times post-pIpC as indicated. Recipient mice were monitored every 4 weeks thereafter by flow cytometry analysis of PB samples using a cocktail of CD45.1 (clone A20), CD45.2, CD11b, B220, CD3ε (clone 145-2C11), and Gr-1 on an LSRII (BD). For secondary transplantation studies, female B6.CD45.1 mice received a sublethal dose of gamma irradiation (600 rads) and were intravenously injected with 106 BM MNCs from moribund primary recipients of Dnmt3aR878H/+ Npm1cA/+ Mx1-Cre R26FlpoER cells. For tertiary transplantation studies, female B6.CD45.1 mice received a sublethal dose of gamma irradiation (600 rads) and were intravenously injected with 5 × 103 sorted MyPro (Lin− Sca-1− c-Kit+) from moribund secondary recipients of Dnmt3aR878H/+ Npm1cA/+ Mx1-Cre R26FlpoER cells.
Analysis of moribund mice
Mice demonstrating declining health status were sacrificed and PB, spleen, liver, and BM harvested. Differential blood cell counts were obtained from PB using an Advia 120 Hematology Analyzer (Siemens). Single-cell suspensions of PB, spleen, and BM were analyzed by flow cytometry for mature lineage markers and c-Kit on an LSRII. Cytospin preparations of whole BM MNCs were stained with May–Grunwald–Giemsa stain. Liver and spleens were fixed for 24 h in 10% buffered formalin phosphate, embedded in paraffin, and sections were stained with H&E. Images of stained BM, liver, and spleen slides were captured on a Nikon Eclipse E200 upright microscope with SPOT imaging software (v.5.2).
M-IMPACT
Genomic DNA was extracted from isolated BM cells of diseased mice and WT littermates. DNA underwent targeted capture and deep sequencing performed by the MSKCC Integrated Genomics Operation Core using the MSK mouse Integrated Mutation Profiling of Actionable Cancer Targets (M-IMPACT) v1 assay. M-IMPACT is a DNA sequencing assay, which captures and sequences the exons and select introns of 578 known cancer genes, using solution-phase hybridization-based exon capture as described previously [23]. Gene selection, mouse ortholog mapping, and bait design has been described previously [23, 24]. Mouse genomic DNA (250 ng) was used for library construction with molecular barcoding of each sample occurring prior to capture and sequencing. Equimolarly pooled libraries containing captured DNA fragments were subsequently sequenced on one lane of an Illumina HiSeq system for paired end 125/125 reads with intended 500× coverage for all reads. Sequencing analysis was performed by the MSKCC Bioinformatics Core. Processed FASTQ files were mapped to the mouse reference genome mm10 (GRCm38) using BWA-MEM (Burrows-Wheeler Aligner v0.7.12; http://arxiv.org/abs/1303.3997). Resulting files were sorted, grouped, and PCR duplicates identified by MarkDuplicates in PICARD Tools (v1.124; https://github.com/broadinstitute/picard). BAM files were processed through GATK toolkit (v3.2) [25] to perform variant calling in tumor-vs-normal paired mode. Specifically, somatic single-nucleotide variants (SNVs) were called from processed BAM files using muTect (v1.1.7) [26]. Haplotype caller from GATK was used to identify somatic small insertions and deletions (indels) with a custom post-processing script (https://github.com/soccin/Variant-PostProcess). Variant allele frequency (VAF) was determined by calculating the fraction of variant reads for a specific out of the total reads at that location.
Statistical analysis
No sample group randomization or blinding was performed. For overall survival, Log-rank (Mantel–Cox) test was performed on Kaplan–Meier survival curves. Statistical analysis of non-survival data was performed by unpaired student’s t-test, non-parametric one-way ANOVA (Kruskal–Wallis test) followed by Dunn’s multiple comparisons test, or two-way ANOVA followed by Tukey’s multiple comparisons test. For these experiments, a minimum of three samples per condition was chosen to detect statistical differences with >80% power at an error rate of 5%. All statistical tests, including evaluation of normal distribution of data and examination of variance between groups being statistically compared, were assessed using Prism 7 software (GraphPad).
Results
Generation of an independently regulatable Dnmt3aR878H allele
To generate an inducible mouse model of human DNMT3A-mutant (DNMT3AR882H) CH, we engineered a Cre-inducible knockin model of the equivalent Dnmt3aR878H mutation in mice (Fig. 1a, Fig. S1A,B). While the general targeting strategy was similar to two recently published knockin models of Dnmt3aR878H [10, 11], our model has unique advantages of preserving expression of wild-type Dnmt3a prior to Mx1-Cre-mediated recombination (Fig. S1C) and following recombination, the Dnmt3aR878H mutant allele retains endogenous polyA and 3′ UTR elements. Indeed, prior to induction of Mx1-Cre by injection of poly(I:C) (pIpC), there is no observable change in frequency of the hematopoietic stem and progenitor cell compartment in the BM of Dnmt3afl-R878H mice (Fig. S1D). Furthermore, there is no observable change in frequency of stem and progenitor cell subsets including long-term HSCs (LT-HSC), short-term HSCs (ST-HSC), multipotent progenitor cell 2 (MPP2), MPP3, and MPP4 in the BM of Dnmt3afl-R878H mice (Fig. S1E, gating strategy shown in Fig. S1F). Thus, our mice model somatic acquisition of Dnmt3a mutation as it is observed in human CH.
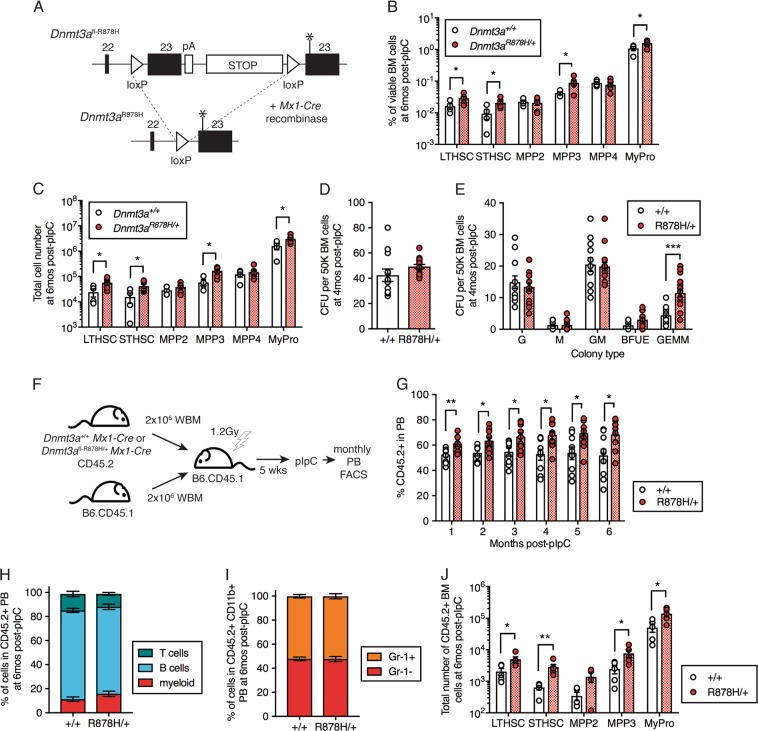
Fig. 1.
