Abstract
Schizophrenia (SCZ) and bipolar disorder (BD) are severe mental disorders associated with cognitive impairment, which is considered a major determinant of functional outcome. Despite this, the etiology of the cognitive impairment is poorly understood, and no satisfactory cognitive treatments exist. Increasing evidence indicates that genetic risk for SCZ may contribute to cognitive impairment, while the genetic relationship between BD and cognitive function remains unclear. Here, we combined large genome-wide association study data on SCZ (n=82,315), BD (n=51,710) and general intelligence (n=269,867) to investigate overlap in common genetic variants using conditional false discovery rate (condFDR) analysis. We observed substantial genetic enrichment in both SCZ and BD conditional on associations with intelligence indicating polygenic overlap. Using condFDR analysis, we leveraged this enrichment to increase statistical power and identified 75 distinct genomic loci associated with both SCZ and intelligence, and 12 loci associated with both BD and intelligence at conjunctional FDR<0.01. Among these loci, 20 are novel for SCZ, and four are novel for BD. Most SCZ risk alleles (61 of 75, 81%) were associated with poorer cognitive performance, whereas most BD risk alleles (9 of 12, 75%) were associated with better cognitive performance. A gene-set analysis of the loci shared between SCZ and intelligence implicated biological processes related to neurodevelopment, synaptic integrity and neurotransmission; the same analysis for BD was underpowered. Altogether, the study demonstrates that both SCZ and BD share genetic influences with intelligence, albeit in a different manner, providing new insights into their genetic architectures.
Introduction
Schizophrenia (SCZ) and bipolar disorder (BD) are severe psychiatric disorders recognized as leading causes of morbidity and mortality in the world.1,2 SCZ and BD share many clinical features,3 including disturbances in mood, thought, perception and social functioning, and they are often accompanied by cognitive impairment.4,5 The cognitive deficits are consistently found to be more severe in SCZ than BD.6–8 A wide range of cognitive domains are affected in both SCZ6,7,9,10 and BD,6,7,11–14 including executive function, verbal learning, processing speed, and memory, as well as general intelligence. The general factor of intelligence, denoted as g, is a latent trait that captures around 40–50% of the shared variance across diverse cognitive abilities.15,16 It is noteworthy, however, that cognitive dysfunction is not a prerequisite of either SCZ or BD, at least in terms of traditional definitions of cognitive dysfunction.8 Many individuals with SCZ and BD perform within the normal range of cognitive functioning,6,9,17 and superior intelligence occurs among individuals with both disorders, although more frequently in BD.18–20 Furthermore, both higher and lower premorbid cognitive performance have been linked to increased BD risk.20–22 In SCZ, however, cognitive underperformance often precedes the onset of psychotic symptoms and subsequent diagnosis of SCZ by many years.8,23,24 Accumulating evidence indicates that intelligence is a major determinant for many socioeconomic and health-related outcomes in the general population.25,26 Additionally, the degree of cognitive underperformance is a key predictor of functional and treatment outcome in both SCZ6,27 and BD.6,28,29 Despite this, there are currently no psychiatric treatments available that effectively amend cognitive impairment,4,5 and the limited mechanistic understanding prevents the development of novel effective therapies.
SCZ, BD and intelligence are complex, heterogeneous phenotypes under strong genetic control, with heritability estimates ranging between 0.6 and 0.8 for SCZ and BD,30 and around 0.5 for cognitive abilities.31 There is abundant genetic overlap between SCZ and BD,32 in accordance with their high degree of clinical overlap.3 Results from family and twin-studies indicate that genetic liabilities of SCZ and cognitive abilities covary,33–35 and molecular genetic studies have implicated rare and common alleles influencing both SCZ and cognitive abilites.36,37 Family studies have found that cognitive impairments are more common among unaffected relatives of patients with BD than in healthy controls,13,38–40 suggesting that genetic susceptibility to BD may also contribute to cognitive dysfunction. Yet, whereas genome-wide association studies (GWAS) analyses have consistently found significant negative genome-wide correlations between SCZ and cognitive abilities (rg ranging between −0.2 to −0.4),41–48 most studies have found no genome-wide correlations between BD and cognitive abilites.41–49 One study did find a significant genome-wide correlation between BD and a measure of memory,41 but the latter shows low test-retest correlation,50 low single nucleotide polymorphism (SNP)-heritability,51 and no genetic correlation with educational attainment,41 suggesting that its genetic architecture may be different from that of other cognitive traits.
