Abstract
Structure-by-structure analysis, in which the brain magnetic resonance imaging (MRI) is parcellated based on its anatomical units, is widely used to investigate chronological changes in morphology or signal intensity during normal development, as well as to identify the alterations seen in various diseases or conditions. The multi-atlas label fusion (MALF) method is considered a highly accurate parcellation approach, and anticipated for clinical application to quantitatively evaluate early developmental processes. However, the current MALF methods, which are designed for neonatal brain segmentations, are not widely available. In this study, we developed a T1-weighted, neonatal, multi-atlas repository and integrated it into the MALF-based brain segmentation tools in the cloud-based platform, MRICloud. The cloud platform ensures users instant access to the advanced MALF tool for neonatal brains, with no software or installation requirements for the client. The web platform by braingps.mricloud.org will eliminate the dependence on a particular operating system (e.g., Windows, Macintosh, or Linux) and the requirement for high computational performance of the user’s computers. The MALF-based, fully-automated, image parcellation could achieve excellent agreement with manual parcellation, and the whole and regional brain volumes quantified through this method demonstrated developmental trajectories comparable to those from a previous publication. This solution will make the latest MALF tools readily available to users, with minimum barriers, and will expedite and accelerate advancements in developmental neuroscience research, neonatology, and pediatric neuroradiology.
Keywords: Brain, MRI, Neonate, Multi-atlas, Parcellation
Introduction
Structure-by-structure analysis, in which the brain is parcellated based on structural units that follow standard ontology in brain anatomy, is widely used to investigate disease-related changes seen on brain MRI scans.1–4 Numerous tools for brain parcellation methods have been proposed in the past and their accuracy has continuously improved, especially in the past decade since the introduction of multi-atlas label-fusion (MALF) algorithms.5–10 The major strength of this approach is that the algorithm relies on a large collection of pre-parcellated images (atlases) as teaching files to achieve high parcellation accuracy against a wide range of anatomical variability, even for diseased brains11,12 and to account for differences in scanner and scan parameters.12,13 This robustness to anatomic variations is beneficial in neurodevelopmental studies; a set of multiple atlases that covers the entire landscape of anatomic variations during brain development can minimize the errors in parcellating structures with anatomical variability. For example, a highly variable structure is the cavum septum pellucidum, which is a cavity between the left and right anterior horns of the lateral ventricles, and is seen in approximately 70% at 36 weeks and 35% at 40 weeks of gestation,14 7% in children under 17 years of age,15 and 1 – 6% in the adult brain.15–18 Images with and without this cavity are topologically different; therefore, transforming an image with the cavity to an image without the cavity is an ill-posed problem.19 Image normalization to a single atlas space is, thus, problematic, especially in studies targeting neonates or infants.
Although MALF-based parcellation is now considered a highly accurate parcellation approach, it works only when a multi-atlas library with accurate parcellation of brain structures is available. In particular, when brains in active development are targeted, the library must cover the age-range of interest and variations in local brain volume and intensity, which is changing during brain development. Existing multi-atlas repositories are available for pediatric and adult brains,20 and recently, for neonatal brains.21–26 The neonatal atlas repositories may be applied to the clinical setting, since they can quantify brain MRIs from infants during early development, which may provide early diagnosis or prediction of neurobehavioral sequelae. However, there are several practical issues in the application to clinical MRIs. First, the existing MALF approach applied to neonatal brain MRIs typically utilizes T2-weighted contrast or a combination of T1- and T2-weighted contrast to drive the algorithm,21–23,27 since T2-weighted scans provide greater contrast than T1-weighted scans in separating the cortical gray matter area from the unmyelinated white matter adjacent to the cortex. Three-dimensional (3D) or thin-slice, high-resolution T2-weighted protocols that are compatible with the advanced image analysis framework have been adopted in ongoing research studies of the neonatal brain.28,29 However, in clinical practice, conventional, two-dimensional, T2-weighted protocols with thicker slices (typically > 2 mm) are still common, while high-resolution, three-dimensional (3D), T1-weighted images are readily available as a routine clinical protocol. Second, the majority of the existing MALF methods require a skull-stripping procedure, which is a challenging task for fully-automated image processing. Third, the application of MALF to neonatal brain imaging was reported by a limited number of institutes that have strong biomedical engineering groups and was supported by programmers and imaging science specialists. For neurobiologists or clinician scientists without such support, the critical barriers are the requirement for knowledge and skills for the implementation of mathematical modeling into the practical image analysis system.
We aimed to break the current barrier in accessibility to MALF-based neonatal image quantification. We integrated the MALF framework with a T1-weighted, neonatal, multi-atlas repository into the cloud-based platform, MRICloud, which has more than 2,000 registered users worldwide (as of November 2018). This integration ensures users instant access to the advanced MALF tool for neonatal brain image analyses, with no requirement for the installation or download of additional software by the client. The web platform at braingps.mricloud.org will eliminate the dependence on a particular operating system (e.g., Windows, Macintosh, or Linux) and the requirement for high computational performance of the user’s computer. This solution will make the latest MALF tools readily available to users with minimum barriers, and will accelerate advancements in developmental neuroscience research, neonatology, and pediatric neuroradiology.
Methods
Participants
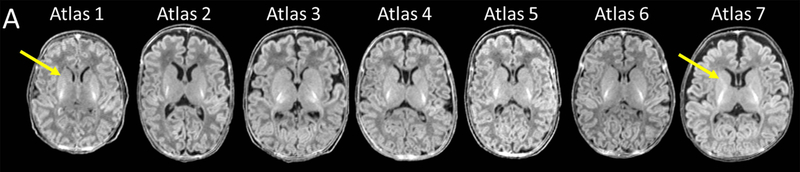
A neonatal brain MRI database from a longitudinal study of early brain development30–32 was used. Written and verbal informed consent were provided by the infants’ parents or legal guardians, in accordance with the Cooperative Institutional Review Board (IRB) of the Queen’s Medical Center, the University of Hawaii, and the Johns Hopkins University. Exclusion criteria for the term-born infants were: prolonged intensive care (>7 days); intracranial hemorrhage; neonatal hypoxic-ischemic encephalopathy; brain infections (toxoplasma, syphilis, varicella-zoster, parvovirus B19, rubella, cytomegalovirus, and herpes); and congenital heart disease or other anomaly; or any chromosomal anomaly. Among 248 MRIs available, the MRIs of term and preterm born neonates (three boys and four girls born during 26 to 41 gestational weeks), scanned during 39 to 46 post-menstrual weeks, were used to create the multi-atlas repository (Table 1 and Fig. 1A). The images were chosen to cover the morphological variability seen in this age-range. Namely, we intentionally selected images based on: (i) the width of the subarachnoid fluid space [from wide (Atlas 7) to narrow (Atlas 1); (ii) the ventricular volume [from large (Atlas 2) to small (Atlas 6)]; (iii) the existence of the cavum septum pellucidum [from prominent (Atlas 4) to absent (Atlases 2 and 3)]; (iv) the myelination status [from partial myelination seen in the posterior limb of the internal capsule and the low-intensity thalamus (Atlases 1 and 6) to myelination seen in both the anterior and posterior limbs of the internal capsule (Atlas 7)]; and (v) the shape [from round (Atlas 1) to dolichocephalic (Atlases 2 and 5)]. The selection was based on agreement between two neurologists who went over the images. One hundred seventy-seven MRIs from term-born neonates that were not used to create the atlases were used for the validation study (Table 2).