Dnmt3aR878H/+ expands HSCs and multipotent progenitor cells. a Schematic diagram of the design of the Dnmt3afl-R878H/+ allele. Asterisk indicates 2633G>A in exon 23 to encode the R878H mutation. b Frequency and c total number of LT-HSC, ST-HSC, MPP2, MPP3, MPP4, and MyPro cells in the BM of +/+ (n = 4) and R878H/+ (n = 6) mice at 6 months post-pIpC. d Total colony-forming units (CFU) and e colony types derived from 50 K BM MNCs isolated from +/+ (n = 6) and R878H/+ (n = 8) mice at 4 months post-pIpC. G (granulocyte), M (macrophage), GM (granulocyte-macrophage), BFUE (burst forming-unit erythroid), GEMM (mixed granulocyte-erythroid-macrophage-megakaryocyte). Results are from three independent experiments. f Experimental design for competitive transplantation of +/+ or fl-R878H/+ BM cells with wild-type B6.CD45.1 BM cells at a 1:1 ratio into lethally irradiated B6.CD45.1 recipient mice followed by pIpC to induce recombination of Dnmt3afl-R878H. g Frequency of donor-derived (CD45.2+)+/+ (n = 9) or R878H/+ (n = 9) cells in PB of recipient mice post-pIpC. Results are from two independent experiments. h Frequency of myeloid (CD11b+), B (B220+), and T (CD3+) cells within donor-derived (CD45.2+) PB and i Gr-1+ and Gr-1− cells within donor-derived myeloid PB at 6 months post-pIpC. j Frequency of LT-HSC, ST-HSC, MPP2, MPP3, and MyPro cells within donor-derived +/+ (n = 5) or R878H/+ (n = 5) BM at 6 months post-pIpC. Results are from two independent experiments. In all graphs, dots represent individual mice, bars indicate mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001
Dnmt3aR878H/+ expands the HSC and MPP compartments
As Dnmt3a mutation is considered to be an early event causing CH, we first investigated the phenotype of our Dnmt3aR878H/+ Mx1-Cre mice. At 6 months following activation of Cre recombinase expression, we observed a significant expansion of both the percentage and total number of viable LT-HSC, ST-HSC, MPP3, and myeloid progenitors (MyPro) in Dnmt3aR878H/+ vs. control Dnmt3a+/+ mice (Fig. 1b, c). To examine the clonogenic potential of Dnmt3aR878H/+ cells, we isolated whole BM cells from control and Dnmt3aR878H/+ mice for colony-forming unit (CFU) assays. We observed similar numbers of total CFU emerging from control and Dnmt3aR878H/+ mice (Fig. 1d). However, there was a significant expansion of multi-lineage, mixed granulocyte/erythroid/macrophage/megakaryocyte (GEMM) colonies (Fig. 1e), consistent with an expansion of multipotent stem and progenitor cells in the BM. Together, this data is generally consistent with human data demonstrating that the HSC compartment is most dramatically altered by the Dnmt3aR882H somatic mutation [8] and existing mouse Dnmt3aR878H models demonstrating expansion of stem and progenitor cells [10, 11].
To test the competitive fitness of Dnmt3aR878H/+ LT-HSCs, we examined their ability to competitively reconstitute the hematopoietic system of an irradiated host. We isolated BM cells from control and Dnmt3aR878H/+ mice that had not yet received pIpC and transplanted them at a 1:1 ratio with wild-type B6.CD45.1 BM cells into lethally irradiated congenic B6.CD45.1 recipient mice (Fig. 1f). At 5 weeks post-transplant, following initial homing and engraftment of transplanted LT-HSCs, all recipients were administered pIpC to induce Mx1-Cre expression and followed by monthly peripheral blood (PB) flow cytometry analysis. We observed that the chimerism (% CD45.2+ cells) in the PB was significantly higher from Dnmt3aR878H/+ cells compared to control Dnmt3a+/+ cells (Fig. 1g), supporting a competitive advantage of Dnmt3aR878H/+ HSCs. To determine whether this chimerism was biased toward production of particular types of mature hematopoietic cells, we examined myeloid (CD11b+), B cell (B220+) and T cell (CD3+) populations, and Gr-1+ and Gr-1− cells (within the myeloid fraction) of donor-derived CD45.2+ PB cells. We observed no differences in mature hematopoietic lineage composition between Dnmt3aR878H/+ and control Dnmt3a+/+ cells (Fig. 1h, i). In contrast, examination of chimerism within the hematopoietic stem and progenitor cell compartment of recipient mice revealed increased numbers of donor-derived LT-HSC, ST-HSC, MPP3, and myeloid-restricted progenitors (MyPro) (Fig. 1j). As this result closely resembles the hematopoietic stem and progenitor cell expansion in non-transplanted mice (Fig. 1c), these data together indicate that this expansion is a cell-intrinsic effect of the Dnmt3aR878H/+mutation.
