Abstract
Background
Programmed death-ligand 1 (PD-L1), a transmembrane protein, binds to the programmed death-1 (PD-1) receptor, and anti-PD-1 therapy enables immune responses against tumors. This study aimed to assess clinical characteristics of PD-L1 expression using immunohistochemistry among Korean patients with lung cancer.
Methods
We retrospectively reviewed the data of patients with pathologically proven lung cancer from a single institution. PD-L1 expression determined by Tumor Proportion Score (TPS) was detected using 22C3 pharmDx (Agilent Technologies) and SP263 (Ventana Medical Systems) assays.
Results
From July 2016 to July 2017, 267 patients were enrolled. The main histologic type was adenocarcinoma (69.3%). Most participants were smokers (67.4%) and had clinical stage IV disease (60.7%). In total, 116 (42%) and 58 (21%) patients had TPS ≥1% and ≥50%, respectively. The patients were significantly older in TPS ≥1% group than in TPS <1% group (64.83±9.38 years vs. 61.73±10.78 years, p=0.014), not in TPS ≥50% cutoff value (64.69 ± 9.39 vs. 62.36 ± 10.51, p= 0.178). Regarding histologic grade, higher proportions of poorly differentiated tumor were observed in the TPS ≥1% (40.8% vs. 25.8%, p=0.020) and TPS ≥50% groups (53.2% vs. 27.2%, p=0.004). Among 34 patients examined with 22C3 and SP263 assays, 27 had positive results in both assays, with a cutoff of TPS ≥1% (r=0.826; 95% confidence interval, 0.736–0.916).
Conclusion
PD-L1 expression, defined as TPS ≥1%, was related to older age and poorly differentiated histology. There was a similar distribution of PD-L1 expression in both 22C3 and SP263 results.
Keywords: Asian Continental Ancestry Group; Patients; Lung Neoplasms; Gene Expression; Carcinoma, Non-Small-Cell Lung
Introduction
Although many treatments have led to improved outcomes, lung cancer remains the leading cause of cancer-related mortality worldwide1,2,3. Immune checkpoint inhibitors provide new treatment options for many cancer patients. Programmed death-ligand 1 (PD-L1) is a transmembrane protein that binds to the programmed death-1 (PD-1) receptor. This interaction leads to inactivation of cytotoxic T cells and downregulation of the immune response, permitting cancer progression and metastasis. Blocking the PD-1/PD-L1 interaction between tumor cells and activated T-cells permits the immune system to remain active4,5,6,7,8. With the introduction of immune checkpoint inhibitors, the treatment for non-small cell lung cancer (NSCLC) is developing rapidly. Anti-PD-1 inhibitors (nivolumab and pembrolizumab) and anti-PD-L1 inhibitors (atezolizumab) have been approved as second-line treatments for patients with PD-L1 expression in advanced NSCLC in the United States, Europe, and Korea. The clinical characteristics of PD-L1 expression must be better understood, to be able to choose effective immune checkpoint inhibitors.
Although there is controversy regarding the use of PD-L1 as a predictive biomarker, until now the expression of PD-L1 had been thought to be the most valuable predictor of the response to anti-PD-1/PD-L1 inhibitors9,10. Many PD-L1 inhibitors have been assessed with different immunohistochemical (IHC) assays using different antibodies, clones, platforms, scoring systems, and cutoff values. For example, the PD-L1 IHC 22C3 pharmDx assay (Agilent Technologies) and SP263 assay (Ventana Medical Systems) are approved as companion diagnostic tests for pembrolizumab. The PD-L1 IHC 28-8 pharmDx assay and SP263 have been used for nivolumab and the PD-L1 IHC SP142 assay for atezolizumab. Because there is a PD-L1 assay for each PD-1/PD-L1 inhibitor, many clinical trials have examined the correlation of PD-L1 expression with clinical outcomes. A blueprint project assessing the analytic comparability among 22C3, 28-8, SP263, and SP142 showed that three assays (excluding SP142) were closely aligned on tumor cell staining11.