Table 1:
Demographics of the infants from which the multi-atlas repository was created.
| Sex | gestational age at birth (weeks) | postmenstrual age at scan (weeks) | birth weight (kg) | birth body length (cm) | |
|---|---|---|---|---|---|
| Atlas 1 | girl | 39.0 | 39.9 | 2.98 | 50.2 |
| Atlas 2 | boy | 33.1 | 41.2 | 1.55 | 40.6 |
| Atlas 3 | girl | 28.6 | 45.0 | 1.20 | 38.1 |
| Atlas 4 | boy | 26.4 | 42.4 | 0.96 | 34.0 |
| Atlas 5 | girl | 28.7 | 41.3 | 1.00 | 36.5 |
| Atlas 6 | boy | 40.9 | 43.2 | 3.07 | 48.3 |
| Atlas 7 | girl | 33.1 | 46.2 | 1.96 | 45.0 |
Fig. 1.
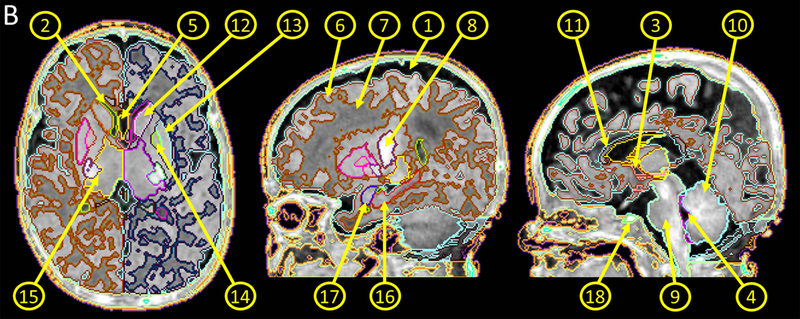
Neonatal multi-atlas repository. A: Images used to create atlases. Images with different features were intentionally selected to account for normal variations in volume, shape, and myelinaton of the brain and the variations seen in the subarachnoid fluid space, ventricles, and the cavum septum pellucidum. Atlas 1 represents a less myelinated brain with the anterior limb of the internal capsule (yellow arrow) more hypointense than the adjacent caudate and putamen. Ongoing myelination is seen in the anterior limb of the internal capsule of Atlas 7 (yellow arrow), which is hyperintense compared to the adjacent caudate and putamen. B: A representative atlas showing the parcellation map overlaid on the image. ①, subarachnoid fluid space; ②, lateral ventricle; ③, 3rd ventricle; ④, 4th ventricle, ⑤, cavum septum pellucidum; ⑥, cortical gray matter; ⑦, white matter; ⑧; myelinated white matter; ⑨; brainstem; ⑩, cerebellum; ⑪, corpus callosum; ⑫, caudate nucleus; ⑬, putamen; ⑭, globus pallidus; ⑮, thalamus; ⑯, hippocampus; ⑰; amygdala; ⑱, pituitary gland.
Table 2:
Demographics of the 177 infants included as the test dataset.
| number of boys and % | 86 | 48 |
| gestational age (weeks), mean and standard deviation | 39.34 | 1.12 |
| postmenstrual age (weeks), mean and standard deviation | 41.59 | 2.28 |
| weight at birth (kg), mean and standard deviation | 3.32 | 0.42 |
| body length at birth (cm), mean and standard deviation | 50.75 | 2.47 |
MRI scans
The whole-head, 3D, T1-weighted images were acquired using a 3.0 Tesla Siemens TIM Trio scanner (Siemens Medical Solutions, Erlangen, Germany) equipped with a 12-channel phased-array RF coil for parallel imaging. The infants were scanned without sedation. A vacuum immobilization mat (Noras MRI Products, Hoechberg, Germany) was used to minimize infant motion, and earmuffs were used to attenuate the scanner noise. Images were acquired with a three-dimensional (3D), magnetization-prepared, rapid gradient-echo (MPRAGE) sequence, with TE/TI/TR of 4.15/1400/3200 ms, a flip angle of 7°, an imaging matrix of 176 × 256 × 160, and 1 mm isotropic resolution.
Creation of a multi-atlas repository
The seven T1-weighted images were rigidly transformed to the JHU-neonate atlas33 and manually parcellated by board-certified neurologists and an image analysis specialist. The deep gray matter structures identified in our previous studies of neonatal brains33–35 were readily identified in these neonate brain images using T1-weighted contrasts. We manually parcellated these structures on each atlas, using the JHU-neonate-SS as a reference. After the deep gray matter structures were defined, we identified the cortical surface and the ventricles. Between the brain surface and the deep gray matter structures, we located the cerebral cortex and the white matter areas. We parcellated these areas based on intensity thresholding, followed by manual corrections. For these manual parcellation procedures, ROIEditor (www.Mristudio.org) was used, with inspection of all three slice orientations. A total of 38 anatomical structures were parcellated, as shown in Fig, 1 and listed in Table 3. The resultant seven T1-weighted atlases with corresponding parcellation maps were in the same space: an imaging matrix of 220 × 280 × 220 with 0.6 mm isotropic resolution.
Table. 3.