Generation of an independently regulatable Npm1cA allele
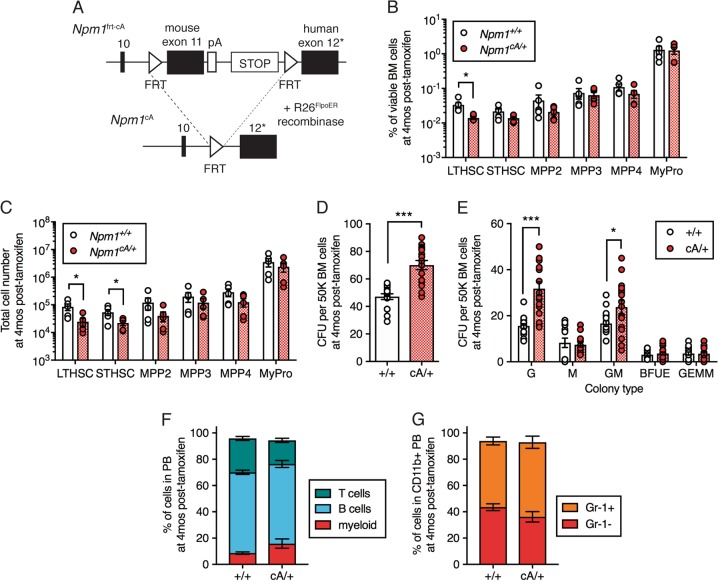
To develop an independently regulatable cooperating mutation that may synergize with Dnmt3aR878H to drive AML, we generated a Flp-inducible knockin mouse model of humanized Npm1cA (Fig. 2a, Fig. S2A–C). Mutations involving the NPM1 are found in one-third of all AML cases [20]. NPM1 mutations (referred to as NPM1c) disrupt tryptophan residues and generate an additional nuclear export signal leading to aberrant localization of NPM1c in the cytoplasm [27]. Our general targeting strategy was similar to a published knockin model of humanized Npm1flox-cA [28]; however, our model has a unique advantage of being Flp recombinase-inducible by the use of FRT sites rather than loxP sites. This strategy allows for independent regulation of the Dnmt3a and Npm1 mutant alleles. We utilized a Rosa26-knockin tamoxifen-inducible FlpoER mouse strain [22] to induce expression of Npm1cA following administration of tamoxifen. To compare alterations in the hematopoietic stem and progenitor cell compartment driven by Npm1cA as a single mutation with the Dnmt3aR878H/+ phenotype, we examined the frequency and total number of stem and progenitor cells in Npm1cA/+ mice at 4 months post-tamoxifen injection. This revealed a significant decrease in frequency of LT-HSC (Fig. 2b) as well as total numbers of LT-HSC and ST-HSC (Fig. 2c) in Npm1cA/+ compared to Npm1+/+ control mice. To examine the clonogenic potential of Npm1cA/+ cells, we isolated whole BM cells from control and Npm1cA/+ mice at 4 months post-tamoxifen injection for CFU assays. We observed a significant increase in the total number of colonies formed (Fig. 2d) and expansion in both granulocyte (G) and granulocyte-macrophage (GM) colony types (Fig. 2e). This Npm1cA/+ in vitro phenotype is in contrast to the Dnmt3aR878H/+ BM, which demonstrated an expansion of the multi-lineage GEMM colony type (Fig. 1e). Lastly, as Npm1cA/+ has been demonstrated to cause myeloproliferation and AML with 30% penetrance late in life (median survival 617 days) [28], we examined mature hematopoietic cell lineage composition in the PB in our Npm1cA/+ mice at 4 months post-tamoxifen injection. We observed no alteration in frequency of mature hematopoietic lineages in Npm1cA/+ mice compared to controls (Fig. 2f, g), suggesting that at the time point at which we observe changes in the hematopoietic stem and progenitor cell compartment and clonogenicity of BM cells, there is no overt peripheral myeloproliferation.
Fig. 2.
Npm1cA/+ depletes HSCs and expands myeloid colony-forming cells. a Schematic diagram of the design of the Npm1frt-cA/+ allele. Asterisk indicates frameshift mutation to create a humanized mutant exon 12 (p.W288fs*12). b Frequency and c total number of LT-HSC, ST-HSC, MPP2, MPP3, MPP4, and MyPro cells in the BM of +/+ (n = 5) and cA/+ (n = 5) mice at 4 months post-tamoxifen. d Total CFU and e colony types derived from 50K BM MNCs isolated from +/+ (n = 8) and cA/+ (n = 8) mice at 4 months post-tamoxifen. Results are from four independent experiments. f Frequency of myeloid, B, and T cells within PB and g Gr-1+ and Gr-1− cells within myeloid PB at 4 months post-tamoxifen in +/+ (n = 6) and cA/+ (n = 12) mice. In all graphs, dots represent individual mice, bars indicate mean ± s.e.m. *P < 0.05; ***P < 0.001
Dnmt3aR878H/+Npm1cA/+ mice develop a lethal myeloproliferative disorder
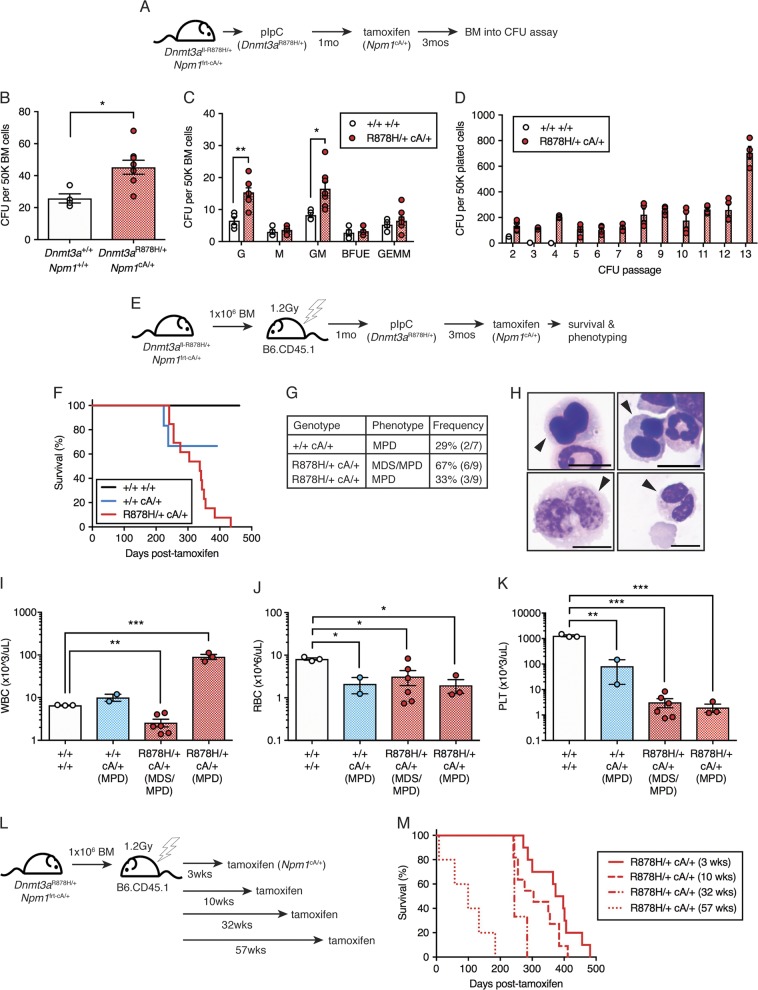
We then crossed the Npm1frt-cA/+ mice with the Dnmt3afl-R878H/+ Mx1-Cre model, to generate compound Dnmt3afl-R878H/+ Npm1frt-cA/+ Mx1-Cre R26FlpoER mice. We induced expression of the Dnmt3aR878H mutation first then followed this with induction of Npm1cA expression three months later to allow development of CH in the interim period (Fig. 3a). We first examined the clonogenic potential of BM cells isolated from these Dnmt3aR878H/+ Npm1cA/+ mice. We observed a significant increase in total CFU from Dnmt3aR878H/+ Npm1cA/+ BM compared to control (Fig. 3b). Assessing colony morphology revealed a significant increase in granulocyte (G) and granulocyte-macrophage (GM) colony types (Fig. 3c). Serial replating of Dnmt3aR878H/+ Npm1cA/+ colonies revealed robust proliferative and clonogenic potential for at least 13 passages (Fig. 3d), suggesting unlimited in vitro self-renewal potential.
