Abstract
Single cell RNA-sequencing (scRNA-seq) technologies are increasingly being applied to reveal cellular heterogeneity in kidney development and disease. In just the last year, multiple scRNA-seq datasets have been generated from kidney organoids, developing mouse and human kidney, adult kidney and kidney cancer. The data generated enables a much deeper understanding of biological processes within and between cells. It has also elucidated unforeseen cell lineage relationships, defined the presence of off-target cell types in kidney organoids and revealed a diverse inflammatory response in a human kidney allograft undergoing rejection. This review summarizes the recent rapid progress in scRNA-seq of the kidney and outlines future directions for single cell technologies as applied to the kidney.
Keywords: Next generation sequencing, transcriptomics, single cell, informatics, organoids, stem cell
Introduction to scRNAseq Technology and Methodologies
Single cell RNA sequencing (scRNAseq) has emerged as a powerful tool for exploring the transcriptional heterogeneity of large cell populations by measuring gene transcription in individual cells. This rapidly emerging technology enables the detection and characterization of specific cell populations as they relate to health and disease. There are a number of methodologies that have been used for scRNAseq and all of them share a similar procedure: single cell or single nucleus isolation, cell lysis and RNA capture, reverse transcription and amplification, library generation, and next-generation sequencing (NGS) [1]. Several technologies that are currently in use include 10X Chromium, Drop-Seq, InDrops and Microwell-Seq [2–5]. The strengths and weaknesses of these methods have been recently reviewed and our discussion will focus on their application to kidney development and disease [6, 7]. To date, scRNAseq has been applied to healthy kidney, developing kidney, several kidney disease states and to kidney organoids generated from pluripotent stem cells. These studies have led to an improved understanding of the mechanisms that lead to kidney injury and the pathways critical for nephrogenesis.
Characterization of mature cell populations in the mouse kidney
Recent efforts have utilized scRNAseq to create an atlas of the mouse kidney. Park et al. used 10X Chromium to generate 57,979 single cell transcriptomes in healthy mouse kidneys. By mapping expression of murine homologs of genes associated with both Mendelian inherited disease and expressed quantitative trait loci identified from genome-wide association studies, they provided strong evidence that these disease-relevant genes are usually expressed in only a single cell type [8, 9]. This provides important clues as to the mechanism by which these risk alleles might ultimately cause kidney disease. The genes were associated with a broad phenotypic spectrum that included nephrotic syndrome, renal tubular acidosis, nephrolithiasis, and hypertension. The group also performed cell trajectory analysis with pseudo time reconstitution to identify a novel cell type intermediate between principal cells and intercalated cells. They hypothesized that Notch signaling plays an important role in the transition between intercalated and principal cells and may be an important mediator of kidney fibrosis. Principal cells predominantly express Notch2 receptor, whereas intercalated cells express Notch ligand, suggesting that principal cells are the recipients of Notch signaling in the collecting duct. Initiation of Notch signaling with an inducible transgenic mouse model demonstrated that the proportion of cells expressing the principal cell marker, aquaporin 2 (Aqp2), increased and the proportion of cells expressing the intercalated cell V-ATPase (Atp6v1b1) decreased. A mouse model of CKD showed that there is a loss of normal alternation with a relative increase in Aqp2-positive cells and a decrease in Atp6v1b1-positive cells consistent with increased Notch signaling [8]. These data suggest Notch-mediated signaling in the distal nephron may play an important role in CKD as it regulates the transition between cell types.