Results of the volume measurement and the leave-one-out cross validation.
| Manual-mean | Manual-SD | MALF-mean | MALF-SD | Dice-mean | Dice-SD | Jaccard-distance-mean | Jaccard-distance-SD | ICC | ICC 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gray Matter_L | 116661 | 14110 | 120976 | 15413 | 0.84 | 0.02 | 0.27 | 0.02 | 0.80 | 0.28 | to | 0.96 |
| Gray Matter_R | 119456 | 14248 | 124866 | 16072 | 0.84 | 0.02 | 0.27 | 0.03 | 0.73 | 0.12 | to | 0.95 |
| White Matter_L | 62898 | 5179 | 59336 | 1974 | 0.82 | 0.03 | 0.31 | 0.04 | 0.19 | −0.55 | to | 0.79 |
| White Matter_R | 62893 | 6037 | 58790 | 2327 | 0.81 | 0.03 | 0.32 | 0.04 | 0.14 | −0.59 | to | 0.77 |
| WM bright area_L | 627 | 286 | 375 | 107 | 0.58 | 0.12 | 0.59 | 0.11 | −0.24 | −0.78 | to | 0.56 |
| WM bright area_R | 581 | 234 | 346 | 87 | 0.60 | 0.15 | 0.55 | 0.14 | −0.36 | −0.83 | to | 0.46 |
| Corpus Callosum_L | 1154 | 384 | 1201 | 165 | 0.66 | 0.13 | 0.50 | 0.12 | 0.02 | −0.66 | to | 0.71 |
| Corpus Callosum_R | 1187 | 417 | 1295 | 261 | 0.64 | 0.15 | 0.51 | 0.14 | −0.03 | −0.69 | to | 0.69 |
| Caudate_L | 1194 | 295 | 1254 | 225 | 0.80 | 0.04 | 0.33 | 0.05 | 0.73 | 0.12 | to | 0.95 |
| Caudate_R | 1220 | 393 | 1208 | 190 | 0.77 | 0.05 | 0.38 | 0.06 | 0.43 | −0.35 | to | 0.87 |
| Putamen_L | 1233 | 129 | 1121 | 198 | 0.70 | 0.11 | 0.45 | 0.11 | 0.47 | −0.29 | to | 0.88 |
| Putamen_R | 1145 | 105 | 1011 | 104 | 0.73 | 0.07 | 0.43 | 0.08 | 0.11 | −0.61 | to | 0.75 |
| Globus Pallidus_L | 424 | 64 | 283 | 92 | 0.57 | 0.11 | 0.60 | 0.09 | −0.65 | −0.92 | to | 0.09 |
| Globus Pallidus_R | 385 | 70 | 234 | 71 | 0.57 | 0.09 | 0.60 | 0.08 | −0.67 | −0.92 | to | 0.07 |
| Thalamus_L | 4125 | 350 | 4194 | 322 | 0.88 | 0.01 | 0.21 | 0.02 | 0.73 | 0.12 | to | 0.95 |
| Thalamus_R | 4081 | 391 | 4132 | 378 | 0.87 | 0.03 | 0.23 | 0.04 | 0.63 | −0.08 | to | 0.92 |
| Hippocampus_L | 986 | 109 | 1039 | 63 | 0.76 | 0.03 | 0.39 | 0.04 | 0.51 | −0.25 | to | 0.89 |
| HIppocampus_R | 827 | 150 | 846 | 94 | 0.68 | 0.07 | 0.48 | 0.07 | 0.55 | −0.20 | to | 0.90 |
| Amygdala_L | 377 | 56 | 337 | 34 | 0.79 | 0.03 | 0.35 | 0.04 | 0.37 | −0.41 | to | 0.85 |
| Amygdala_R | 396 | 48 | 363 | 40 | 0.74 | 0.07 | 0.41 | 0.08 | 0.24 | −0.52 | to | 0.80 |
| Pituitary Gland | 90 | 50 | 89 | 18 | 0.66 | 0.26 | 0.47 | 0.25 | 0.12 | −0.60 | to | 0.76 |
| Cerebellum_L | 14009 | 2758 | 14235 | 2363 | 0.91 | 0.02 | 0.17 | 0.03 | 0.95 | 0.77 | to | 0.99 |
| Cerebellum_R | 13904 | 2622 | 13987 | 2556 | 0.90 | 0.04 | 0.18 | 0.05 | 0.97 | 0.83 | to | 0.99 |
| Brain Stem_L | 4020 | 518 | 3955 | 339 | 0.89 | 0.01 | 0.20 | 0.02 | 0.76 | 0.19 | to | 0.95 |
| Brain Stem_R | 3733 | 341 | 3795 | 484 | 0.89 | 0.02 | 0.20 | 0.03 | 0.27 | −0.50 | to | 0.82 |
| Subarachnoidal Space | 94344 | 33295 | 93903 | 25060 | 0.80 | 0.07 | 0.33 | 0.09 | 0.94 | 0.71 | to | 0.99 |
| Lateral Ventricle_L | 2923 | 1363 | 3333 | 1171 | 0.82 | 0.05 | 0.30 | 0.07 | 0.91 | 0.60 | to | 0.98 |
| Lateral Ventricle_R | 2597 | 1447 | 2906 | 1006 | 0.79 | 0.09 | 0.34 | 0.11 | 0.88 | 0.52 | to | 0.98 |
| 3rd Ventricle | 344 | 157 | 410 | 141 | 0.79 | 0.12 | 0.34 | 0.14 | 0.85 | 0.41 | to | 0.97 |
| 4th Ventricle | 550 | 207 | 594 | 157 | 0.77 | 0.07 | 0.37 | 0.08 | 0.83 | 0.35 | to | 0.97 |
| Cavum Septum Pellucidu | 166 | 271 | 143 | 152 | 0.63 | 0.23 | 0.51 | 0.19 | 0.83 | 0.35 | to | 0.97 |
L: left, R: right, WM: white matter, Manual-mean: mean of the local volume in mm3, obtained from manually drawn regions-of-interest (ROI). Manual-SD: standard deviation (SD) of the local volume in mm3, obtained from manually drawn ROI. MALF-mean: mean of the local volume in mm3, obtained from the multi-atlas label-fusion (MALF) method. MALF-SD: SD of the local volume in mm3, obtained from the MALF method. Dice-mean: mean of the Dice coefficient, obtained from comparisons between manually drawn ROI and MALF-based ROI. Dice-SD: SD of the Dice coefficient, obtained from comparisons between manually drawn ROI and MALF-based ROI. Jaccard-distance-mean: mean of the Jaccard distance, obtained from comparisons between manually drawn ROI and MALF-based ROI. Jaccard-distance -SD: SD of the Jaccard-distance, obtained from comparisons between manually drawn ROI and MALF-based ROI. ICC: intraclass correlation, obtained between volumes measured on manually drawn ROI or measured on MALF-based ROI. ICC 95% CI: 95% confidence interval of the ICC.