Fig. 3.
Dnmt3aR878H/+ followed by Npm1cA/+ causes development of lethal MPD. a Experimental design for induction of Dnmt3aR878H/+ driving clonal hematopoietic expansion prior to induction of Npm1cA/+, followed by in vitro CFU assay. b Total CFU and c colony types derived from 50K BM MNCs isolated from control (n = 4) or R878H/+ cA/+ (n = 8) mice at 3 months post-tamoxifen. Results are from 3 independent experiments. (d) CFU derived from 50K BM MNCs isolated from control (n = 2) and R878H/+ cA/+ (n = 4) mice, serially passaged for up to 13 passages. Results are from two independent experiments. e Experimental design for non-competitive transplantation of fl-R878H/+ frt-cA/+ BM into lethally irradiated recipient mice, followed by pIpC induction of R878H/+ and clonal hematopoietic expansion prior to tamoxifen induction of cA/+. f Overall survival of mice transplanted with 106 BM cells from control (n = 3), cA/+ only (n = 7) or R878H/+ cA/+ (n = 13) mice. Results are from three independent experiments. g Description and penetrance of phenotypes in moribund mice following transplant with cA/+ only or R878H/+ cA/+ BM cells. h Representative Giemsa-stained BM cytospins (100×) from R878H/+ cA/+ MDS/MPD showing dysplastic cells (arrows). Scale bars are 10 μm. i WBC count, j RBC count, and k PLT count in moribund mice (control, n = 3; cA/+ MPD, n = 2; R878H/+ cA/+ MDS/MPD, n = 6; R878H/+ cA/+ MPD, n = 3). l Experimental design for tamoxifen induction of cA/+ at various times following induction of R878H/+. m Overall survival of mice transplanted with 106 BM cells with latency between R878H/+ and cA/+ mutations of 3 weeks (n = 10), 10 weeks (n = 11), 32 weeks (n = 3), or 57 weeks (n = 5). Results are from three independent experiments. In all graphs, dots represent individual mice, bars indicate mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001
To determine whether this enhanced clonogenic potential was reflective of transformation, we transplanted BM from control Dnmt3a+/+ Npm1+/+, single mutant Npm1frt-cA/+ or double mutant Dnmt3afl-R878H/+ Npm1frt-cA/+ mice into lethally irradiated recipient mice, administered pIpC at 1-month post-transplant, tamoxifen at 3 months post-pIpC, and followed the natural life course of the recipients (Fig. 3e). Importantly, this experimental design restricted expression of Dnmt3aR878H and Npm1cA exclusively to the hematopoietic compartment. We observed that within a 440-day period, 100% of Dnmt3aR878H/+ Npm1cA/+ recipient mice died while only 29% of single mutant Npm1cA/+ recipient mice died (Fig. 3f). The phenotypes at the time of death comprised two major groups; 67% (6 out of 9) Dnmt3aR878H/+ Npm1cA/+ mice had mixed myelodysplastic syndrome and myeloproliferative disorder (MDS/MPD) while 33% (3 out of 9) Dnmt3aR878H/+ Npm1cA/+ mice and 29% (2 out of 7) single mutant Npm1cA/+ mice had MPD (Fig. 3g). The single mutant Npm1cA/+ mice developed MPD with survival times of ~7.5–8 months post-tamoxifen, demonstrating that this mutation is capable of causing myeloproliferation beyond our original 4-month post-tamoxifen sampling (Fig. 2f).
An extensive characterization of Dnmt3aR878H/+ Npm1cA/+ MDS/MPD, Dnmt3aR878H/+ Npm1cA/+ MPD and single mutant Npm1cA/+ MPD was performed to contrast phenotypic similarities and differences. The Dnmt3aR878H/+ Npm1cA/+ MDS/MPD phenotype was characterized by erythroid and myeloid dysplasia (Fig. 3h), pancytopenia including significantly decreased white blood cell (WBC) (Fig. 3i), red blood cell (RBC) (Fig. 3j), and platelet (PLT) counts (Fig. 3k), increased PB myeloid cell production (Fig. S3A), and myeloid cell infiltration into the spleen and liver (Fig. S3C,E). The Dnmt3aR878H/+ Npm1cA/+ MPD phenotype was characterized by significantly increased WBC count (Fig. 3i), significantly decreased RBC and PLT counts (Fig. 3j, k), significantly increased spleen weight (Fig. S3D), increased PB myeloid cell production with monocyte bias (Fig. S3A,B), and myeloid (monocyte) cell infiltration into the spleen and liver (Fig. S3C–F). The single mutant Npm1cA/+ MPD phenotype was distinguished from Dnmt3aR878H/+ Npm1cA/+ MPD phenotype by lower WBC count (Fig. 3i), lower spleen weight (Fig. S3D) and greater monocyte bias in myeloid PB cells (Fig. S3B). Together, these data demonstrate that acquiring the Npm1cA mutation on pre-existing Dnmt3aR878H/+ CH causes progression to MDS/MPD or MPD.
To demonstrate one application of independently regulatable disease alleles, we examined whether the duration of pre-existing Dnmt3aR878H/+ CH would impact disease development or survival of mice. Npm1cA/+ was induced at 3, 10, 32, or 57 weeks post-recombination of Dnmt3aR878H/+ (Fig. 3l). We observed that longer latency between acquisition of Dnmt3a and Npm1 mutations resulted in a significantly shorter survival time post-tamoxifen (3 vs. 10 weeks, P = 0.067; 3 vs. 32 weeks, ***P = 0.0009; 3 vs. 57 weeks, ***P < 0.0001) (Fig. 3m). This suggests that a greater length of time with Dnmt3a-mutant clonal expansion increases the risk of progression to malignancy.