Here we aimed to provide further insights into the genetic relationship between SCZ, BD and intelligence by analyzing summary data from recent large GWAS on SCZ (n=82,315),52 BD (n=51,710),49 and general intelligence (n=269,867).44 In these GWAS, 108 genomic loci were associated with SCZ at the genome-wide significance level,52 30 loci were associated with BD,49 and 205 loci were associated with intelligence.44 Among the loci, 24 were jointly associated with SCZ52 and intelligence,44 with SCZ risk linked to poorer cognitive performance at 18 loci. There was a significant moderate negative genetic correlation between SCZ and intelligence (rg=–0.21, p= 3.82 × 10−17).44 Four genome-wide significant loci were jointly associated with BD49 and intelligence,44 with BD risk linked to poorer cognitive performance at three loci. In line with prior studies,41–48 the genetic correlation between BD and intelligence was non-significant (rg=−0.05, p=0.08).49 Despite the success of these GWAS to uncover trait-associated variants, large fractions of the polygenic architectures underlying SCZ, BD, and intelligence still remain to be uncovered.44,49,52 To identify additional common genetic variants jointly influencing these phenotypes, we applied a conditional false discovery rate (condFDR) approach.53,54 This method builds on an empirical Bayesian statistical framework, and combines GWAS summary data to increase statistical power to detect SNPs that did not reach genome-wide significance.53,54 The condFDR approach does not require a genetic correlation to improve discovery, but leverages systematic co-localization of SNP associations to prioritize likely pleiotropic SNPs.55 To our knowledge, there are no previous conditional GWAS studies comparing BD and intelligence, while a recent condFDR study identified 21 genomic loci shared between SCZ and different cognitive abilities, where most (18 out of 21) SCZ risk alleles were associated with poorer cognitive performance.36 Applying the same statistical approach to larger GWAS samples,44,49,52 we here investigated polygenic overlap between SCZ, BD and intelligence.
Methods
Participant Samples
We obtained GWAS results in the form of summary statistics (p-values and z-scores).44,49,52 Data on SCZ and BD were acquired from the Psychiatric Genomics Consortium. The SCZ dataset consisted of 49 case control samples (34,241 cases with SCZ or schizoaffective disorder and 45,604 controls) and 3 family based association studies (1,235 parent affected-offspring trios).52 The BD dataset consisted of 20,352 cases and 31,358 controls from 32 cohorts.49 Among the cases, 14,879 individuals had a diagnosis of BD type I (BD1), 3,421 had BD type II (BD2), 977 had schizoaffective disorder, bipolar type (SAB), and the remaining unspecified BD.49 Data on general intelligence were based on 269,867 individuals drawn from 14 cohorts, primarily consisting of data from the UK Biobank (n=195,653).44 The cohorts contributing to the intelligence GWAS either calculated Spearman’s g or used a primary measure of fluid intelligence that correlates highly with g.44,56 All GWAS investigated in the present study were approved by the local ethics committees, and informed consent was obtained from all participants. The Norwegian Institutional Review Board for the South-East Norway Region has evaluated the current protocol and found that no additional institutional review board approval was needed because no individual data were used. For details, see Supplementary Methods and the original publications.44,49,52
Statistical analysis
To provide a visual pattern of overlap in SNP associations, we constructed conditional quantile-quantile (Q-Q) plots. The conditional Q-Q plots compare the association with a primary trait across all SNPs and within SNPs strata determined by their association with the secondary trait. Pleiotropic enrichment exists if the proportion of SNPs associated with a phenotype increases as a function of the strength of the association with a secondary phenotype. In conditional Q-Q plots, this enrichment is visualized as successive leftward deflections from the null distribution, and can be directly interpreted in terms of the true discovery rate (1-FDR).53–55 To improve the discovery of genetic variants associated with SCZ, BD and intelligence, we applied a condFDR statistical framework.53–55 The condFDR is an extension of the standard FDR, that re-ranks the test-statistics of a primary phenotype based on a conditional variable, in this case the strength of the association with a secondary phenotype.53–55 Inverting the roles of primary and secondary phenotypes provides the inverse condFDR value. The conjunctional FDR (conjFDR), defined in turn as the maximum of the two condFDR values, provides a conservative estimate of the FDR for association with both phenotypes. P-values were corrected for inflation using a genomic inflation control procedure.53 All code used for carrying out the described analyses is available upon request from the corresponding author. All analysis was performed after excluding SNPs in the major extended histocompatibility complex and 8p23.1 regions. For details, see Supplementary Methods.
Genomic loci definition
We defined independent genomic loci according to the FUMA57 protocol. FUMA is an online platform for functional mapping of genetic variants (http://fuma.ctglab.nl/).57 First, we identified independent significant SNPs as SNPs with condFDR<0.01 and independent from each other at r2<0.6. A subset of these in approximate linkage equilibrium with each other at r2<0.1 were then selected as lead SNPs. To define distinct genomic loci, we merged any physically overlapping lead SNPs (LD blocks<250kb apart). The borders of the genomic loci were defined by identifying all SNPs in LD (r2≧0.6) with one of the independent significant SNPs in the locus. The region containing all of these candidate SNPs was considered to be a single independent genomic locus. All LD information was calculated from the 1000 Genomes Project reference panel.58 We evaluated the directional effects of the loci shared between SCZ, BD and intelligence by comparing their z-scores and odds ratios.
Functional annotation
Using FUMA,57 we functionally annotated all candidate SNPs in the genomic loci with a condFDR or conjFDR value <0.10 having an LD r2≧0.6 with one of the independent significant SNPs. SNPs were annotated with Combined Annotation Dependent Depletion (CADD)59 scores, which predict how deleterious the SNP effect is on protein structure/function, RegulomeDB60 scores, which predict likelihood of regulatory functionality, and chromatin states, which predict transcription/regulatory effects from chromatin states at the SNP locus.61,62 We also identified previously reported GWAS associations in the NHGRI-EBI catalog63 overlapping with the identified loci. Moreover, we used FUMA57 to evaluate gene ontology (GO)64 gene-set enrichment for the genes nearest the identified shared loci. Finally, we used data from the genotype tissue expression (GTEx) resource,65 to determine gene expression and assess expression quantitative trait locus (eQTL) functionality of likely regulatory lead SNPs. All analyses were corrected for multiple comparisons. For details, see Supplementary Methods.