MALF algorithm
We applied an existing system that allows users to submit their images (target image) and to receive parcellation maps generated through the MALF procedure. There are two crucial components in the algorithms: image registration and the multi-atlas label fusion process. In the registration step, seven atlases are transformed to the target image using Large Deformation Diffeomorphic Metric Mapping (LDDMM).36–38 This process provides seven parcellated maps on the target space, each of which was derived from each atlas transformed to the target space. The seven parcellated maps are then fused through the atlas fusion step, in which local weighted voting with the joint label fusion technique, developed by the Penn Image Computing and Science Laboratory (PICSL),39,40 was adopted to generate a parcellation map for each target image. The algorithm incorporates correlations between the parcellation errors produced by any two atlases to reduce bias in the atlas set. Namely, the optimal voting weights were obtained by minimizing the expected label difference between the consensus label (obtained from weighted voting) and the label of the target image. To increase robustness against registration errors, the algorithm integrates the probabilistic correspondence model, in which each patch within the searching area is weighted by the estimated probability of the correct match.41,42 This algorithm was adopted because of its superiority in accurate delineation of anatomical boundaries and in removing atlases with inaccurate co-registration to the target image, compared to other label-fusion algorithms based on independent voting weight estimation or that relying on a single-candidate corresponding patch.39
The MRICloud platform
The MRICloud is a cloud-based architecture for neuroimage analysis tools through the web. Representational State Transfer (REST)-ful web application programming interfaces (API) are accessible from any platform that is capable of secured Hypertext Transfer Protocol (HTTP) communications. Our MALF tool was implemented in the MRICloud through the API. For the hardware, the MRICloud has been utilizing the Extreme Science and Engineering Environment (XSEDE) Computational Anatomy Science Gateway (https://www.xsede.org/gateways-overview) at the Texas Advanced Computing Center Stampede Cluster and supercomputing resources at the Maryland Advanced Research Computing Center (MARCC). The core algorithms of our MALF image analysis pipeline have been implemented to take full advantage of these computing resources with highly paralleled, multi-threaded CPU and GPU codes.
Evaluation
Leave-one-out cross-validation was performed to investigate the parcellation accuracy when the multi-atlas repository was implemented in the MALF algorithm. The Dice coefficient43 was used to evaluate the overlap, and the Jaccard distance was used to evaluate the distance between manual and automated anatomical labels. The intra-class correlation (ICC) was used to evaluate the consistency between manual and automated volume measures. The ICC calculation for a single rating was performed on an open script “icc” that runs on R (https://cran.r-project.org/web/packages/irr/index.html).
Application to test datasets
The fully-automated MALF tool was applied to two test datasets (Table 2) to quantify the volumes of the brain structures listed in Table 3. Whether the developmental trajectories obtained by our MALF method were congruent with a previous study that utilized multi-contrast (T1- and T2-weighted) MRI44 was qualitatively investigated. To account for the difference in structural boundary definition used in our approach (definition based on the JHU-neonate-SS atlas) and that used in the previous morphometry study,44 the measured regional volumes were adjusted based on the equation: (adjusted volume) = (volume measured by MALF) x (MRV24 x MTVMALF)/ (MTV24 x MRVMALF), where MRV24 represents mean of the 24 infants’ regional volume measured according to manual delineation,44 MRVMALF represents the mean of the seven atlases’ regional volume used in our study, MTV24 represents the mean of the 24 infants’ total brain volume from the prior study,44 and MTVMALF represents the mean of the seven atlases’ total brain volume used in our study. Only total brain volume and regional volumes of the cerebellum, caudate, and putamen were compared, because the developmental trajectories of these structures were elaborated with the mean and the 5th – 95th percentiles in the prior study.44
Results
Multi-atlas repository
The atlases with structure labels are shown in Fig. 1. Note that extra-axial structures, such as the skull and scalp were also parcellated to avoid the necessity of the skull-stripping step that most of the existing MALF procedures require. The defaced version (facial features of the head MRIs were removed) of the atlas repository is available through our website (http://lbam.med.jhmi.edu/).
Web interface
The MRICloud web interface was built so that the target images can be uploaded instantly with a selection of “UH-JHU-neonate_38Labels_7atlases” from the dropdown menu. The tool accepts images in Analyze 7.5 format. For the users who process clinical images in the Digital Imaging and Communications in Medicine (DICOM) format, we provide a DICOM to Analyze converter. The time required to process one image ranged from 20 minutes to six hours, depending on the number of images submitted at the same time from multiple users.
Leave-one-out cross-validation
Representative images comparing the parcellation map based on manual and MALF-based automated parcellations are demonstrated in Fig. 2. The mean and standard deviation (SD) of the measured volume of each structure, the mean Dice coefficient and the SD, and the mean Jaccard distance and the SD, are listed in Table 3, along with the ICC and the 95% confident interval (CI). In general, most of the structures indicated an excellent Dice coefficient (> 0.7)45 and Jaccard distance (< 0.5), except for the structures with a volume less than 170 mm3 [the pituitary gland and the cavum septum pellucidum], thin structures with a minimum thickness at the perpendicular slice of less than 3 mm [the corpus callosum and the myelinated WM] or structures with vague contrast to define the boundary [the globus pallidus]. The ICC of the measured volume between that based on the MALF and on the gold standard (manual delineation) varied depending on the type of structure, from excellent [subarachnoid space = 0.94, cerebellum = 0.95 (left), 0.97 (right), lateral ventricles = 0. 91 (left), 0.88 (right)], 3rd ventricle = 0.85, 4th ventricle = 0.83, cavum septum pellucidum = 0.83, cortical gray matter = 0.80 (left), 0.73 (right)], where the boundaries were mostly defined by the CSF space, to poor [corpus callosum = 0.02 (left), −0.03 (right), myelinated WM = −0.24 (left), −0.36 (right) and the globus pallidus = −0.65 (left), −0.67 (right)], where the boundary was vague on the T1-weighted image of the neonatal brain (Fig. 3).