Although there are advantages to unbiased, whole-organ scRNAseq, rare cell types and specific populations may be difficult to capture without enrichment. The glomerulus, specific interstitial populations and minor nephron segments all might be lost in whole organ scRNA-seq. Karaiskos employed DropSeq to generate a comprehensive atlas of the mouse glomerulus [10]. By isolating glomeruli prior to sequencing, they were able to capture nearly 13,000 cells spread among the 3 known glomerular cell types. Their methodology allowed a comprehensive characterization of glomerular endothelial cells, podocytes, and mesangial cells, which would have otherwise been extremely difficult to identify if scRNAseq had been applied to whole kidney cortex. The increased number of cells led to the identification of novel podocyte (Wsb2) and mesangial cell markers (Pde3a) and glomerular endothelial subpopulations (Jag1, Fbln5, Cxcl1, Cldn5).
scRNAseq of Human and Mouse Kidney in Health and Disease
In only the last year, several high-profile publications have used scRNAseq to identify and characterize cell populations within the mature kidney. Sivakamasundari et al. used Chromium single cell expression (10x Genomics) to identify 27 cell types in a library of 22,469 cells from a healthy human kidney [11]. They leveraged their large library size to identify subpopulations of intercalated and endothelial cells that expressed genes associated with human disease. They defined a novel subpopulation of glomerular endothelial cells (GECs) that co-express claudin 5 (CLDN5) and plasmalemma vesicle associated protein (PLVAP). CLDN5 encodes a membrane protein and integral component of tight junctions and PLVAP is a marker of endothelial diaphragms in fenestrae. Neither of these genes is known to be expressed in GECs although CLDN5 expression is seen in afferent-efferent arteriolar endothelial cells [12]. Sivakamasundari et al. demonstrated expression of CLND5 in GECs and their data indicates that there is a previously unrecognized subpopulation of fenestrated glomerular endothelial cells. A subset of the CLDN5+ cells co-express PLVAP and alternative modes of transcellular transport mediated by caveolin 1 (CAV1) and aquaporin 1 (AQP1), however, the functional significance of this subpopulation is unknown.
Sivakamasundari et al. also highlighted the strength of scRNAseq in the discovery of novel intercellular signaling networks. [11] Endothelial cell-mediated recruitment of pericytes is regulated by interaction of platelet derived growth factor subunit B (PDGFB) with its receptor, PDGFRB, and blockade of PDGFRB decreases fibrosis and capillary rarefaction in a unilateral ureteral obstruction (UUO) model of fibrosis in the mouse kidney [13, 14]. However, a recent scRNAseq study by Chen et al. implicated the interaction between KIT protooncogene receptor tyrosine kinase (KIT) its ligand, KITLG in the distal nephron [15]. Application of scRNAseq enabled the characterization of both signaling networks and raises the possibility that KIT-KITLG signaling principal and intercalated cells may play an important role in their survival and proliferation.
Gillies et al. investigated the transcriptional profiles of glomeruli and tubulointerstitium in 187 patients from the NEPTUNE study with nephrotic syndrome using whole genome sequencing (WGS) and scRNAseq [16]. The patient population ranged in age from 17 to 56 and included 51 pediatric patients with a mixture of minimal change disease, focal and segmental glomerulosclerosis, and other histopathologic diagnoses. They identified a large number of SNVs that are associated with differential expression of glomerular and tubulointerstitial genes (eQTLs) and were able to associate them with specific cell populations. Phospholipase C gamma 2 (PLCG2) and vacuolar protein sorting 33b (VPS33B) are 2 genes that had highly significant glomerular-specific eQTLs. A rare missense variant in PLCG2 has been previously implicated as a candidate risk locus in steroid sensitive nephrotic syndrome (SSNS) and mutations in VPS33B cause arthrogryposis, renal dysfunction, and cholestasis (ARC) syndrome [17, 18]. Gillies et al. utilized their glomerular and tubulointerstitial eQTLs to better interpret known single nucleotide polymorphisms (SNPs) associated with CKD in GWAS studies. A compelling SNP that they identified in their tubulointerstitial eQTL dataset (rs12917707) is upstream of uromodulin and is associated with increased UMOD expression.