Results
We observed successive increments of SNP enrichment for SCZ and BD as a function of the significance of the associations with intelligence (Figure 1). This indicates polygenic overlap between the phenotypes. The reverse conditional Q-Q plots demonstrate consistent enrichment in intelligence given associations with SCZ and BD (Supplementary Figure 1). To increase statistical power, we leveraged this pleiotropic enrichment using condFDR analysis and re-ranked SCZ and BD SNPs conditional on their association with intelligence, and vice versa. At condFDR<0.01, we identified 236 loci associated with SCZ and 48 loci associated with BD conditional on their association with intelligence (Supplementary Tables 1–2). Next, we identified 337 loci associated with intelligence conditional on SCZ and 283 loci conditional on BD at condFDR<0.01 (Supplementary Tables 3–4). Altogether, we identified 138 SCZ loci, 31 BD loci, and 165 intelligence loci that were not identified in the original GWAS,44,49,52 demonstrating the improved power for SNP discovery gained by combining GWAS in a condFDR framework.
Figure 1.
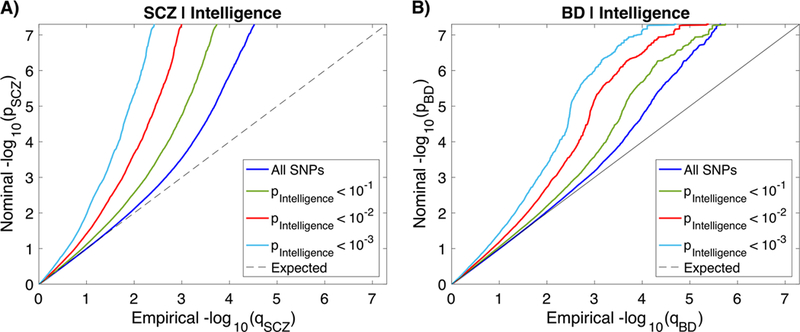
Polygenic overlap between schizophrenia (SCZ), bipolar disorder (BD) and intelligence. Conditional Q-Q plots of nominal versus empirical –log10 p-values (corrected for inflation) in A) SCZ and B) BD below the standard GWAS threshold of p<5×10−8 as a function of significance of association with intelligence, at the level of p ≤ 0.1, p ≤ 0.01, p ≤ 0.001, respectively. The blue lines indicate all SNPs. The dashed lines indicate the null hypothesis.
A total of 75 distinct genomic loci were jointly associated with SCZ and intelligence at conjFDR<0.01 (Figure 2A; Supplementary Table 5). 42 of these loci were not identified in the original SCZ GWAS.52 22 of the 42 were however reported in other SCZ GWAS according to the NHGRI-EBI catalog (Supplementary Table 6), yielding a total number of 20 novel SCZ risk loci among the shared loci.63 Further, 37 of the top lead SNPs in these loci were associated with BD at p<0.05. As denoted by the sign of the effect sizes, most of the SCZ risk alleles (61 out of 75; Supplementary Table 5) were associated with cognitive underperformance, corroborating prior findings.36 We also identified 12 distinct loci shared between BD and intelligence at conjFDR<0.01 (Figure 3A; Supplementary Table 7). Eight of these were not identified in the original BD GWAS.49 Four of the eight had however been identified in prior BD GWAS (Supplementary Table 8), yielding a total of four novel BD risk loci among the shared loci.63 Eight of the lead SNPs in these loci were associated with SCZ at p<0.05. Although SNPs near SRPK2 on chromosome 7 reached genome-wide significance in both GWAS on BD49 and intelligence,44 no SNPs in this locus were jointly associated with these phenotypes at conjFDR<0.01 but rs9655780 came close (conjFDR=0.012; Supplementary Table 9). At the shared loci, 9 out of 12 BD risk alleles were associated with higher cognitive performance (Supplementary Table 7). However, among the shared loci at conjFDR<0.05, only 40 out of 79 (51%) loci had concordant effects on BD risk and intelligence (Supplementary Table 9). To visualize the distribution of the shared variants, we constructed ‘conjFDR Manhattan plots’ where all SNPs without pruning are shown, and the independent significant lead SNPs are encircled in black (Figures 2A and 3A).
Figure 2.