In previous studies, several clinicopathologic variables have been found to be correlated with PD-L1 expression, such as age, histology, degree of differentiation, and lymph node metastasis12,13. One study reported that the expression of PD-L1 was significantly higher for women, never smokers, and in adenocarcinoma14. To clarify these discordant study results, the present study aimed to investigate the clinical and pathologic characteristics of PD-L1 expression in Korean patients with lung cancer who might be candidates for immunotherapy. We also examined the correlation between PD-L1 IHC 22C3 and SP263 data to assess the possibility of using these PD-L1 tests interchangeably.
Materials and methods
1. Patients and materials
We retrospectively reviewed the clinical and pathologic data of patients with pathologically proven lung cancer from a single institution between July 2016 and July 2017. A total of 267 patients with formalin-fixed, paraffin-embedded tissue samples were included. All data were gathered in accordance with the amended Declaration of Helsinki, following approval of the study by an independent hospital institutional review board (IRB approval number: CNUHH-2018-020). The need for written informed consent was waived because of the retrospective design of the study.
2. Immunohistochemistry
PD-L1 expression was detected using qualitative ihc staining with the in vitro diagnostic PD-L1 IHC 22C3 pharmDx test on the Dako Autostainer (Agilent Technologies, Santa Clara, CA; Dako, Carpinteria, CA, USA) and the in vitro diagnostic PD-L1 IHC SP263 test on the Ventana BenchMark platform (Ventana Medical Systems, Tucson, AZ, USA) by one pathologist. The 22C3 pharmDx assay was performed for all patients, and both assays were carried out for 34 patients.
For the 22C3 pharmDx assay, sections were stained with anti-PD-L1 22C3 mouse monoclonal primary antibody using the EnVision FLEX visualization system on a Dako Autostainer Link 48 system with negative reagent controls and cell line run controls, as described in the PD-L1 IHC 22C3 pharmDx package insert11,15. Deparaffinization, rehydration, and target retrieval was performed with a 3-in-1 procedure using PT Link. Following peroxidase blocking, specimens were incubated with monoclonal mouse primary antibody to PD-L1 or the negative control reagent. Specimens were then incubated with Mouse Linker, followed by incubation with a ready-to-use visualization reagent consisting of secondary antibody molecules and horseradish peroxidase molecules coupled to a dextran polymer backbone. The enzymatic conversion of the subsequently added chromogen results in the precipitation of a visible reaction product at the site of the antigen. The color of the chromogenic reaction is modified using a chromogen enhancement reagent; the specimen may then be counterstained and cover slipped. Results were interpreted using a light microscope.
For SP263 assay, sections were stained with anti-PD-L1 SP263 rabbit monoclonal primary antibody and a matched rabbit immunoglobulin G-negative control, using an OptiView DAB IHC Detection Kit (Ventana Medical Systems) on the BenchMark ULTRA automated staining platform11,16.
Detection and quantification of the percentage of immunoreactive tumor cells was performed according to the manufacturer's recommendations. Briefly, neoplastic cells were considered positive when any cell membrane staining was present, ignoring pure cytoplasmic immunoreaction. Staining of immune cells was also disregarded17. PD-L1 protein expression was determined using the Tumor Proportion Score (TPS), which is the percentage of viable tumor cells showing partial or complete membrane staining15. A specimen was considered PD-L1 positive with ≥50% of viable tumor cells exhibiting membrane staining at any intensity (i.e., TPS ≥50%); a weak PD-L1-positive result is 1%≤TPS<50%. We designated two categories according to cutoffs of TPS 1% or 50%.
3. Statistical analyses
Clinical characteristics and associations with biomarkers were examined by comparing differences using the Fisher exact test or independent sample t test, as appropriate. To evaluate the relationship between PD-L1 expression levels ascertained with the 22C3 and SP263 assays, we investigated 34 samples using precision analysis with Pearson's concordance correlation coefficient. A p-value of <0.05 was considered significant. Statistical analysis was performed using IBM SPSS version 23 (IBM Corp., Armonk, NY, USA).