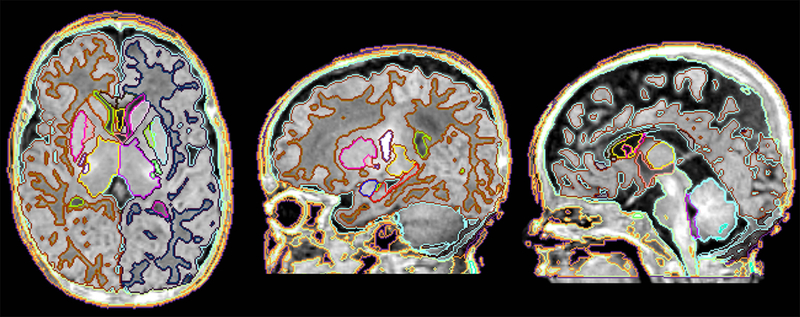
Fig. 2.
Result of the fully-automated image parcellation based on the MALF algorithm. The result was comparable to that based on the gold-standard manual parcellation (Fig. 1B).
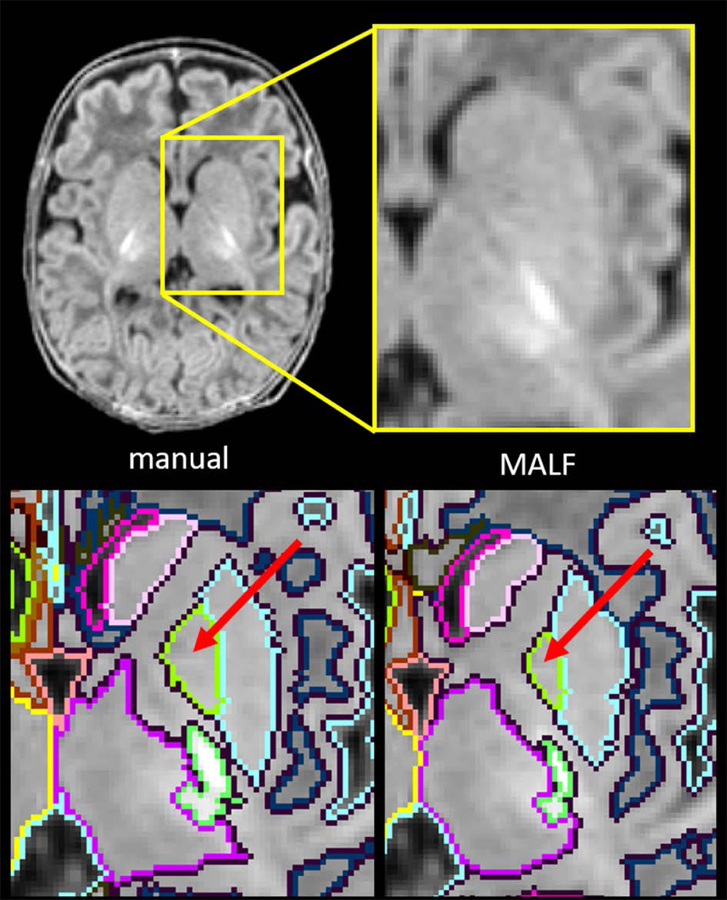
Fig. 3.
Comparison between manual parcellation (lower left) and the MALF-based image parcellation (lower right). The deep brain area, surrounded by the yellow rectangle (upper left) with the magnified view (upper right), indicated that the contrast between the globus pallidus (red arrows) and the surrounding structures was obscure.
Normal developmental curves of early brain development
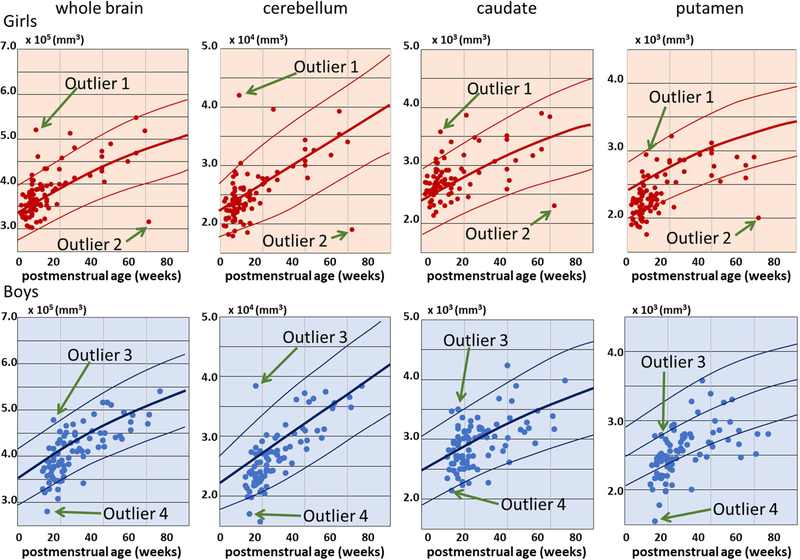
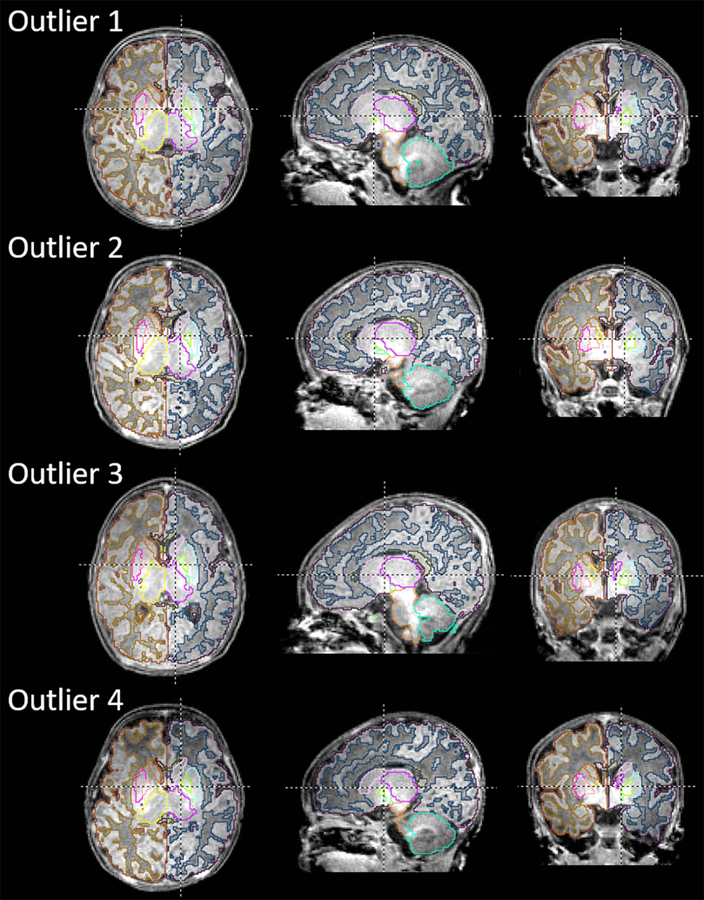
Total brain volume and adjusted volumes of representative structures (cerebellum, caudate, and putamen) quantified through the MALF tool were plotted against the postmenstrual age (PMA) for each infant, as shown in Fig. 4, which clearly demonstrated the chronologically increasing volumes in these structures. The results were comparable with those reported from the previous publication. Note that most of the measured volumes were plotted within the 5 – 95% range of the normal growth trajectories reported in the previous study,44 although a systematic downward bias (approximately 400 mm3 smaller than those reported previously44) was observed in the putamen. There were several infants in whom local brain volumes, as well as the total brain volume, were large or small for their age. Some of these infants are labeled as Outliers 1 – 4 in Fig. 4 and demonstrated in Fig. 5, showing that brain parcellation was appropriately performed even for these outliers.