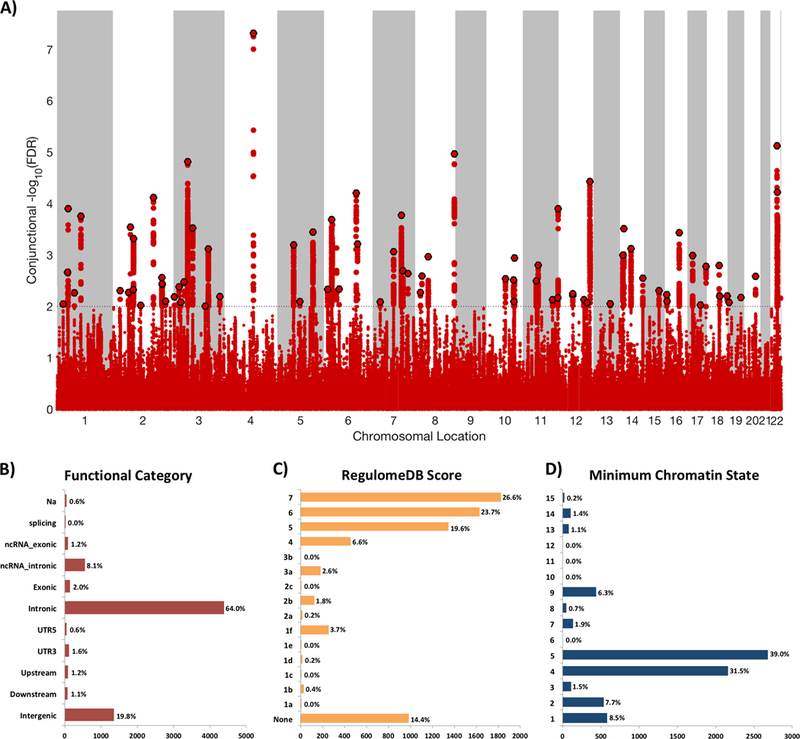
A) Common genetic variants jointly associated with schizophrenia (n=82,315) and intelligence (n=269,867) at conjunctional false discovery rate (conjFDR) <0.01. Manhattan plot showing the -log10 transformed conjFDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal line represents the threshold for significant shared associations (conjFDR<0.01, ie -log10(conjFDR)>2.0). Independent lead SNPs are encircled in black. The significant shared signal in the major histocompatibility complex region (chr6:25119106–33854733) is represented by one independent lead SNP. Further details are provided in Supplementary Table 5. B) Distribution of functional consequences of SNPs in the shared genomic risk loci. C) Distribution of RegulomeDB score for SNPs in shared genomic loci, with a low score indicating a higher likelihood of having a regulatory function. D) The minimum chromatin state across 127 tissue and cell types for SNPs in shared genomic loci, with lower states indicating higher accessibility and states 1–7 referring to open chromatin states.
Figure 3.
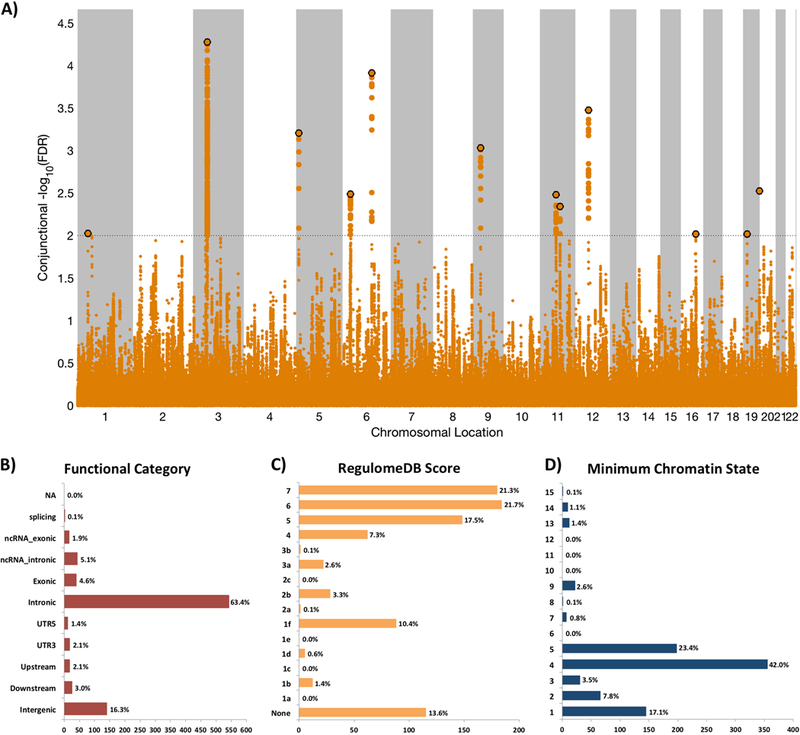
A) Common genetic variants jointly associated with bipolar disorder (n=59,315) and intelligence (n=269,867) at conjunctional false discovery rate (conjFDR) <0.01. Manhattan plot showing the -log10 transformed conjFDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal line represents the threshold for significant shared associations (conjFDR<0.01, ie -log10 (conjFDR)>2.0). Independent lead SNPs are encircled in black. Further details are provided in Supplementary Table 7. B) Distribution of functional consequences of SNPs in the shared genomic risk loci. C) Distribution of RegulomeDB score for SNPs in shared genomic loci, with a low score indicating a higher likelihood of having a regulatory function. D) The minimum chromatin state across 127 tissue and cell types for SNPs in shared genomic loci, with lower states indicating higher accessibility and states 1–7 referring to open chromatin states.