Dnmt3aR878H/+Npm1cA/+ MPD progresses to AML upon transplant
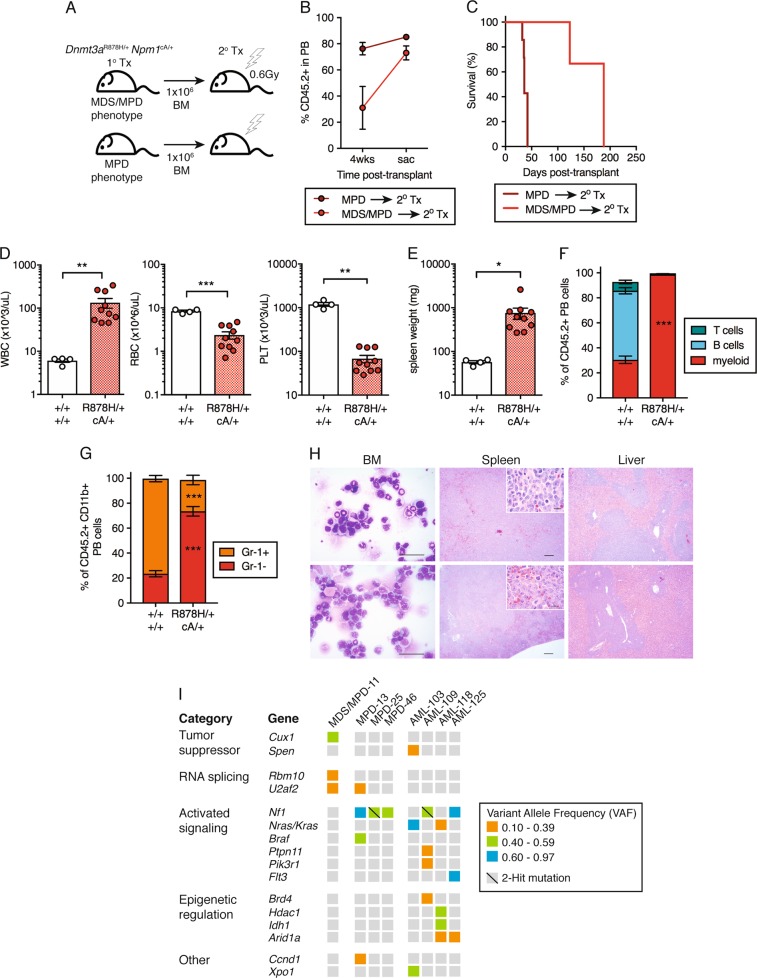
While human NPM1 mutations are most often observed in de novo AML, several studies have shown that NPM1 mutations can be present in MDS or MDS/myeloproliferative neoplasm (MPN) prior to blast counts reaching the threshold for AML diagnosis [29, 30]. This suggests Dnmt3aR878H/+ Npm1cA/+ MDS/MPD or MPD might have potential for further progression to AML. To determine whether Dnmt3aR878H/+ Npm1cA/+ MPDs were transplantable to recipient mice and to test whether they are capable of further progression to AML, we isolated BM cells from primary Dnmt3aR878H/+ Npm1cA/+ transplant recipients with either the MDS/MPD or MPD phenotypes and transplanted these cells into sublethally irradiated secondary recipient mice (Fig. 4a). We assessed donor cell engraftment (% CD45.2+) at 4 weeks post-transplant as well as when these mice were moribund. This analysis revealed that MPD samples achieved high levels of engraftment in secondary recipients almost immediately post-transplant (Fig. 4b). In contrast, MDS/MPD samples had initially low levels of engraftment in secondary recipients but dramatically expanded to achieve high levels of engraftment at the time of sacrifice. There was a significant difference between the survival of mice transplanted with MDS/MPD donors (median survival 188 days) vs. MPD donors (median survival 36 days) (Fig. 4c). However, these both represent a dramatic disease acceleration compared to the primary recipient animals (Fig. 3f).
Fig. 4.
Dnmt3aR878H/+ Npm1cA/+ MPD progresses to AML following transplantation. a Experimental design for non-competitive secondary transplantation of Dnmt3aR878H/+ Npm1cA/+ BM MNCs from primary recipient mice with MDS/MPD or MPD phenotypes. b Frequency of donor-derived (CD45.2+) cells in PB of secondary recipient mice at 4 weeks post-transplant and at time of sacrifice. Dots represent mean ± s.e.m. (MPD, n = 7; MDS/MPD, n = 3). Results are from three independent experiments. c Overall survival of secondary transplant-recipient mice (MPD, n = 7; MDS/MPD, n = 3). d WBC, RBC, and PLT counts and e spleen weights of moribund mice (control, n = 4; +/+ cA/+, n = 10). f Frequency of myeloid, B, and T cells within the donor-derived CD45.2+ fraction in PB and g Gr-1+ and Gr-1− cells within donor-derived myeloid PB of moribund mice (control, n = 3; +/+ cA/+, n = 10). h Representative Giemsa-stained BM cytospins (far left; 40×, scale bars are 40 μm) and H&E-stained spleen (center; 4×, scale bars are 100 μm) (inset: 40×, scale bars are 40 μm) and liver sections (far right; 4×, scale bars are 100 μm) from moribund recipient mice. i Somatic, nonsynonymous mutations in individual genes and sets of genes, grouped into five categories. Orange boxes indicate mutations at VAF 0.10-0.39; green boxes, VAF 0.40–0.59; blue boxes, VAF 0.60–0.97. In all graphs unless otherwise specified, dots represent individual mice, bars indicate mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001
We observed that 100% of secondary transplant recipient mice succumbed to acute myeloid leukemia (AML), regardless of whether they were derived from an MDS/MPD or MPD primary donor. All Dnmt3aR878H/+ Npm1cA/+ recipients showed significantly increased WBC count and decreased RBC and PLT counts (Fig. 4d), increased spleen weight (Fig. 4e), complete myeloid cell dominance in the PB with monocyte bias (Fig. 4f, g), >20% blast cells in the BM and vast infiltration of blast cells in the spleen and liver (Fig. 4h). Together these data suggest that Dnmt3aR878H/+ Npm1cA/+ MDS/MPD and MPD can both progress to AML.