Results
1. Baseline demographics
A total of 267 patients were included in this study (Table 1); median age was 64 years (range, 24–82 years). Most patients were male (n=191, 71.5%) and former (n=83, 31.3%) or current smokers (n=97, 36.3%). The most frequent histologic type was adenocarcinoma (n=185, 69.3%), followed by squamous cell carcinoma (n=73, 27.7%) and small cell lung cancer (n=5, 1.9%). Most patients had clinical stage IV disease (n=162, 60.7%). Some patients were analyzed for the presence of epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) rearrangement: the result showed 65 patients (24.3%) with EGFR mutation and 17 (6.4%) with ALK rearrangement. Most specimens were obtained by small biopsy (n=192, 71.9%), followed by operation (n=71, 26.6%). The histologic grade included moderately differentiated (n=76, 28.5%), poorly differentiated (n=74, 27.7%), and well differentiated (n=54, 20.2%).
Table 1. Baseline demographics.
| Characteristic | No. (%) (n=267) |
|---|---|
| Age, median (range), yr | 64 (24–82) |
| Sex | |
| Male | 191 (71.5) |
| Female | 76 (28.5) |
| Smoking history | |
| Never | 87 (32.6) |
| Former | 83 (31.3) |
| Current | 97 (36.3) |
| Histologic type | |
| Adenocarcinoma | 185 (69.3) |
| Squamous cell carcinoma | 73 (27.7) |
| Others* | 8 (3) |
| Biopsy method | |
| Operation | 71 (26.6) |
| Small biopsy† | 192 (71.9) |
| Cytology | 4 (1.5) |
| Histologic grade | |
| Well differentiated | 54 (20.2) |
| Moderately differentiated | 76 (28.5) |
| Poorly differentiated | 74 (27.7) |
| Others‡ | 23 (8.6) |
| EGFR mutation | |
| Wild type | 153 (57.3) |
| Mutated | 65 (24.3) |
| Not checked | 58 (17.6) |
| ALK FISH | |
| Negative | 93 (34.8) |
| Positive | 17 (6.4) |
| Not checked | 157 (58.8) |
*Other types consisting of small cell lung cancer (5 cases) and large cell lung cancer (3 cases). †Small biopsy specimens were collected via bronchoscopy, endobronchial ultrasound-transbronchial needle aspiration, or transthoracic needle biopsy of the lung. ‡“Others” included specimens for which histologic grade could not be checked.
EGFR: epidermal growth factor receptor; ALK: anaplastic lymphoma kinase; FISH: fluorescence in situ hybridization.
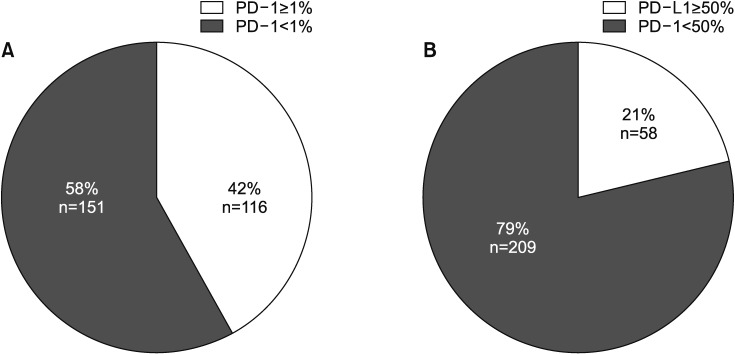
2. Characteristics of patients with PD-L1 expression
PD-L1 was evaluated in 267 patients, categorized using cutoffs of TPS 1% or 50% (Table 2, Figure 1). There were 116 (42%) and 58 (21%) patients with TPS ≥1% and ≥50%, respectively. The correlation between age and PD-L1 expression was not significant (p=0.097). But the patients in the TPS ≥1% group were older than those in the TPS <1% group (64.83±9.38 vs. 61.73±10.78, p=0.014). Furthermore, the patients in the TPS ≥50% group were older than those in the TPS <50% group, but this was not statically significant (64.69±9.39 vs. 62.36±10.51, p=0.178). The group with TPS ≥1% included patients with adenocarcinoma (n=78, 67.8%) and squamous cell carcinoma (n=34, 29.6%) histology. The TPS ≥50% group also included patients with adenocarcinoma (n=42, 72.4%) and squamous cell carcinoma (n=13, 22.4%). The rate of poorly differentiated pathology was significantly higher in patients with TPS ≥1% (40.8% vs. 25.8%, p=0.020) and TPS ≥50% (53.2% vs. 27.2%, p=0.004) than their counterparts. There were no differences with respect to smoking, EGFR mutation, ALK rearrangement status, or biopsy site.