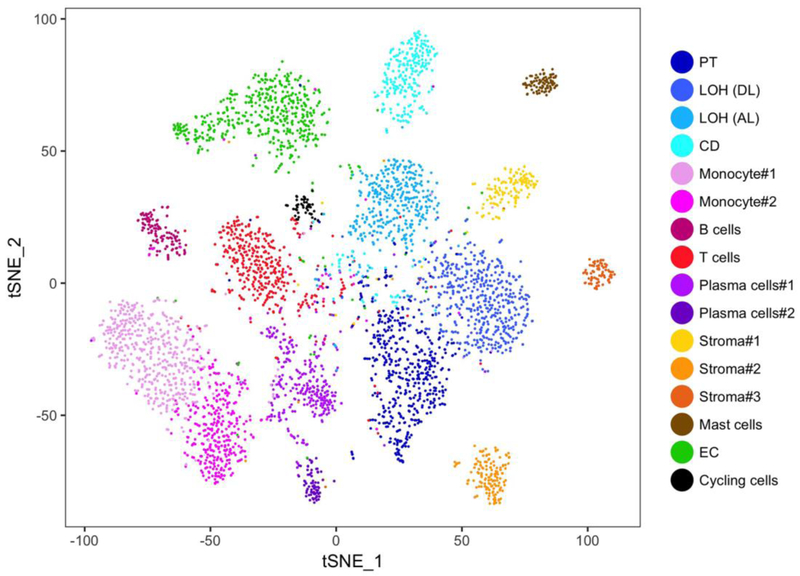
Wu and Malone performed scRNAseq on a single human kidney allograft biopsy with mixed cellular and antibody-mediated rejection [19]. Application of scRNAseq to kidney allografts enables the detection and characterization of infiltrating immune cells and endothelial cell injury. They used the InDrops system to generate ~5000 single cell libraries consisting of 16 cell types (Figure 1). Endothelial cells were further separated into 3 groups based on the expression of Ig-associated and angiogenesis-associated genes. Further characterization of the endothelial targets of donor specific antibodies (DSAs) may help differentiate between antibodies that mediate rejection and those that do not. Monocyte infiltration in kidney allografts is associated with kidney allograft dysfunction and Wu et al. identified 2 monocyte clusters based on expression of FCGR3A (CD16) [19].. Notably, infiltration by CD16+ cells is associated with allograft rejection and this monocytic cell population expressed ABCA1, which is a sterol efflux transporter found in activated dendritic cells [20, 21]. These data indicate that monocytes may undergo a phenotypic transition within the kidney during allograft rejection. Receptor ligand analysis of their dataset detected 14 receptor-ligand pairs. Fibroblasts, pericytes, and myofibroblasts expressed the chemokine CXCL12, which is a key regulator of immune cell recruitment via its interaction with CXCR4 on leukocytes. Additionally, expression of KIT and KITLG was detected in mast cells and collecting duct epithelium, respectively. This study provides a proof of principle for the application of single cell transcriptomics to characterize, and ultimately aid in diagnostics of, human kidney biopsies [1].
Figure 1. Comprehensive single-cell RNA sequencing of an allograft biopsy reveals diverse kidney and immune cell types. T-distributed stochastic neighborhood embedding.
(tSNE) plot of cell clusters identified from a human kidney allograft biopsy based on the expression of highly variable genes [19]. PT proximal tubule, LOH (DL) loop of henle, distal limb, LOH (AL) loop of henle, ascending limb, CD collecting duct, Mono monocyte, EC endothelial cells. Figure courtesy of Haojia Wu, PhD.
Der et al. applied scRNAseq to human skin and kidney biopsies to demonstrate a conserved IFN signature present in multiple cell subtypes [22]. Although their sample size was relatively small, they were able to demonstrate that the IFN signature correlated with histologic evidence of chronicity, proteinuria, and immunoglobulin deposition in the glomerulus. Future studies should aim to define a transcriptional signature that has prognostic value beyond the ISN/RPS pathologic classification of lupus nephritis.