To examine clonality of recombination of Dnmt3afl-R878H and Npm1frt-cA alleles in mice with MDS/MPD, MPD and AML, we performed a BM CFU assay and picked individual colonies for PCR. We observed that in MDS/MPD, 82% of colonies had recombination of both Dnmt3aR878H and Npm1cA (Fig. S3G). In MPD and AML, 100% of colonies had recombination of both Dnmt3aR878H and Npm1cA. This demonstrates similar levels of recombination in Dnmt3aR878H/+ Npm1cA/+ MDS/MPD, MPD, and AML. To further examine additional acquired somatic mutations, we performed somatic mutational analysis of BM from MDS/MPD, MPD and AML samples using a custom targeted sequencing panel querying 578 known cancer genes (MSK-Mouse IMPACT; M-IMPACT) (Table S2). We observed that Dnmt3aR878H/+ Npm1cA/+ MDS/MPD was associated with mutations in the tumor suppressor Cux1, previously shown to drive MDS and MDS/MPD [31] and in the RNA splicing genes Rbm10 and U2af2 (Fig. 4i, Table S3). Mutations in key factors of the spliceosome are known to occur frequently in CH and MDS [32]. Development of Dnmt3aR878H/+ Npm1cA/+ MPD was associated with mutations activating the Ras/Raf/MAPK pathway (including Nf1, Nras, Kras, and Braf) at high VAF, consistent with known function of these mutations [33, 34]. Progression to Dnmt3aR878H/+ Npm1cA/+ AML was also associated with these same mutations, but with the addition of mutations activating the tyrosine phosphatase SHP2 (Ptpn11), PI3K (Pik3r1), or Flt3 (Flt3) signaling as well as mutations involved in epigenetic regulation including Brd4, Hdac1, Idh1, and Arid1a. Many of these genes are recurrently mutated in human AML [35–37], and demonstrate additional mutations are required to cooperate with mutant Dnmt3a and Npm1 to promote transformation to MDS/MPD, MPD and AML.
Dnmt3aR878H/+Npm1cA/+ AML mice have decreased HSC and increased myeloid progenitors
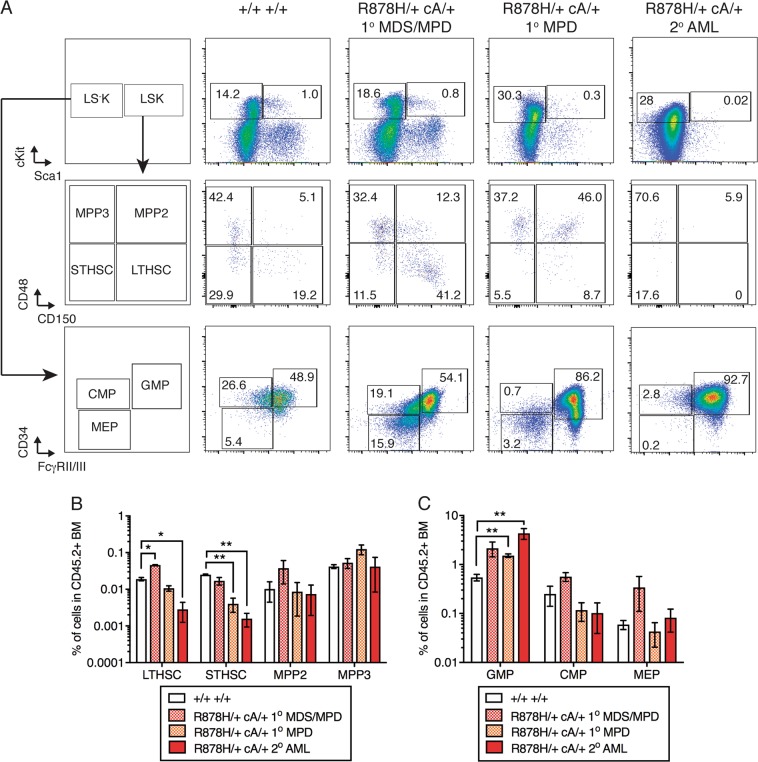
To determine how Dnmt3aR878H/+ CH progresses to MPD and AML with acquisition of Npm1cA mutation, we examined the BM stem and progenitor cell compartment of primary and secondary recipient mice transplanted with Dnmt3aR878H/+ Npm1cA/+ cells (representative gating shown in Fig. 5a). We observed a significant increase in LTHSC frequency in the Dnmt3aR878H/+ Npm1cA/+ MDS/MPD group and decrease in LT-HSC frequency in the Dnmt3aR878H/+ Npm1cA/+ AML group compared to control transplanted mice (Fig. 5b). In addition, we observed a significant decrease in ST-HSC frequency in the Dnmt3aR878H/+ Npm1cA/+ MPD and AML groups compared to control transplanted mice. Strikingly, we observed that granulocyte-macrophage progenitors (GMP) were significantly expanded in the Dnmt3aR878H/+ Npm1cA/+ MPD and AML groups vs. the control group (Fig. 5c). Together, these data suggest that Npm1cA transformation of Dnmt3aR878H/+ CH is accompanied by a shift in the stem and progenitor cell compartment towards expansion of myeloid-restricted progenitor cells at the expense of HSCs.
Fig. 5.
Mice with Dnmt3aR878H/+ Npm1cA/+ AML show expansion of granulocyte-macrophage progenitor cells. a Representative flow cytometric gating for hematopoietic stem and progenitor cells in recipient mice transplanted with Dnmt3a+/+ Npm1+/+ or Dnmt3aR878H/+ Npm1cA/+ with MDS/MPD, MPD, or AML phenotypes. b Frequency of LT-HSC, ST-HSC, MPP2, and MPP3 and c GMP, CMP, and MEP in donor-derived CD45.2+ BM cells (control, n = 2; R878H/+ cA/+ MDS/MPD, n = 2; R878H/+ cA/+ MPD, n = 3; R878H/+cA/+ AML, n = 4). Results are from two independent experiments
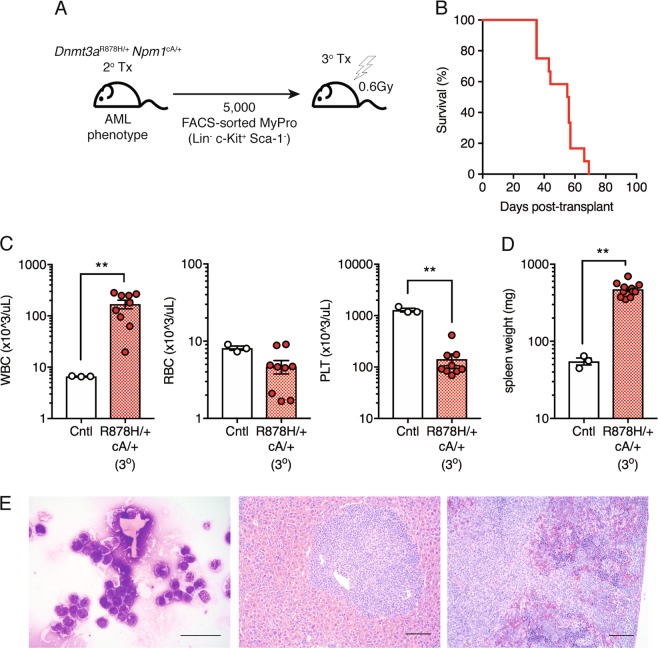
Dnmt3aR878H/+Npm1cA/+ AML is propagated by myeloid-restricted progenitors
Within Dnmt3aR878H/+ Npm1cA/+ AML, the shift in the stem and progenitor cell compartment towards expansion of myeloid-restricted progenitor cells suggests that these cells may represent tumor-propagating cells capable of transferring disease to recipient mice. To test this, we performed a tertiary transplant of sorted myeloid progenitors (Lin− c-Kit+ Sca-1−) (Fig. 6a). We observed that 100% of tertiary transplant recipient mice succumbed to acute myeloid leukemia (AML) with a median survival of 56 days (Fig. 6b), similar to the rate and penetrance of AML development in secondary transplant recipients (Fig. 4c). This is evidenced in all recipients by significantly increased WBC and decreased PLT (Fig. 6c) counts, increased spleen weight (Fig. 6d), >20% blast cells in the BM and vast infiltration of blast cells in the spleen and liver (Fig. 6e). Together, these data support diagnosis of AML in secondary recipient animals and demonstrate that myeloid-restricted progenitors are leukemia stem cells (LSCs) capable of propagating Dnmt3aR878H/+ Npm1cA/+ AML.