Fig. 4.
Volume measures obtained from the MALF-based image parcellation. Each plot represents each infant. Upper row: volume of girls’ brains. Lower row: volume of boys’ brains. The measured structures were (from left to right): the whole brain parenchyma; the cerebellum; the caudate; and the putamen. The three solid lines overlaid on each scattergram represent the 95th percentile, the 50th percentile, and the 5th percentile of the volume measures, reported by44 (adapted from the eFigures 3 and 6 with written permission).
Fig. 5.
Images with a brain volume more than the 95th percentile (Outliers 1 and 3) or less than the 5th percentile (Outliers 2 and 4). The structural boundaries were automatically generated through the MALF-based image parcellation and overlaid on the corresponding images.
Discussion
Although MALF-based parcellation has become a common practice during the past decade, multi-atlas repositories for the neonatal brain that are available for public use are still scarce.19 Over the past two decades, we have built multiple atlases and created atlas libraries with more than 200 atlases that provide coverage between 2 to 95 years of age20 (https://braingps.mricloud.org/atlasrepo). This study is an extension of our effort toward building user-friendly image analysis tools for even younger-age subjects, which is becoming more prevalent and in higher demand, both in research and in clinical practice.
The leave-one-out cross validation was applied to evaluate the accuracy of the MALF-based, fully-automated image parcellation. Note that the validation was performed on seven atlases that were intentionally selected to cover major developmental and anatomical variations seen in this age-range. This variation was disadvantageous for the accuracy measurement because none of the six images were similar to the selected image. Nevertheless, the MALF-based parcellation did achieve excellent agreement45 with the manual parcellation, as shown by the high Dice coefficient and the ICC, and low Jaccard distance. The result was comparable to an existent label propagation and fusion method based on T1-weighted contrast, which demonstrated a Dice coefficient, averaged across regions, of 0.81.46 We also tested the applicability of the MALF-based parcellation on the cohort of 177 typically developing term-born infants, and found that the local brain volumes measured were comparable to those based on T2-weighted contrast or a combination of T1- and T2-weighted contrasts. However, the effect of including a T2-weighted image, compared to the T1-weighted image only, on the accuracy of image parcellation is yet to be investigated. Ongoing infant brain imaging projects, such as the baby connectome project and the development of the human connectome project,28,29 might provide opportunities to investigate the effects of adding T2-weighted contrast to the parcellation accuracy.
We are aware of several weaknesses of the MALF-based method. The Dice coefficient was lower and the Jaccard distance was higher for small (< 170 mm2) or thin (thickness < 3mm) structures, compared to other structures, because a small shift in the structural boundary (e.g., one pixel) of such structures has a substantial impact on the Dice coefficient and the Jaccard distance. We are also aware that the ICC was affected by the clarity of the structural boundaries. This issue was typically seen is the putamen, of which the medial boundary with the internal capsule and the lateral boundary with the putamen are vague, and the boundary definition is dependent on a priori knowledge about the brain anatomy (Fig. 3). For such structures, the ICC between the MALF approach and manual delineation is difficult to evaluate for two reasons. First, the parcels generated by the MALF approach are less accurate compared to the structures with clear anatomical boundaries, because the image contrast is used for both image registration and label fusion steps. Second, the manual parcel delineation is less stable for such structures, compared to structures with clear anatomical boundaries. Another potential issue is the age range to which the MALF-based method can be reliably applied. Although the result indicated that the seven atlases cover the anatomical features of the ages 37 – 47 weeks PMA, the upper- and lower-limits are still unknown. The creation of a tool that covers a wider age-range is particularly important to quantify images from a longitudinal cohort study; therefore, continuous effort is needed to create atlases outside this age range and to create an algorithm to incorporate an age to select the appropriate set of atlases.
The major motivation that drove this project is the demand from clinician scientists who wanted to quantify their patients’ clinical 3D- T1-weighted images, scanned at the infantile period, to see correlations with the prognosis at school age or later. In such a study, they need to analyze legacy images (typically scanned more than 10 years ago) when only T1-weighted anatomical images were available for a volumetric study. We implemented the MALF tool on the MRICloud because it offers several substantial advantages, which are particularly suitable for clinician scientists with limited computational resources and time available for image analysis. First, the web-based interface provides a platform-free environment, which means that users do not have to spend time downloading, updating, or compiling the programs in order to perform the image processing. Second, the users do not need to own a high-end computer to process their images, because the computational resources are in the cloud: the system monitors the data queues, send the data to available supercomputers, and utilizes CPU/GPU resources. Third, the cloud platform provides users with novel or updated algorithms, with minimal changes in the image-submission interface with which they are familiar. Image processing algorithms are rapidly developing, and it is highly possible that existing algorithms will become outdated in the near future. For the majority of clinician scientists, it is not practical to keep getting used to the new tools that become available every month. The use of a cloud system is beneficial for both users and developers because incorporating additional algorithms from other labs is straightforward. For example, one of the core technologies that support this study is the MALF technology developed by PICSL (Picsl_upenn), which was among the most accurate image segmentation algorithms demonstrated at the 2012 workshop organized by the Medical Image Computing and Computer-Assisted Intervention (MICCAI) society, the Multi-Atlas Labeling workshop, and the Neonatal Brain Segmentation 2012 (NeoBrainS12) challenge. Although the Picsl_upenn has been rigorously tested for the past six years, and is, therefore, highly reliable, recent advancements in this field may improve that MALF algorithm or provide an even better algorithm that uses a multi-atlas repository as a training dataset to parcellate target images. An example of this approach was demonstrated in the Grand Challenge on MR Brain Segmentation (MRBrainS18) at MICCAI 2018 (mrbrains18.isi.uu.nl). The MRICloud platform allows developers to implement the latest algorithms through the API whenever needed.