Functional annotation57 of all SNPs having a conjFDR value<0.10 in the loci shared between SCZ and intelligence (n=6853; Figures 2B–D) demonstrated that these were mostly intronic and intergenic, while 2.0% were exonic (Supplementary Table 10). Of the 75 top lead SNPs in the loci shared between SCZ and intelligence, 40 were located inside a protein-coding gene and 11 inside a non-coding RNA (Supplementary Table 5). Of the 75 top lead SNPs, two SNPs (rs11695125, rs1805645) had CADD scores above 12.37, the threshold suggested to signify deleteriousness,59 and one SNP (rs5751191) had a RegulomeDB60 score of 1f suggesting that it was likely affecting binding sites (Supplementary Table 5). We followed up this finding using the GTEx database,65 and found that rs5751191 was significantly associated with eQTL functionality in different brain regions for genes CYP2D6, NAGA, WBP2NL and RP4–669P10.16 (Supplementary Table 11). Using GTEx65 data, we found that the genes nearest the 75 shared loci were significantly overexpressed in multiple brain regions (Supplementary Figures 2–3). GO gene-set analysis for these genes revealed 32 significantly associated biological processes, the most strongly associated being “regulation of neuron differentiation”, “regulation of cell development”, “neurogenesis”, “modulation of synaptic transmission”, and “regulation of receptor binding” (Supplementary Table 12). Further, the genes were significantly associated with 10 cellular component gene-sets, including “neuronal projection”, “the synapse”, and “the anchored part of membranes”, as well as six molecular function gene sets, the most strongly associated being “GABA receptor binding” (Supplementary Table 12).
We also functionally annotated all SNPs having a conjFDR value<0.10 in the loci shared between BD and intelligence (n=846; Figures 3B–D). 4.6% were exonic and most of the others were intronic or intergenic (Supplementary Table 13). Of the 12 top lead SNPs in the loci shared between BD and intelligence, seven were located inside a protein-coding gene and one inside a non-coding RNA (Supplementary Table 7). One of the top SNPs (rs60144015; intronic variant within FOXO6) had a CADD score of 18.65 suggesting deleteriousness (Supplementary Table 7). The genes nearest the 12 shared loci were significantly overexpressed in three tissues, all in the brain, namely “frontal cerebral cortex BA9”, “nucleus accumbens” and “cerebral cortex” (Supplementary Figures 2 and 4). Gene-set analysis identified one biological process significantly associated with these genes: “long chain fatty acid metabolic process” (Bonferroni-corrected p-value 0.015; data not shown). No gene-sets for cellular components or molecular functions were significantly associated with these genes.
Discussion
In the current study, we analyzed large GWAS datasets on SCZ, BD and intelligence44,49,52 to gain insights into their shared genetic basis. First, we showed that common genetic variants associated with SCZ and BD are enriched for associations with intelligence (Figure 1). Using conjFDR analysis we leveraged this pleiotropic enrichment and identified 75 genomic loci jointly associated with SCZ and intelligence (Figure 2A) and 12 genomic loci jointly associated with BD and intelligence (Figure 3A). Among the shared loci, 20 are novel SCZ risk loci and four are novel BD risk loci. Altogether, this study indicates that large fractions of the genomic risk architectures underlying SCZ and BD also influence intelligence, albeit in a different manner. This provides new insights into the molecular genetic underpinnings of the altered cognitive performance in these patients groups.6–14,38 Further experimental interrogation of the identified loci may uncover biological insights that can inform the development of novel effective cognitive treatments, which remains to this day a pressing need in psychiatry.4,8
The GWAS power for BD (n=51,710)49 is still trailing that of SCZ (n=82,315),52 which limits the validity of comparing the present findings for the two disorders. Yet, the study strengthens prior genetic evidence44,49 that SCZ and BD differ in their relation to intelligence.8 Whereas most identified SCZ risk alleles (81%) were associated with lower cognitive performance (Supplementary Table 5), most BD risk alleles (75%) were associated with better cognitive performance (Supplementary Table 7). The discrepant results may thus highlight unique genetic effects underlying SCZ and BD, contrasting their otherwise high degree of genetic overlap.32 Yet, 8 of 12 lead SNPs associated with both BD and intelligence were also associated with SCZ at p<0.05, and 37 of 75 lead SNPs associated with both SCZ and intelligence were also associated with BD at p<0.05. Firstly, these results suggest that many of the genetic variants do not exclusively influence SCZ or BD, although they may be more specific to one of the disorders. Secondly, the consistent effects between the disorders support the validity of the present findings, even though the genetic correlation between BD and SCZ is not perfect (rg=0.7049).