Significant advances have been made in applying scRNAseq technology to the study of renal development and neoplasia. Young et al. generated 72,501 single cell transcriptomes to compare Wilms tumor, clear cell renal cell carcinoma (ccRCC), and papillary renal cell carcinoma (pRCC) with fetal, pediatric, adolescent, and adult kidneys [23]. Their study was able to show that Wilm’s tumor closely resembles normal fetal ureteric bud and primitive vesicle cells. Pseudotime reconstruction revealed a branching point between the development of nephrogenic rests and Wilms tumor cells that was closely related to normal nephrogenesis. In contrast, ccRCC and pRCC were closely related to mature proximal tubular cells and associated with VEGFA signaling in infiltrating macrophages. These data point towards the utility of scRNAseq in establishing lineage-specific precursors in neoplasia and development.
scRNAseq of Developing Human and Mouse Kidney
Lindstrom et al. used scRNAseq to identify cell populations within the nephrogenic niche of a 17-week human kidney [24]. Among 3367 cells, they identified multiple clusters representing nephron progenitors (NPCs), podocyte precursors, distal precursors, maturing cell types of the loop of Henle, and precursors of the proximal and distal tubules. The NPCs (CITED1+, SIX1+, LYPD1+, DAPL1+) were further separated into cells that were primed for differentiation (HEY1+, LYPD1+), and those that were in the process of differentiation (HES1+, LHX1+, PAX8+) depending on their location and time of recruitment to the nephrogenic niche. These data led to the hypothesis that distal NPCs initially express JAG1 and later transition to a SOX9+ JAG1- phenotype and highlight the importance of cellular positional and temporal identity during proximal-distal nephron alignment. A subsequent study by Lindstrom et al investigated the differences in cell specific transcription between human and murine nephrogenesis [25]. Mice have distinct compartments of Six2+ NPCs and Foxd1+ interstitial progenitor cells (IPCs) in contrast to humans, which have significant transcriptional overlap between NPCs and IPCs. By comparing the cell-specific profiles of mouse and human NPCs, they were able to identify species-specific bias in the expression of certain transcripts. Human NPCs are more likely to express H1P1R, UNC5B, LYPD1, ECEL1, WASF3, TNFRSF19, CRABP2, CDH24, DAPL1 and COL9A2 whereas mouse NPCs express Crym, Serpinf1, Slc12a2, Foxd1, Rspo1, and Capn6. Together, these studies demonstrate significant divergence between mouse and human nephrogenesis. Pode-Shakked et al. performed single-cell analysis on in vitro cultures of human fetal kidney cells (hFK) to examine the phenotype of cell subpopulations in early nephrogenesis. [26] They utilized a modified nephron progenitor expansion medium (mNPEM) to prevent hFK dedifferentiation and were able to characterize 3 subpopulations of cells. The cap mesenchyme segregated into induced and uninduced fractions in the NCAM1+ CD133- EpCAM- cells and an additional subpopulation (NCAM1+ CD133- EpCAMdim) transitioning between a mesenchymal and epithelial phenotype appeared in cells cultured in mNPEM. SIX2 expression was identified in this subpopulation, which may represent a cap mesenchyme-derived cell type with both a stem and differentiating phenotype.
Modeling Development with Kidney Organoids
Kidney organoids derived from pluripotent stem cells are increasingly used to model kidney development and disease. However, there are major unanswered questions that remain, including what cell types exist within organoids, their degree of differentiation, and how to improve differentiation protocols. scRNA-seq is well-suited to address all of these open questions, and two recent studies have used single cell transcriptomics to characterize human kidney organoids.
Czerniecki et al. developed a robotic pipeline to efficiently create human kidney organoids in microwell plates [27]. Analysis of the organoids by high content automated imaging and by scRNAseq showed that organoids appropriately differentiated into well-described kidney cell types. The 96- and 384-well formats made it possible to experiment with varying concentrations of the WNT agonist, CHIR99021, to determine which concentration is optimal for directed differentiation. An unexpected finding was that the optimal concentration of CHIR99021 was dependent on which pluripotent stem cell subclone was used to generate kidney organoids despite the fact that all of the subclones were created from the same hPSC cell line. The concentration of CHIR99021 did not appear to affect the proportion of proximal tubule, distal tubule, and podocyte components. However, addition of VEGF increased the number of endothelial cells by approximately 10-fold. These data highlight the importance of organoid culture techniques that significantly impact the cell repertoire. This technology will be useful for optimizing differentiation protocols as well as for assessing kidney-specific toxicity of candidate drugs, for anyone with access to such technology.