One of the major strengths of our approach is the compatibility with high-throughput analysis. The majority of existing automated image parcellation algorithms requires image pre-processing routines, including skull-stripping, a process by which the brain is extracted from a head image, which itself is still an active research field.47–53 The “human-in-the-loop” quality control process is almost always required to ensure the accuracy of the skull-stripping. While such a quality control process is reasonable for scientific research that targets a limited number of images (typically less than several hundred image datasets), it becomes unrealistically time-consuming in high-throughput, big-data analysis. In our approach, the whole head, instead of the whole brain, is parcellated based on the MALF algorithm. This approach is similar to the multi-atlas skull-stripping method,47 which is known as one of the most accurate methods for skull-stripping. Our MALF is suitable for handling big data, such as a clinical database that typically contains a huge amount of legacy data.
For clinical applications, the robustness and flexibility to variations in scan protocols and scanners are important. Contrary to a clinical research study that uses a single scanner and scanning protocol, clinical MRIs contain artefactual heterogeneity caused by variations in the scanner type and differences in imaging protocols. Moreover, clinical MRIs often include brains with substantial pathological changes in volume or intensity, or both, or even mass lesions that may distort the geometry of the brain. Further testing is still needed to evaluate the robustness of our method with regard to the heterogeneity of MRI protocols and scanners and potential pathological changes. In such a validation study, the involvement of multiple institutes is essential. The MRICloud platform is ideal in such a multi-institutional study because the only requirement is access to the web-browser. Using MRIs from multiple institutes and observing the robustness with regard to the differences and the sensitivity to biological variations will be essential in testing the feasibility of our method for large-scale clinical applications.
Acknowledgments
Parts of the preliminary results of this study were presented at the XXIII World Congress of Neurology. This work was made possible by grants R01HD065955, 2K24DA16170, and U54NS056883 from the National Institutes of Health (NIH). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official view of the NIH. The authors are grateful to the families of our research participants, the pediatricians/neonatologists who referred the participants (Dr. Lillian Fujimoto, Dr. Lois Chiu, and Dr. Joseph Hudak), and our dedicated research staff (Steven Buchthal, Eric Cunningham, Daniel Alicata, Heather Johansen, Antonette Hernandez, Robyn Yamakawa, Sara Hayama, Tamara Andres), who assisted with the data collection. We also thank our board-certified neuroradiologist, Dr. Doris Lin, for her radiological reading of the scans, Dr. Kunihiro Matsushita for his statistical advice, and Ms. Mary McAllister for help with manuscript editing. Dr. Miller and Dr. Mori are the co-founders, and Dr. Oishi is a consultant, for Anatomy Works.
Footnotes
Disclosure
The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- 1.Hasan KM, Walimuni IS, Abid H, Datta S, Wolinsky JS, Narayana PA. Human brain atlas-based multimodal mri analysis of volumetry, diffusimetry, relaxometry and lesion distribution in multiple sclerosis patients and healthy adult controls: Implications for understanding the pathogenesis of multiple sclerosis and consolidation of quantitative mri results in ms. J Neurol Sci 2012;313:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshpande R, Chang L, Oishi K. Construction and application of human neonatal dti atlases. Front Neuroanat 2015;9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickie DA, Shenkin SD, Anblagan D, et al. Whole brain magnetic resonance image atlases: A systematic review of existing atlases and caveats for use in population imaging. Front Neuroinform 2017;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irimia A, Wang B, Aylward SR, et al. Neuroimaging of structural pathology and connectomics in traumatic brain injury: Toward personalized outcome prediction. Neuroimage Clin 2012;1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage 2009;46:726–38. [DOI] [PubMed] [Google Scholar]
- 6.Artaechevarria X, Munoz-Barrutia A, Ortiz-de-Solorzano C. Combination strategies in multi-atlas image segmentation: Application to brain mr data. IEEE Trans Med Imaging 2009;28:1266–77. [DOI] [PubMed] [Google Scholar]
- 7.Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. NeuroImage 2010;51:221–7. [DOI] [PubMed] [Google Scholar]
- 8.Pipitone J, Park MT, Winterburn J, et al. Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 2014;101:494–512. [DOI] [PubMed] [Google Scholar]
- 9.Tang X, Yoshida S, Hsu J, et al. Multi-contrast multi-atlas parcellation of diffusion tensor imaging of the human brain. PLoS One 2014;9:e96985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (staple): An algorithm for the validation of image segmentation. IEEE Trans Med Imaging 2004;23:903–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zu C, Wang Z, Zhang D, et al. Robust multi-atlas label propagation by deep sparse representation. Pattern Recognit 2017;63:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Z, He X, Ceritoglu C, et al. Evaluation of cross-protocol stability of a fully automated brain multi-atlas parcellation tool. PLoS One 2015;10:e0133533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erus G, Doshi J, An Y, Verganelakis D, Resnick SM, Davatzikos C. Longitudinally and inter-site consistent multi-atlas based parcellation of brain anatomy using harmonized atlases. Neuroimage 2018;166:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mott SH, Bodensteiner JB, Allan WC. The cavum septi pellucidi in term and preterm newborn infants. J Child Neurol 1992;7:35–8. [DOI] [PubMed] [Google Scholar]
- 15.Pauling KJ, Bodensteiner JB, Hogg JP, Schaefer GB. Does selection bias determine the prevalence of the cavum septi pellucidi? Pediatr Neurol 1998;19:195–8. [DOI] [PubMed] [Google Scholar]
- 16.Gur RE, Kaltman D, Melhem ER, et al. Incidental findings in youths volunteering for brain mri research. AJNR Am J Neuroradiol 2013;34:2021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JJ, Chen CJ, Chang HF, Chen DL, Hsu YC, Chang TP. Prevalence of cavum septum pellucidum and/or cavum vergae in brain computed tomographies of taiwanese. Acta Neurol Taiwan 2014;23:49–54. [PubMed] [Google Scholar]