Fig. 6.
Myeloid-restricted progenitor cells are tumor-propagating cells in Dnmt3aR878H/+ Npm1cA/+ AML. a Experimental design for non-competitive tertiary transplantation of FACS-sorted Dnmt3aR878H/+ Npm1cA/+ BM myeloid progenitors from secondary recipient mice with AML. b Overall survival of tertiary transplant recipient mice (n = 12). Results are from three independent experiments. c WBC, RBC, and PLT counts in moribund mice (control, n = 3; +/+ cA/+, n = 9). d Spleen weights of moribund mice (control, n = 3; +/+ cA/+, n = 11). e Representative Giemsa-stained BM cytospin (far left; 40×, scale bars are 40 μm) and H&E-stained spleen (center; 10×, scale bars are 100 μm) and liver sections (far right; 10×, scale bars are 100 μm) from moribund recipient mice. In all graphs, dots represent individual mice, bars indicate mean ± s.e.m. **P < 0.01
Discussion
Individuals with CH have an increased risk of development of hematologic malignancy, including AML. Currently, no methods exist to distinguish individuals with benign CH from those with pre-malignant CH due to a lack of understanding of the mechanisms by which CH progresses to AML. Improved in vivo models of the progression of CH to AML are needed to identify specific pathways and molecules for development of prognostic tests and therapies to prevent progression to malignancy. In this study, we have developed novel inducible mouse models allowing temporal control of expression of Dnmt3aR878H, a common event in human CH, and Npm1cA, a common cooperating mutation in human AML. We find that acquisition of Npm1cA/+ mutation on the background of pre-existing Dnmt3aR878H/+ CH drives development of MPD, which is transformed to AML upon subsequent transplantation. Furthermore, increasing the duration of Dnmt3aR878H/+ CH increased the aggressiveness of MPD development upon acquisition of Npm1cA/+ mutation and decreased overall survival in mice. Previously, the combination of Dnmt3aR878H/+ and Npm1cA/+ induced simultaneously did not cause lethal hematologic malignancy within a 45-week timeframe [11]. We hypothesize that this is distinct from our current findings based on the timeframe of the observation period of animals and the sequential acquisition of mutations over time with concomitant genetic and epigenetic evolution. Based on our results, we propose that accumulation of cellular and molecular alterations over time, as a consequence of Dnmt3a mutation, synergize with Npm1 mutation to cause more aggressive malignancy. These data are pertinent for considering the timing and importance of follow-up mutational screening in individuals with CH.
In our CH model, we find that heterozygous Dnmt3aR878H alone is sufficient to cause expansion of phenotypic and functional HSCs and MPPs, and confer a competitive advantage of LTHSCs over wild-type LTHSCs in vivo. Integrating this phenotype with published models of Dnmt3a knockout, which demonstrate specific expansion of LTHSCs [9], or Dnmt3aR878H mutation, which demonstrate expansion in both multipotent and myeloid progenitors downstream of the LTHSC [10, 11], suggest that the Dnmt3aR878H mutation is distinct from Dnmt3a loss. Dnmt3aR878H can confer impaired differentiation but not the complete differentiation block that is observed upon Dnmt3a loss [16], and Dnmt3aR878H HSCs, unlike Dnmt3a−/− HSCs [9, 16], do not appear to have overt lineage bias in the context of competitive transplantation.
In experiments examining the hematopoietic stem and progenitor cell compartment at different stages along the trajectory of CH to MPD and finally to AML, we revealed that Npm1cA/+ drives progressive transition of expansion of the HSC and MPP compartments in CH to the myeloid-restricted progenitor cell compartment in AML. Furthermore, these myeloid-restricted progenitor cells represent LSCs capable of propagating AML in transplanted recipient mice. Seminal work examining the pre-leukemic state in AML has suggested that the cell of origin acquiring DNMT3A mutation is an HSC, which causes an expanded pool of HSCs and downstream progenitors, within which additional mutations including NPM1c are acquired to drive progression to AML [8]. These findings pointed to GMPs and/or mixed lineage progenitors (MLP) as the likely populations in which NPM1c was acquired. This model is consistent with our findings that progression of Dnmt3a-mutant CH to Dnmt3a- and Npm1-mutant AML is accompanied by a depletion of the expanded pool of HSCs observed in CH and a shift towards an expanded pool of myeloid-restricted progenitors that possess LSC activity. These data also provide important context for interpretation of a previously published report demonstrating that conditional knockin of Dnmt3aR878H initiates AML in the absence of other engineered mutations after a long latency [10]. This study found that LTHSCs decreased and myeloid lineage-restricted progenitors were expanded in their Dnmt3a-mutant AML mice. Based on these data taken together, we propose that a decline in HSC frequency and shift toward myeloid-restricted progenitor expansion in the HSPC compartment may serve as a biomarker for risk or early progression of CH to AML. Our data suggest that distinct cooperating mutations, in addition to Npm1, drive progression of Dnmt3a-mutant CH to MDS/MPD, MPD and AML. Mutations in the tumor suppressor Cux1 and RNA splicing factor mutations are associated with MDS/MPD, while strong selection for mutations activating Ras/Raf/MAPK signaling are invariably associated with progression to MPD. Furthermore, transformation to AML is accompanied by selection for additional mutations in signaling molecules and/or epigenetic regulatory factors.