Wu et al. compared kidney organoid differentiation protocols by scRNAseq and introduced a novel method for inhibiting the growth and differentiation of off-target neuronal cell types [19]. They compared the Morizane [28] and Takasato [29] protocols for differentiation of kidney organoids from hPSC and showed that these protocols have significant differences in cellular composition. They concluded that the Morizane protocol is best suited for investigating glomerular biology because it has a larger proportion of podocytes. In contrast, the Takasoto protocol has a larger proportion of tubulointerstitium. Both protocols resulted in the production of substantial numbers of off-target neural cell populations, which was prevented by the inhibition of BDNF-NTRK2 signaling. Notably, neither protocol had an identifiable population of ureteric bud-derived cells, which ultimately differentiate into the collecting duct system. Interconnection between the developing nephron and the collecting duct is a significant hurdle in kidney organoid development and may require separate cell populations. Taguchi et al. used separate protocols for the induction of metanephric mesenchyme and ureteric bud precursors followed by recombination, which significantly improved collecting duct architecture [30].
Limitations of scRNA-seq
There are several notable limitations to scRNA-seq methodologies. Depending on sample preparation methods, certain cell populations may not be accurately represented due to cell dissociation bias [19]. Cell dissociation typically relies on enzymatic digestion of dense kidney matrix, which compromise cell viability and induce artifactual transcriptional stress responses [31]. Newer sample preparation methods which can be performed entirely on ice like single nucleus RNA-seq (snRNAseq) [32], have partially resolved this problem, but it remains challenging to capture specific cell types particularly in inflamed or fibrotic tissues (Parker and Humphreys, personal observations) Furthermore, spatial information is lost after cell dissociation. This is particularly important when considering paracrine signaling networks and receptor ligand interactions, which may be restricted to a microenvironmental niche.
The informatics infrastructure required to analyze scRNAseq data remains a significant practical barrier to widespread adoption of this transformative technology. The initial data processing stages involve alignment and clustering of thousands of cells and are typically performed in a high performance computing environment on a university cluster. Trends in scRNA-seq include the analysis of ever larger numbers of single cell transcriptomes (100,000 transcriptomes is no longer unusual for a single publication). This requires considerable computational power and not all researchers are likely to have access either to adequately rapid computational power or the bioinformatics expertise necessary to implement the pipelines. Downstream data analysis generally relies on differential gene expression but can be extended to numerous additional techniques including pseudotemporal ordering of cells to reveal lineage relationships, receptor-ligand analyses, transcription factor networks and even copy number variations that arise in cancerous cells. Performing more advanced analyses generally requires a commitment to learn a programming language such as R.
Future Directions
As the cost of scRNAseq decreases it will become more accessible to the research community and enable the creation of larger datasets. Recent studies have shown that by increasing the number of cells analyzed, rare cell types and subpopulations are more easily detectable. Identification of cell subpopulations may become increasingly important as scRNAseq is used to measure transcriptional changes in injury and repair. Current scRNA-seq protocols can measure only more abundant cellular transcripts, typically around 1,500 unique genes per cell. [33] This represents only about 10% of the total pool of cellular mRNA. It is very likely that in the near future protocols will be developed for improved gene detection and we predict that detection of 3,000 – 5,000 unique genes per cell will be routine within a few years.
Preparation methods will have to be optimized to reduce cell dissociation bias and improve assay efficiency in difficult sample types. Current methods exhibit a lower yield in the presence of chronic changes like interstitial fibrosis. Approaches like snRNAseq may mitigate this drawback but do not entirely resolve the problem.
Novel methods like single cell transposase-accessible chromatin sequencing (ATAC-seq) can measure chromatin accessibility and a cell’s epigenetic landscape [34]. Similarly, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) can measure individual cell surface protein levels and cellular mRNAs simultaneously. This approach can theoretically measure a nearly unlimited number of proteins simultaneously – unlike fluorescence-activated cell sorting [35]. In the future these single cell approaches will increasingly be combined – single cell multi-omics - in order to reveal more profound biological insight than any single technology used alone [36].