Table 2. Clinical and pathological characteristics of the 267 patients with PD-L1 expression.
| Characteristic | PD-L1 | p-value | PD-L1 | p-value | ||
|---|---|---|---|---|---|---|
| TPS ≥1% (n=116) | TPS <1% (n=151) | TPS ≥50% (n=58) | TPS <50% (n=209) | |||
| Age, yr | 64.83±9.38 | 61.73±10.78 | 0.014 | 64.69±9.39 | 62.36±10.51 | 0.178 |
| Sex | 0.278 | 0.743 | ||||
| Male | 87 (75.0) | 104 (68.9) | 43 (74.1) | 148 (70.8) | ||
| Female | 29 (25.0) | 47 (31.1) | 15 (25.9) | 61 (29.2) | ||
| Smoking | 0.102 | 0.191 | ||||
| Never smoker | 35 (30.2) | 52 (34.4) | 19 (32.8) | 68 (32.5) | ||
| Ex-smoker | 44 (37.9) | 39 (25.8) | 23 (39.7) | 60 (28.7) | ||
| Current smoker | 37 (31.9) | 60 (39.7) | 16 (27.6) | 81 (38.8) | ||
| Histologic type | 0.082 | 0.012 | ||||
| SQC | 34 (29.6) | 38 (25.3) | 13 (22.4) | 59 (28.5) | ||
| ADC | 78 (67.8) | 107 (71.3) | 42 (72.4) | 143 (69.1) | ||
| Others* | 3 (2.6) | 5 (3.3) | 3 (4.1) | 5 (2.4) | ||
| Biopsy method | 0.853 | 0.298 | ||||
| Operation | 29 (25.0) | 42 (27.8) | 13 (22.4) | 58 (27.8) | ||
| Small biopsy† | 85 (73.3) | 107 (70.0) | 43 (74.1) | 149 (71.3) | ||
| Cytology | 2 (1.7) | 2 (1.7) | 2 (3.4) | 2 (1.0) | ||
| Histologic grade | 0.020 | 0.004 | ||||
| Well differentiated | 16 (15.5) | 38 (30.6) | 5 (10.6) | 49 (27.7) | ||
| Moderately differentiated | 33 (32.0) | 43 (34.7) | 12 (25.5) | 64 (35.6) | ||
| Poorly differentiated | 42 (40.8) | 32 (25.8) | 25 (53.2) | 49 (27.2) | ||
| Others‡ | 12 (11.7) | 11 (8.9) | 5 (10.6) | 18 (10.0) | ||
| EGFR mutation (n=218) | 0.901 | 0.888 | ||||
| Wild type | 62 (67.4) | 91 (72.8) | 33 (67.3) | 120 (71.4) | ||
| Mutated | 30 (32.7) | 34 (27.2) | 16 (32.3) | 48 (28.6) | ||
| ALK FISH (n=110) | 0.793 | 0.234 | ||||
| Negative | 44 (83.0) | 49 (86.0) | 23 (76.7) | 70 (87.5) | ||
| Positive | 9 (17.0) | 8 (14.0) | 7 (23.7) | 10 (12.5) | ||
Values are presented as mean±SD or number (%).
*Other types included small cell lung cancer (5 cases) and large cell lung cancer (3 cases). †Small biopsy specimens were collected via bronchoscopy, endobronchial ultrasound-trans-bronchial needle aspiration, or transthoracic needle biopsy of the lung. ‡“Others” included specimens for which histologic grade could not be checked.
PD-L1: programmed death-ligand 1; TPS: Tumor Proportion Score; SQC: squamous cell carcinoma; ADC: adenocarcinoma; EGFR: epidermal growth factor receptor; ALK: anaplastic lymphoma kinase; FISH: fluorescence in situ hybridization; SD: standard deviation.
Figure 1. Prevalence of programmed death-ligand 1 (PD-L1) expression using two cutoff points, in all patients. (A) The rate of PD-L1 expression was 42% with cutoff Tumor Proportion Score (TPS) 1%. (B) The rate of PD-L1 expression was 21% with cutoff TPS 50%. PD-1: programmed death-1.