To our knowledge, the finding of polygenic overlap between BD and intelligence is novel (Figure 1B, Figure 3A). Prior investigations did not find any genetic correlation between BD and cognitive abilities using LD score regression and polygenic risk scores.41–49 However, these methods are unable to detect genetic overlap if there are no consistent effect directions among the overlapping SNPs.66,67 Indeed, at the 79 loci associated with both BD and intelligence at a lower significance threshold (conjFDR<0.05, Supplementary Table 9), only 51% of BD risk alleles were associated with higher cognitive performance. This balanced mixture of directional effects complies with the non-significant genetic correlation between the phenotypes,44,49 indicating that a substantial polygenic component underlying BD risk also influences intelligence. The converging genetic data does not provide an explanation for the cognitive impairments associated with BD,6,7,11–14,38 suggesting that environmental factors or undetected rare and common genetic variants may also play a role. Our study further dissects the well-established polygenic overlap between SCZ and cognitive abilities,36,41–47 strengthening the hypothesis that common genetic variance may contribute to cognitive dysfunction in SCZ. It is nevertheless noteworthy that SCZ risk alleles were associated with higher cognitive performance at almost one fifth (~19%) of the shared loci. Overall, our findings suggest that the genetic relationship between SCZ, BD and intelligence is more complex than what is expressed by their genetic correlations,44,49 which may help explain the diverse cognitive performance within these patient groups.6,9,17–22
Phenotypic refinement32 may further illuminate the genetic relationship between BD, SCZ and intelligence. For example, patients with BD without a history of psychosis showed milder cognitive deficits than those with a history of psychosis,7 while BD1 is associated with more severe cognitive deficits than BD2.14 In the BD GWAS presently analyzed, 73% of cases had BD1, 17% of cases had BD2 and 5% of cases had SAB.49 However, larger GWAS samples are required to clarify any genetic differences underlying these subtypes. Note that the genetic effects on intelligence were determined in individuals representative of the normal population.44 Hence, the experimental design of the conjFDR approach ensures that the cognitive effects here linked to BD and SCZ risk alleles are not attributable to confounding known to bias neuropsychological assessment of these patient groups, such as high symptomatic load or medication.68 Although some participants in the intelligence GWAS44 likely suffered from psychiatric disorders, their contribution would not bias the results as they represent a minor fraction of the total GWAS sample.44
The genes nearest the 75 loci shared between SCZ and intelligence and the 12 loci shared between BD and intelligence were significantly overexpressed in human brain regions (Supplementary Figures 2–4). Although these genes are not necessarily the genes by which the genetic variants exert their phenotypic effect, the findings support the importance of brain- expressed genes in the shared genetic etiology underlying SCZ, BD and intelligence. The gene-set enrichment analysis implicated 32 biological processes significantly associated with the joint loci between SCZ and intelligence, converging on processes related to neurodevelopment, synaptic integrity and neurotransmission (Supplementary Table 10). These processes are previously linked to SCZ risk5,37,69 and intelligence.44,56 In line with these results, the gene-set analysis for cellular components revealed significant associations for neuronal projections, the synapse, and the anchored part of membranes, among others (Supplementary Table 12). The most strongly associated gene-set for molecular functions was “GABA receptor binding”, suggesting that inhibitory signaling may be affected. Intriguingly, we identified several loci shared between SCZ and intelligence previously associated with subcortical brain volumes (at DPP4, SPATS2L, NEK4, FOXO3, and DCC),70,71 providing plausible genetic links between SCZ, intelligence and brain structure formation. At all of these loci, SCZ risk was associated with poorer cognitive performance. We identified four loci shared between BD and intelligence that had not reached genome-wide significance in the BD GWAS49 analyzed but were identified in prior BD GWAS (at SUMO2P2,72 TENM4/ODZ4,73,74 RHEBL1,72,73 and NFX73; Supplementary Table 8), supporting their role in BD risk. Experimental follow-up studies are needed to determine the causal variants underlying the shared associations detected here, and to detail how these variants individually and collectively affect brain function and development.
In conclusion, our study demonstrates polygenic overlap between intelligence and the psychiatric disorders SCZ and BD, providing new insights into their common genetic basis. Owing to the well-powered GWAS44,49,52 investigated and their large degree of overlapping associations, the number of shared loci identified here substantially exceeds that of prior condFDR studies.36,53,54,70 Yet, the substantial pleiotropic enrichment observed suggests that many more loci shared between SCZ, BD and intelligence will be identified as GWAS samples get larger. This may have profound implications for understanding, and potentially treating, the cognitive abnormalities associated with these psychiatric disorders.
Supplementary Material
Acknowledgments
We thank the Schizophrenia and Bipolar Disorder Working Groups of the Psychiatric Genomics Consortium for access to data, and the consortia for making available their GWAS summary statistics, and the many people who provided DNA samples. We gratefully acknowledge support from the Research Council of Norway (262656, 248984, 248778, 223273) and KG Jebsen Stiftelsen; ABCD-USA Consortium (5U2 4DA041123).
Footnotes
Conflict of interest
Dr. Andreassen has received a speaker’s honorarium from Lundbeck. The other authors have no conflicts of interest to declare.