Mutations in DNMT3A are common in the general population and increase with aging. With next-generation sequencing becoming more and more routine as a part of general medical care, we need better predictive tools to assess who is at risk for progression from CH to MDS, MPN or AML in addition to development of new preventative therapeutic strategies. Addressing a number of these pressing basic and translational research questions regarding the evolution of CH to AML will now be possible by utilizing the in vivo models we have developed. For example, different stressors and environmental factors (age, tobacco use, prior radiation therapy) have been hypothesized to facilitate emergence of clinically relevant phenotypes of the DNMT3A mutation [38]. Our models will not only permit prospective testing of the effects of stress or environmental factors on the Dnmt3a-mutant CH phenotype, but also whether effects on CH ultimately change the risk of progression to malignancy upon acquisition of a cooperating mutation in Npm1. Furthermore, orthogonal regulation of the Dnmt3a and Npm1 mutations will permit interrogation of the underlying biological mechanisms by which these mutations interact to cause malignancy, including hematopoietic cell-intrinsic interactions (ex. chromatin regulation, DNA repair) and cell-extrinsic interactions (ex. alterations in the BM cytokine environment). Lastly, this study serves as a proof-of-principle that utilizing an inducible, dual-recombinase system is a feasible and translationally relevant strategy to model combinations of somatic mutations in CH, pre-leukemia and hematologic malignancy, and to model clonal evolution more broadly in different malignant contexts.
Supplementary information
Acknowledgements
This work was supported by National Institutes of Health (NIH), National Cancer Institute (NCI) grant R21CA184851 (J.J.T.), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R56DK112947 and R01DK118072 (J.J.T.), NCI Cancer Core Grant P30CA034196, Nathan Shock Center Grant P30AG038070 and Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) T32HD007065 (K.Y. and J.M.S.). This work was also supported by The V Foundation V Scholar award (J.J.T.) and grants from the Maine Cancer Foundation (J.J.T.). Additionally, this work is supported in part by NCI grant R35CA197594 and NIH Office of the Director grant U54OD020355 (both to R.L.L.). K.Y. is supported by an American Society of Hematology (ASH) Scholar Award and the Pyewacket Fund at The Jackson Laboratory. L.A.M. is supported by a Leukemia & Lymphoma Society (LLS) Fellow Award. We acknowledge the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, the Memorial Sloan Kettering Cancer Center (MSKCC) Bioinformatics Core, and the use of the Integrated Genomics Operation Core, funded by MSKCC Support Grant (NCI P30CA008748). We thank Nicole Dean, Tara Murphy, Eraj Khokhar, Kai Cheng, and Judy Morgan for technical help, experimental and laboratory support, Kevin Mills and members of the Trowbridge laboratory for helpful discussion and critical comments, and Will Schott for cell sorting.
Author contributions
L.O.G., D.E.B. and J.J.T. designed and generated the mouse models. M.A.L., R.K.B., E.E., K.Y. and J.J.T. designed experiments. M.A.L., R.K.B., E.E., L.A.M. and K.Y. performed and analyzed experiments. R.K.S. analyzed and interpreted experiments. R.L. provided critical reagents. M.A.L., J.M.S. and J.J.T. wrote the manuscript. R.K.B., L.O.G., E.E., L.A.M., K.Y., D.E.B., R.L.L. and R.K.S. edited the manuscript.
Compliance with ethical standards
Conflict of interest
R.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics, and Isoplexis. He receives research support from and consulted for Celgene and Roche, research support from Prelude Therapeutics, and has consulted for Novartis and Gilead. He has received honoraria from Lilly and Amgen for invited lectures. The remaining authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41375-018-0368-6) contains supplementary material, which is available to authorized users.
References
- 1.Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22:157–70. doi: 10.1016/j.stem.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung SS, Park CY. Aging, hematopoiesis, and the myelodysplastic syndromes. Blood Adv. 2017;1:2572–8. doi: 10.1182/bloodadvances.2017009852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373:35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–47. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci USA. 2014;111:E5401–10. doi: 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111:2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai YJ, Wang YY, Huang JY, Xia L, Shi XD, Xu J, et al. Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc Natl Acad Sci USA. 2017;114:5237–42. doi: 10.1073/pnas.1703476114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–95. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Wang YY, Dai YJ, Zhang W, Zhang WN, Xiong SM, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci Usa. 2014;111:2620–5. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer SE, Qin T, Muench DE, Masuda K, Venkatasubramanian M, Orr E, et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Discov. 2016;6:501–15. doi: 10.1158/2159-8290.CD-16-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;30:363–5. doi: 10.1016/j.ccell.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2015;125:619–28. doi: 10.1182/blood-2014-08-594564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong M, Park HJ, Celik H, Ostrander EL, Reyes JM, Guzman A, et al. Loss of Dnmt3a Immortalizes hematopoietic stem cells in vivo. Cell Rep. 2018;23:1–10. doi: 10.1016/j.celrep.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schonhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20:1340–7. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–7. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 22.Lao Z, Raju GP, Bai CB, Joyner AL. MASTR: a technique for mosaic mutant analysis with spatial and temporal control of recombination using conditional floxed alleles in mice. Cell Rep. 2012;2:386–96. doi: 10.1016/j.celrep.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pronier E, Bowman RL, Ahn J, Glass J, Kandoth C, Merlinsky TR, et al. Genetic and epigenetic evolution as a contributor to WT1-mutant leukemogenesis. Blood. 2018;132:1265–78. doi: 10.1182/blood-2018-03-837468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 28.Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43:470–5. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin P, Falini B. Acute myeloid leukemia with recurrent genetic abnormalities other than translocations. Am J Clin Pathol. 2015;144:19–28. doi: 10.1309/AJCP97BJBEVZEUIN. [DOI] [PubMed] [Google Scholar]
- 30.Schnittger S, Bacher U, Haferlach C, Alpermann T, Dicker F, Sundermann J, et al. Characterization of NPM1-mutated AML with a history of myelodysplastic syndromes or myeloproliferative neoplasms. Leukemia. 2011;25:615–21. doi: 10.1038/leu.2010.299. [DOI] [PubMed] [Google Scholar]
- 31.An N, Khan S, Imgruet MK, Gurbuxani SK, Konecki SN, Burgess MR, et al. Gene dosage effect of CUX1 in a murine model disrupts HSC homeostasis and controls the severity and mortality of MDS. Blood. 2018;131:2682–97. doi: 10.1182/blood-2017-10-810028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30:989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birnbaum RA, O’Marcaigh A, Wardak Z, Zhang YY, Dranoff G, Jacks T, et al. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–95. doi: 10.1016/S1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 34.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120:3397–406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou HA, Chou WC, Lin LI, Chen CY, Tang JL, Tseng MH, et al. Characterization of acute myeloid leukemia with PTPN11 mutation: the mutation is closely associated with NPM1 mutation but inversely related to FLT3/ITD. Leukemia. 2008;22:1075–8. doi: 10.1038/sj.leu.2405005. [DOI] [PubMed] [Google Scholar]
- 36.Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, et al. Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia. 2016;30:2430. doi: 10.1038/leu.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. [DOI] [PMC free article] [PubMed]
- 38.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21:374–82.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.