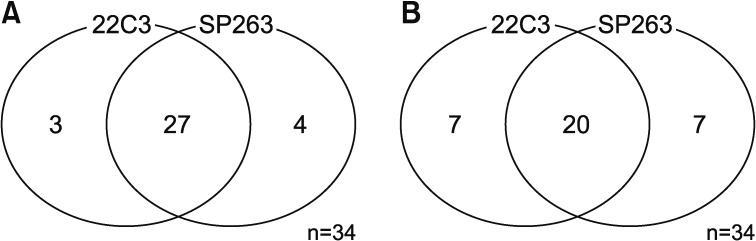
3. Relationship between 22C3 and SP263
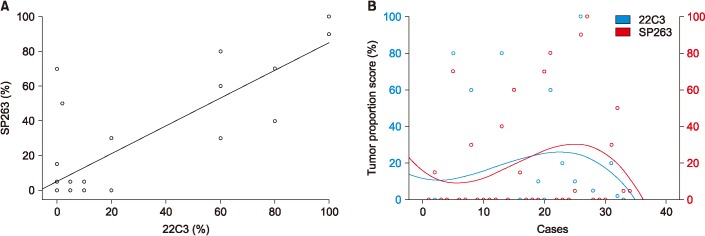
Among the 34 patients analyzed using both the 22C3 and SP263 assays, 27 patients had positive results in both assays, with a cutoff of 1% or higher (Figure 2A) and 20 patients had positive with a cutoff of 50% or higher (Figure 2B). There was a similar distribution between the 22C3 and SP263 assays assays (r=0.826; 95% confidence interval, 0.736–0.916) (Figure 3A). But it didn't show a complete correlation between two assays for each patient (Figure 3B).
Figure 2. Prevalence of programmed death-ligand 1 (PD-L1) expression using the 22C3 pharmDx and SP263 assays. (A) Among the 34 patients analyzed with both the 22C3 and SP263 assays, 27 patients gave positive PD-L1 results for both tests, with a cutoff of Tumor Proportion Score (TPS) ≥1%. (B) Those positive results were found for 20 patients, with a cutoff of TPS ≥ 50%.
Figure 3. Relationship between programmed death-ligand 1 (PD-L1) immunohistochemical 22C3 pharmDx and SP263 assay. (A) Scatter diagrams illustrating the relationship between expression levels with respect to 22C3 pharmDx and SP263 assays. Each point in the diagram indicates the percentage of positive tumor cells in the 22C3 (x-axis) and SP263 (y-axis) assays. A significantly high relationship was observed between the 22C3 and SP263 assays (r=0.826; 95% confidence interval, 0.736-0.916). (B) Analytic comparison of the percentage of tumor proportion score by case for each assay. Data points expressed the scores on each case (blue points for 22C3 pharmDx and red points for SP263 assay).
Discussion
In this study, we explored the clinicopathologic characteristics of PD-L1-positive Korean patients with lung cancer. For all enrolled patients, we performed PD-L1 IHC assays using monoclonal mouse anti-PD-1, clone 22C3; PD-L1 expression was described using TPS. A total 42% of patients were PD-L1 positive using a cutoff of TPS ≥1%; this proportion was 21% with a cutoff of TPS ≥50%. Patients in the PD-L1-positive (cutoff TPS ≥1%) group were significantly older than those in the PD-L1-negative group. But in cutoff TPS ≥ 50% group, the patients were older than those in the TPS <50% group, but this was not statically significant. This seems to be due to the small sample size of the TPS ≥50% group to obtain statistical significance. The most frequent histologic grade was poorly differentiated type in both the TPS ≥1% and TPS ≥50% groups.