Supplementary information is available at MP’s website
References
- 1.Wahlbeck K, Westman J, Nordentoft M, Gissler M, Laursen TM. Outcomes of Nordic mental health systems: life expectancy of patients with mental disorders. Br J Psychiatry 2011; 199(6): 453–458. [DOI] [PubMed] [Google Scholar]
- 2.Nordentoft M, Wahlbeck K, Hallgren J, Westman J, Osby U, Alinaghizadeh H et al. Excess mortality, causes of death and life expectancy in 270,770 patients with recent onset of mental disorders in Denmark, Finland and Sweden. PloS one 2013; 8(1): e55176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keshavan MS, Morris DW, Sweeney JA, Pearlson G, Thaker G, Seidman LJ et al. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale. Schizophr Res 2011; 133(1–3): 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet 2016; 387(10027): 1561–1572. [DOI] [PubMed] [Google Scholar]
- 5.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet 2016; 388(10039): 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green MF. Cognitive impairment and functional outcome in schizophrenia and bipolar disorder. J Clin Psychiatry 2006; 67 Suppl 9: 3–8; discussion 36–42. [PubMed] [Google Scholar]
- 7.Simonsen C, Sundet K, Vaskinn A, Birkenaes AB, Engh JA, Faerden A et al. Neurocognitive dysfunction in bipolar and schizophrenia spectrum disorders depends on history of psychosis rather than diagnostic group. Schizophrenia bulletin 2011; 37(1): 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA psychiatry 2013; 70(10): 1107–1112. [DOI] [PubMed] [Google Scholar]
- 9.Keefe RS, Fenton WS. How should DSM-V criteria for schizophrenia include cognitive impairment? Schizophrenia bulletin 2007; 33(4): 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickinson D, Ragland JD, Gold JM, Gur RC. General and specific cognitive deficits in schizophrenia: Goliath defeats David? Biological psychiatry 2008; 64(9): 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourne C, Aydemir O, Balanza-Martinez V, Bora E, Brissos S, Cavanagh JT et al. Neuropsychological testing of cognitive impairment in euthymic bipolar disorder: an individual patient data meta-analysis. Actapsychiatrica Scandinavica 2013; 128(3): 149–162. [DOI] [PubMed] [Google Scholar]
- 12.Kurtz MM, Gerraty RT. A meta-analytic investigation of neurocognitive deficits in bipolar illness: profile and effects of clinical state. Neuropsychology 2009; 23(5): 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arts B, Jabben N, Krabbendam L, van Os J. Meta-analyses of cognitive functioning in euthymic bipolar patients and their first-degree relatives. Psychological medicine 2008; 38(6): 771–785. [DOI] [PubMed] [Google Scholar]
- 14.Simonsen C, Sundet K, Vaskinn A, Birkenaes AB, Engh JA, Hansen CF et al. Neurocognitive profiles in bipolar I and bipolar II disorder: differences in pattern and magnitude of dysfunction. Bipolar Disord 2008; 10(2): 245–255. [DOI] [PubMed] [Google Scholar]
- 15.Johnson W, Bouchard TJ, Krueger RF, McGue M, Gottesman II. Just one g: consistent results from three test batteries. Intelligence 2004; 32(1): 95–107. [Google Scholar]
- 16.Ree MJ, Earles JA. The Stability of G across Different Methods of Estimation. Intelligence 1991; 15(3): 271–278. [Google Scholar]
- 17.Palmer BW, Heaton RK, Paulsen JS, Kuck J, Braff D, Harris MJ et al. Is it possible to be schizophrenic yet neuropsychologically normal? Neuropsychology 1997; 11(3): 437–446. [DOI] [PubMed] [Google Scholar]
- 18.Vaskinn A, Ueland T, Melle I, Agartz I, Andreassen OA, Sundet K. Neurocognitive Decrements are Present in Intellectually Superior Schizophrenia. Frontiers in psychiatry 2014; 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacCabe JH, Brebion G, Reichenberg A, Ganguly T, McKenna PJ, Murray RM et al. Superior intellectual ability in schizophrenia: neuropsychological characteristics. Neuropsychology 2012; 26(2): 181–190. [DOI] [PubMed] [Google Scholar]
- 20.Gale CR, Batty GD, McIntosh AM, Porteous DJ, Deary IJ, Rasmussen F. Is bipolar disorder more common in highly intelligent people? A cohort study of a million men. Molecular psychiatry 2013; 18(2): 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiihonen J, Haukka J, Henriksson M, Cannon M, Kieseppa T, Laaksonen I et al. Premorbid intellectual functioning in bipolar disorder and schizophrenia: results from a cohort study of male conscripts. The American journal of psychiatry 2005; 162(10): 1904–1910. [DOI] [PubMed] [Google Scholar]
- 22.MacCabe JH, Lambe MP, Cnattingius S, Sham PC, David AS, Reichenberg A et al. Excellent school performance at age 16 and risk of adult bipolar disorder: national cohort study. Br J Psychiatry 2010; 196(2): 109–115. [DOI] [PubMed] [Google Scholar]
- 23.Fusar-Poli P, Deste G, Smieskova R, Barlati S, Yung AR, Howes O et al. Cognitive functioning in prodromal psychosis: a meta-analysis. Archives of general psychiatry 2012; 69(6): 562–571. [DOI] [PubMed] [Google Scholar]
- 24.Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RS, Murray RM et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. The American journal of psychiatry 2010; 167(2): 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deary IJ. Intelligence. Annu Rev Psychol 2012; 63: 453–482. [DOI] [PubMed] [Google Scholar]
- 26.Mackenbach JP, Stirbu I, Roskam AJ, Schaap MM, Menvielle G, Leinsalu M et al. Socioeconomic inequalities in health in 22 European countries. The New England journal of medicine 2008; 358(23): 2468–2481. [DOI] [PubMed] [Google Scholar]
- 27.Mohamed S, Rosenheck R, Swartz M, Stroup S, Lieberman JA, Keefe RS. Relationship of cognition and psychopathology to functional impairment in schizophrenia. The American journal of psychiatry 2008; 165(8): 978–987. [DOI] [PubMed] [Google Scholar]
- 28.Wingo AP, Harvey PD, Baldessarini RJ. Neurocognitive impairment in bipolar disorder patients: functional implications. Bipolar Disord 2009; 11(2): 113–125. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Aran A, Vieta E, Torrent C, Sanchez-Moreno J, Goikolea JM, Salamero M et al. Functional outcome in bipolar disorder: the role of clinical and cognitive factors. Bipolar Disord 2007; 9(1–2): 103–113. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373(9659): 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 2015; 47(7): 702–709. [DOI] [PubMed] [Google Scholar]