The remarkably rapid adoption of scRNA-seq methods by kidney researchers and the resultant work summarized here suggests that there is much more to come, and brings to mind Winston Churchill’s words describing England’s first battle victory in World War 2: “Now this is not the end. It is not even the beginning of the end. But it is, perhaps, the end of the beginning” [37].
Footnotes
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Malone AF, Wu H, Humphreys BD (2018) Bringing Renal Biopsy Interpretation Into the Molecular Age With Single-Cell RNA Sequencing. Semin Nephrol 38:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng gX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler Td, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall- Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg Hj, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH (2017) Massively parallel digital transcriptional profiling of single cells. Nature Comm 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA (2015) Highly Parallel Genome- wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G (2018) Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172:1091–1107 e1017. [DOI] [PubMed] [Google Scholar]
- 6.Hedlund E, Deng Q (2018) Single-cell RNA sequencing: Technical advancements and biological applications. Mol Aspects Med 59:36–46. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, Humphreys BD (2017) The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int 92:1334–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Susztak K (2018) Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360:758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys BD (2018) Mapping kidney cellular complexity. Science 360:709–710. [DOI] [PubMed] [Google Scholar]
- 10.Karaiskos N, Rahmatollahi M, Boltengagen A, Liu H, Hoehne M, Rinschen M, Schermer B, Benzing T, Rajewsky N, Kocks C, Kann M, Muller RU (2018) A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. J Am Soc Nephrol 29:2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivakamasundari V, Bolisetty M, Sivajothi S, Bessonett S, Ruan D, Robson P (2017) Comprehensive Cell Type Specific Transcriptomics of the Human Kidney. bioRxiv doi: 10.1101/238063. [DOI] [Google Scholar]
- 12.Morita K, Sasaki H, Furuse M, Tsukita S (1999) Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramsson A, Lindblom P, Betsholtz C (2003) Endothelial and nonendothelial sources of PDGF-B regulate pericyte recruitment and influence vascular pattern formation in tumors. J Clin Invest 112:1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SL, Chang FC, Schrimpf C, Chen YT, Wu CF, Wu VC, Chiang WC, Kuhnert F, Kuo CJ, Chen YM, Wu KD, Tsai TJ, Duffield JS (2011) Targeting endothelium-pericyte cross talk by inhibiting VEGF receptor signaling attenuates kidney microvascular rarefaction and fibrosis. Am J Pathol 178:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Lee JW, Chou CL, Nair AV, Battistone MA, Paunescu TG, Merkulova M, Breton S, Verlander JW, Wall SM, Brown D, Burg MB, Knepper MA (2017) Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci U S A 114:E9989–E9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillies CE, Putler R, Menon R, Otto E, Yasutake K, Nair V, Hoover P, Lieb D, Li S, Eddy S, Fermin D, McNulty MT, Nephrotic Syndrome Study N, Hacohen N, Kiryluk K, Kretzler M, Wen X, Sampson MG (2018) An eQTL Landscape of Kidney Tissue in Human Nephrotic Syndrome. Am J Hum Genet 103:232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, Kale A, Gipson D, Srivastava T, Lin JJ, Chand D, Hunley TE, Brophy PD, Bagga A, Sinha A, Rheault MN, Ghali J, Nicholls K, Abraham E, Janjua HS, Omoloja A, Barletta GM, Cai Y, Milford DD, O’Brien C, Awan A, Belostotsky V, Smoyer WE, Homstad A, Hall G, Wu G, Nagaraj S, Wigfall D, Foreman J, Winn MP, Mid-West Pediatric Nephrology C (2015) HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol 26:1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gissen P, Johnson CA, Morgan NV, Stapelbroek JM, Forshew T, Cooper WN, McKiernan PJ, Klomp LW, Morris Aa, Wraith JE, McClean P, Lynch SA, Thompson RJ, Lo B, Quarrell OW, Di Rocco M, Trembath RC, Mandel H, Wali S, Karet FE, Knisely AS, Houwen RH, Kelly DA, Maher ER (2004) Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet 36:400–404. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, Gaut JP, Humphreys BD (2018) Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol 29:2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Bosch TPP, Hilbrands LB, Kraaijeveld R, Litjens NHR, Rezaee F, Nieboer D, Steyerberg EW, van Gestel JA, Roelen DL, Clahsen-van Groningen MC, Baan CC, Rowshani AT (2017) Pretransplant Numbers of CD16(+) Monocytes as a Novel Biomarker to Predict Acute Rejection After Kidney Transplantation: A Pilot Study . Am J Transplant 17:2659–2667. [DOI] [PubMed] [Google Scholar]
- 21.Vereyken EJ, Kraaij MD, Baan CC, Rezaee F, Weimar W, Wood KJ, Leenen PJ, Rowshani AT (2013) A shift towards pro-inflammatory CD16+ monocyte subsets with preserved cytokine production potential after kidney transplantation. PLoS One 8:e70152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, Wang T, Jordan N, Bornkamp N, Nwaukoni J, Martinez J, Goilav B, Buyon JP, Tuschl T, Putterman C (2017) Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight doi: 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, Collord G, Botting RA, Popescu DM, Loudon KW, Vento- Tormo R, Stephenson E, Cagan A, Farndon SJ, Del Castillo Velasco- Herrera M, Guzzo C, Richoz N, Mamanova L, Aho T, Armitage JN, Riddick ACP, Mushtaq I, Farrell S, Rampling D, Nicholson J, Filby A, Burge J, Lisgo S, Maxwell PH, Lindsay S, Warren AY, Stewart GD, Sebire N, Coleman N, Haniffa M, Teichmann SA, Clatworthy M, Behjati S (2018) Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindstrom NO, De Sena Brandine G, Tran T, Ransick A, Suh G, Guo J, Kim AD, Parvez RK, Ruffins SW, Rutledge EA, Thornton ME, Grubbs B, McMahon JA, Smith AD, McMahon AP (2018) Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev Cell 45:651–660 e654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstrom NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, Ransick A, Parvez RK, Thornton ME, Basking L, Grubbs B, McMahon JA, Smith AD, McMahon AP (2018) Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. J Am Soc Nephrol 29:806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pode-Shakked N, Gershon R, Tam G, Omer D, Gnatek Y, Kanter I, Oriel S, Katz G, Harari-Steinberg O, Kalisky T, Dekel B (2017) Evidence of In Vitro Preservation of Human Nephrogenesis at the Single-Cell Level. Stem Cell Reports 9:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czerniecki SM, Cruz NM, Harder JL, Menon R, Annis J, Otto EA, Gulieva RE, Islas LV, Kim YK, Tran LM, Martins TJ, Pippin JW, Fu H, Kretzler M, Shankland SJ, Himmelfarb J, Moon RT, Paragas N, Freedman BS (2018) High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 22:929–940 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morizane R, Bonventre JV (2017) Generation of nephron progenitor cells. and kidney organoids from human pluripotent stem cells. Nat Protoc 12:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, Little MH (2014) Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol 16:118–126. [DOI] [PubMed] [Google Scholar]
- 30.Taguchi A, Nishinakamura R (2017) Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell 21:730–746 e736. [DOI] [PubMed] [Google Scholar]
- 31.Adam M, Potter AS, Potter SS (2017) Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144:3625–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bb Lake, Codeluppi S Yung YC, Gao D, Chun J, Kharchenko PV, Linnarsson S, Zhang K (2017) A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep 7:6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W (2017) Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell 65:631–643 e634. [DOI] [PubMed] [Google Scholar]
- 34.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ (2015) Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chappell L, Russell AJC, Voet T (2018) Single-Cell (Multi)omics Technologies. Annu Rev Genomics Hum Genet 19:15–41. [DOI] [PubMed] [Google Scholar]
- 37.Churchill W, Eade C (1943) The end of the beginning; war speeches . Little, Brown and Company, Boston. [Google Scholar]