- 18.Macpherson P, Teasdale E. Ct demonstration of a 5th ventricle--a finding to ko boxers? Neuroradiology 1988;30:506–10. [DOI] [PubMed] [Google Scholar]
- 19.Oishi K, Chang L, Huang H. Baby brain atlases. Neuroimage 2018. [DOI] [PMC free article] [PubMed]
- 20.Wu D, Ma T, Ceritoglu C, et al. Resource atlases for multi-atlas brain segmentations with multiple ontology levels based on t1-weighted mri. Neuroimage 2016;125:120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gousias IS, Edwards AD, Rutherford MA, et al. Magnetic resonance imaging of the newborn brain: Manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage 2012;62:1499–509. [DOI] [PubMed] [Google Scholar]
- 22.Blesa M, Serag A, Wilkinson AG, et al. Parcellation of the healthy neonatal brain into 107 regions using atlas propagation through intermediate time points in childhood. Front Neurosci 2016;10:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander B, Murray AL, Loh WY, et al. A new neonatal cortical and subcortical brain atlas: The melbourne children’s regional infant brain (m-crib) atlas. Neuroimage 2017;147:841–51. [DOI] [PubMed] [Google Scholar]
- 24.Otsuka Y, Chang L, Skranes J, Ernst T, Oishi K. Neonatal brain mri multi-atlas repository for automated image quantification. XXIII World Congress of Neurology. Kyoto, Japan; 2017. [Google Scholar]
- 25.Dong P, Guo Y, Shen D, Wu G. Multi-atlas and multi-modal hippocampus segmentation for infant mr brain images by propagating anatomical labels on hypergraph. Patch Based Tech Med Imaging (2015) 2015;9467:188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Wang L, Shi F, Lin W, Shen D. Multi-atlas based simultaneous labeling of longitudinal dynamic cortical surfaces in infants. Med Image Comput Comput Assist Interv 2013;16:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanroma G, Benkarim OM, Piella G, et al. Learning to combine complementary segmentation methods for fetal and 6-month infant brain mri segmentation. Comput Med Imaging Graph 2018;69:52–9. [DOI] [PubMed] [Google Scholar]
- 28.Makropoulos A, Robinson EC, Schuh A, et al. The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. Neuroimage 2018;173:88–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howell BR, Styner MA, Gao W, et al. The unc/umn baby connectome project (bcp): An overview of the study design and protocol development. Neuroimage 2019;185:891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akazawa K, Chang L, Yamakawa R, et al. Probabilistic maps of the white matter tracts with known associated functions on the neonatal brain atlas: Application to evaluate longitudinal developmental trajectories in term-born and preterm-born infants. NeuroImage 2016;128:167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang L, Akazawa K, Yamakawa R, et al. Delayed early developmental trajectories of white matter tracts of functional pathways in preterm-born infants: Longitudinal diffusion tensor imaging data. Data Brief 2016;6:1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D, Chang LD, Akazawa K, et al. Mapping the critical gestational age at birth that alters brain development in preterm-born infants using multi-modal mri. Neuroimage 2017;149:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oishi K, Mori S, Donohue PK, et al. Multi-contrast human neonatal brain atlas: Application to normal neonate development analysis. NeuroImage 2011;56:8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oishi K, Faria AV, Yoshida S, Chang L, Mori S. Quantitative evaluation of brain development using anatomical mri and diffusion tensor imaging. Int J Dev Neurosci 2013;31:512–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Chang L, Ceritoglu C, et al. A bayesian approach to the creation of a study-customized neonatal brain atlas. NeuroImage 2014;101:256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granander U, Miller MI. Computational anatomy: An emerging discipline. Statistical computing and graphics newsletter 1996;7:3–8. [Google Scholar]
- 37.Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans Med Imaging 1997;16:864–77. [DOI] [PubMed] [Google Scholar]
- 38.Miller M, Banerjee A, Christensen G, et al. Statistical methods in computational anatomy. Stat Methods Med Res 1997;6:267–99. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Pouch A, Takabe M, et al. Multi-atlas segmentation with robust label transfer and label fusion. Inf Process Med Imaging 2013;23:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell 2013;35:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coupe P, Manjon JV, Fonov V, Pruessner J, Robles M, Collins DL. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. Neuroimage 2011;54:940–54. [DOI] [PubMed] [Google Scholar]
- 42.Asman AJ, Landman BA. Non-local staple: An intensity-driven multi-atlas rater model. Med Image Comput Comput Assist Interv 2012;15:426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dice LR. Measures of the amount of ecological association between species. Ecology 1945;26:297–302. [Google Scholar]
- 44.Holland D, Chang L, Ernst TM, et al. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol 2014;71:1266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in mr images: Method and validation. IEEE Trans Med Imaging 1994;13:716–24. [DOI] [PubMed] [Google Scholar]
- 46.Gousias IS, Hammers A, Counsell SJ, et al. Magnetic resonance imaging of the newborn brain: Automatic segmentation of brain images into 50 anatomical regions. PLoS One 2013;8:e59990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-atlas skull-stripping. Acad Radiol 2013;20:1566–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. Neuroimage 2001;13:856–76. [DOI] [PubMed] [Google Scholar]
- 49.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002;17:143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 2011;30:1617–34. [DOI] [PubMed] [Google Scholar]
- 51.Mikheev A, Nevsky G, Govindan S, Grossman R, Rusinek H. Fully automatic segmentation of the brain from t1-weighted mri using bridge burner algorithm. J Magn Reson Imaging 2008;27:1235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahapatra D Skull stripping of neonatal brain mri: Using prior shape information with graph cuts. J Digit Imaging 2012;25:802–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadananthan SA, Zheng W, Chee MW, Zagorodnov V. Skull stripping using graph cuts. Neuroimage 2010;49:225–39. [DOI] [PubMed] [Google Scholar]