- 32.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell 2018; 173(7): 1705–1715 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toulopoulou T, Goldberg TE, Mesa IR, Picchioni M, Rijsdijk F, Stahl D et al. Impaired intellect and memory: a missing link between genetic risk and schizophrenia? Archives of general psychiatry 2010; 67(9): 905–913. [DOI] [PubMed] [Google Scholar]
- 34.Glahn DC, Almasy L, Blangero J, Burk GM, Estrada J, Peralta JM et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2007; 144B(2): 242–249. [DOI] [PubMed] [Google Scholar]
- 35.Fowler T, Zammit S, Owen MJ, Rasmussen F. A population-based study of shared genetic variation between premorbid IQ and psychosis among male twin pairs and sibling pairs from Sweden. Archives of general psychiatry 2012; 69(5): 460–466. [DOI] [PubMed] [Google Scholar]
- 36.Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y et al. Identification of Genetic Loci Jointly Influencing Schizophrenia Risk and the Cognitive Traits of Verbal-Numerical Reasoning, Reaction Time, and General Cognitive Function. JAMA psychiatry 2017; 74(10): 1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen MJ, O’Donovan MC. Schizophrenia and the neurodevelopmental continuum:evidence from genomics. World Psychiatry 2017; 16(3): 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bora E, Yucel M, Pantelis C. Cognitive endophenotypes of bipolar disorder: a meta-analysis of neuropsychological deficits in euthymic patients and their first-degree relatives. Journal of affective disorders 2009; 113(1–2): 1–20. [DOI] [PubMed] [Google Scholar]
- 39.Glahn DC, Almasy L, Barguil M, Hare E, Peralta JM, Kent JW Jr., et al. Neurocognitive endophenotypes for bipolar disorder identified in multiplex multigenerational families. Archives of general psychiatry 2010; 67(2): 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill SK, Reilly JL, Keefe RS, Gold JM, Bishop JR, Gershon ES et al. Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B- SNIP) study. The American journal ofpsychiatry 2013; 170(11): 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Molecular psychiatry 2016; 21(11): 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trampush JW, Yang ML, Yu J, Knowles E, Davies G, Liewald DC et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Molecular psychiatry 2017; 22(3): 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill WD, Davies G, Group CCW, Liewald DC, McIntosh AM, Deary IJ. Age-Dependent Pleiotropy Between General Cognitive Function and Major Psychiatric Disorders. Biological psychiatry 2016; 80(4): 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 2018; 50(7): 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 2017; 49(7): 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebers DT, Pirooznia M, Seiffudin F, Musliner KL, Zandi PP, Goes FS. Polygenic Risk of Schizophrenia and Cognition in a Population-Based Survey of Older Adults. Schizophrenia bulletin 2016; 42(4): 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brainstorm Consortium. Analysis of shared heritability in common disorders of the brain. Science 2018; 360(6395). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun 2018; 9(1): 2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahl E, Forstner A, McQuillin A, Ripke S, , Ophoff R et al. Genomewide association study identifies 30 loci associated with bipolar disorder. bioRxiv 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyall DM, Cullen B, Allerhand M, Smith DJ, Mackay D, Evans J et al. Cognitive Test Scores in UK Biobank: Data Reduction in 480,416 Participants and Longitudinal Stability in 20,346 Participants. PloS one 2016; 11(4): e0154222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Molecular psychiatry 2016; 21(6): 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. American journal of human genetics 2013; 92(2): 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu JZ, Hov JR, Folseraas T, Ellinghaus E, Rushbrook SM, Doncheva NT et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet 2013; 45(6): 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schork AJ, Wang Y, Thompson WK, Dale AM, Andreassen OA. New statistical approaches exploit the polygenic architecture of schizophrenia--implications for the underlying neurobiology. Curr Opin Neurobiol 2016; 36: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat Rev Neurosci 2010; 11(3): 201–211. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8(1): 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015; 526(7571): 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46(3): 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research 2012; 22(9): 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518(7539): 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016; 48(5): 481–487. [DOI] [PubMed] [Google Scholar]
- 63.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E et al. The new NHGRI- EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic acids research 2017; 45(D1): D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25(1): 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017; 550(7675): 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47(11): 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460(7256): 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moritz S, Klein JP, Desler T, Lill H, Gallinat J, Schneider BC. Neurocognitive deficits in schizophrenia. Are we making mountains out of molehills? Psychological medicine 2017; 47(15): 2602–2612. [DOI] [PubMed] [Google Scholar]
- 69.Devor A, Andreassen OA, Wang Y, Maki-Marttunen T, Smeland OB, Fan CC et al. Genetic evidence for role of integration of fast and slow neurotransmission in schizophrenia. Molecular psychiatry 2017; 22(6): 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smeland OB, Wang Y, Frei O, Li W, Hibar DP, Franke B et al. Genetic Overlap Between Schizophrenia and Volumes of Hippocampus, Putamen, and Intracranial Volume Indicates Shared Molecular Genetic Mechanisms. Schizophrenia bulletin 2018; 44(4): 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N et al. Common genetic variants influence human subcortical brain structures. Nature 2015; 520(7546): 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Human molecular genetics 2016; 25(15): 3383–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N et al. A genome- wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Molecular psychiatry 2018; 23(3): 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43(10): 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