In previous studies, several clinicopathologic variables were found to be correlated with PD-L1 expression, but there were discordant results among these studies. Mu et al.12 evaluated 109 patients with adenocarcinoma and estimated the association between expressed PD-L1 and tumor histologic type, degree of differentiation, and lymph node metastasis. Those authors found that higher expression of PD-L1 was related to adenocarcinoma histology and overall survival12. Velcheti et al.18 explored cases of NSCLC including 340 cases in Greece and 204 cases from Yale University in the U.S. They found that advanced stage and squamous cell carcinoma were correlated with high PD-L1 expression. Cooper et al.13 analyzed 678 tissue samples from patients with stage I-III NSCLC and 52 paired nodal metastases. The authors showed that younger patients and those with poor differentiation were associated with high PD-L1 expression; there was no association with sex, tumor size, stage, nodal status, or EGFR or KRAS mutation status13. Azuma et al.14 evaluated 164 specimens of surgically resected NSCLC and found that the expression of PD-L1 was significantly higher in women, never smokers, and patients with adenocarcinoma. These inconsistent results suggest that PD-L1 expression might differ by race or residence, and it may be necessary to analyze the clinical characteristics of each group.
We also examined the relationship between IHC assays in 34 patients and found a similar distribution of PD-L1 expression between the 22C3 pharmDx and SP263 assays. Although there has been some controversy regarding use of PD-L1 as a predictive biomarker, the expression of PD-L1 has been thought to be the most valuable predictor of response to anti-PD-1/PD-L1 inhibitors9,10. Many PD-L1 inhibitors have been assessed with different IHC assays using different antibodies, clones, platforms, scoring systems, and cutoff values11. For example, the 22C3 pharmDx and SP263 assay are approved as companion diagnostic tests for pembrolizumab. The 28-8 pharmDx and SP263 assay have been used for nivolumab and the SP142 assay for atezolizumab.
Because there is an PD-L1 assay for each PD-1/PD-L1 inhibitor, many clinical trials have examined the correlation of PD-L1 expression with clinical outcomes. A blueprint project assessed analytic comparability among the 22C3, 28-8, SP263, and SP142 assays using 39 NSCLC tumor samples. The result showed that three assays (excluding SP142) were closely aligned on tumor cell staining11. In a harmonized trial, 15 lung cancer resection specimens were used to assess interobserver concordance and PD-L1 IHC patterns using the 28-8, 22C3, SP142, and SP263 assays. The result showed that carcinoma cells can be reproducibly scored and there were no differences in interobserver concordance among the tested assays19. However, the scoring of immune cells yielded low concordance rates and might require specific standardization19. A French multicenter harmonization study showed that the 28-8, 22C3, and SP263 assays were comparable20. Another study assessed the concordance between PD-L1 assays using 493 formalin-fixed, paraffin-embedded archival NSCLC samples; the assays showed similar patterns of tumor membrane staining with high concordance between the SP263, 22C3, and 28-8 assays21. And the Italian multicenter comparison study of the 22C3 and SP263 assays found high correlation between PD-L1 IHC expression with the two tests17. In short, all these results suggest that the 22C3 and SP263 assays could be used interchangeably. Our study showed a similar distribution but not complete concordance between assays. This might be due to the small number of subjects who performed both assays simultaneously.
There are some limitations to the present study. First, as mentioned above, this is a retrospective study from a single institution with a small sample size. Second, because most specimens were archival tissue, the PD-L1 IHC results might differ from those of fresh tissue. However, this situation may reflect real world clinical practice in that not all patients are able to undergo rebiopsy before immunotherapy. Third, we could not perform both 22C3 and SP263 assays in all patients; therefore, the interpretation that there is a high correlation between the two tests should be made with caution. Further large-scale clinical studies should be performed to investigate the clinicopathologic variables of PD-L1 in NSCLC.
In conclusion, this retrospective study demonstrated that Korean patients with lung cancer who had PD-L1 expression (TPS ≥1%) were older and had poorly differentiated histology. In patients with these characteristics, use of the PD-L1 IHC assay should be actively considered. We also found that the 22C3 pharmDx and SP263 assays could be used interchangeably. However, a large-scale multicenter study is needed to better understand PD-L1 expression in lung cancer.
Acknowledgments
This work was supported by a clinical research grant from Chonnam National University Hwasun Hospital 2017.
Footnotes
- Conceptualization: Oh IJ, Park HY.
- Methodology: Oh IJ, Choi YD.
- Formal analysis: Park HY, Oh IJ, Choi YD.
- Data curation: Park HY, Oh IJ, Kho BG, Kim TO, Shin HJ, Park CK, Kim YC.
- Software: Park HY.
- Validation: Oh IJ.
- Investigation: Park HY, Oh IJ, Park CK, Kim YC, Kwon YS, Kim YI, Lim SC.
- Writing - original draft preparation: Park HY.
- Writing - review and editing: Oh IJ, Park CK, Kim YC.
- Approval of final manuscript: all authors.
Conflicts of Interest: No potential conflict of interest relevant to this article was reported.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Park JY, Jang SH. Epidemiology of lung cancer in Korea: recent trends. Tuberc Respir Dis. 2016;79:58–69. doi: 10.4046/trd.2016.79.2.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kweon SS. Updates on cancer epidemiology in Korea, 2018. Chonnam Med J. 2018;54:90–100. doi: 10.4068/cmj.2018.54.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen DS, Irving BA, Hodi FS. Molecular pathways: next-generation immunotherapy: inhibiting programmed death-ligand 1 and programmed death-1. Clin Cancer Res. 2012;18:6580–6587. doi: 10.1158/1078-0432.CCR-12-1362. [DOI] [PubMed] [Google Scholar]
- 8.Hall RD, Gray JE, Chiappori AA. Beyond the standard of care: a review of novel immunotherapy trials for the treatment of lung cancer. Cancer Control. 2013;20:22–31. doi: 10.1177/107327481302000105. [DOI] [PubMed] [Google Scholar]
- 9.Grigg C, Rizvi NA. PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction. J Immunother Cancer. 2016;4:48. doi: 10.1186/s40425-016-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the Blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. 2017;12:208–222. doi: 10.1016/j.jtho.2016.11.2228. [DOI] [PubMed] [Google Scholar]
- 12.Mu CY, Huang JA, Chen Y, Chen C, Zhang XG. High expression of PD-L1 in lung cancer may contribute to poor prognosis and tumor cells immune escape through suppressing tumor infiltrating dendritic cells maturation. Med Oncol. 2011;28:682–688. doi: 10.1007/s12032-010-9515-2. [DOI] [PubMed] [Google Scholar]
- 13.Cooper WA, Tran T, Vilain RE, Madore J, Selinger CI, Kohonen-Corish M, et al. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer. 2015;89:181–188. doi: 10.1016/j.lungcan.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol. 2014;25:1935–1940. doi: 10.1093/annonc/mdu242. [DOI] [PubMed] [Google Scholar]
- 15.Roach C, Zhang N, Corigliano E, Jansson M, Toland G, Ponto G, et al. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non-small-cell lung cancer. Appl Immunohistochem Mol Morphol. 2016;24:392–397. doi: 10.1097/PAI.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebelatto MC, Midha A, Mistry A, Sabalos C, Schechter N, Li X, et al. Development of a programmed cell death ligand-1 immunohistochemical assay validated for analysis of non-small cell lung cancer and head and neck squamous cell carcinoma. Diagn Pathol. 2016;11:95. doi: 10.1186/s13000-016-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchetti A, Barberis M, Franco R, De Luca G, Pace MV, Staibano S, et al. Multicenter comparison of 22C3 PharmDx (Agilent) and SP263 (Ventana) assays to test PD-L1 expression for NSCLC patients to be treated with immune check-point inhibitors. J Thorac Oncol. 2017;12:1654–1663. doi: 10.1016/j.jtho.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheel AH, Dietel M, Heukamp LC, Johrens K, Kirchner T, Reu S, et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol. 2016;29:1165–1172. doi: 10.1038/modpathol.2016.117. [DOI] [PubMed] [Google Scholar]
- 20.Adam J, Rouquette I, Damotte D, Badoual C, Danel C, Damiola F, et al. PL04a.04: multicentric French harmonization study for PD-L1 IHC testing in NSCLC. J Thorac Oncol. 2017;12(1 Suppl):S11–S12. doi: 10.1093/annonc/mdy014. [DOI] [PubMed] [Google Scholar]
- 21.Ratcliffe MJ, Sharpe A, Midha A, Barker C, Scott M, Scorer P, et al. Agreement between programmed cell death ligand-1 diagnostic assays across multiple protein expression cutoffs in non-small cell lung cancer. Clin Cancer Res. 2017;23:3585–3591. doi: 10.1158/1078-0432.CCR-16-2375. [DOI] [PubMed] [Google Scholar]