Abstract
The WW and C2 domain containing 1 (WWC1) gene encodes a protein named WWC1 (or KIBRA), which is involved in the Hippo signaling pathway. WWC1 is often lost in triple‐negative breast cancer and has been shown to suppress tumor metastasis. In this study, 470 breast cancer patients were recruited and WWC1 expression in the tumor samples was measured with quantitative reverse transcriptase PCR. Associations of WWC1 expression with breast cancer survival were analyzed using the Cox proportional hazards regression model and Kaplan–Meier survival analysis. The relationship between WWC1 expression and methylation was evaluated in a dataset from The Cancer Genome Atlas. Using our microarray data on gene expression and the Ingenuity Pathway Analysis, we predicted the WWC1‐associated signaling pathways in breast cancer. Our results showed that low expression of WWC1 was significantly associated with advanced‐stage diseases, high‐grade tumors, and estrogen receptor‐ or progesterone receptor‐negative status. Compared to those with high expression, patients with low WWC1 had higher risk of breast cancer relapse [hazard ratio (HR) = 2.06, 95% confidence interval (CI): 1.26–3.37] and higher risk of death (HR = 2.76, 95% CI: 1.51–5.03). The association with relapse‐free survival remained significant after adjustment for disease stage, tumor grade, and hormone receptor status and was replicated in a public dataset. Analysis of high‐throughput gene expression data indicated that WWC1 was involved in the Hippo signaling pathway. Online data also suggested that DNA methylation was inversely associated with WWC1 expression. The study confirmed that low WWC1 expression was associated with aggressive breast cancer and poor survival outcomes.
Keywords: breast cancer, methylation, overall survival, relapse‐free survival, WWC1
Abbreviations
- EMT
epithelial‐to‐mesenchymal transition
- ER
estrogen receptor
- HR
hazard ratio
- IPA
Ingenuity Pathway Analysis
- OS
overall survival
- PR
progesterone receptor
- qRT‐PCR
quantitative reverse transcriptase PCR
- RFS
relapse‐free survival
- TCGA
The Cancer Genome Atlas
- TNBC
triple‐negative breast cancer
- WWC1
WW and C2 domain containing 1
Breast cancer is the most common malignancy and a leading cause of cancer death among women worldwide. The malignancy accounts for 25% of all cancer cases and 15% of all cancer deaths. In developed countries, these figures are even higher, about 50% of all cancer cases and 38% of cancer deaths 1. Breast cancer is a heterogeneous disease 2, and accurately characterizing breast tumor at molecular and genetic levels is important in identifying sensitive and responsive targets for treatment. Triple‐negative breast cancer (TNBC) is a group of aggressive tumors with poor prognosis. Nearly 70% of TNBC had a deletion in chromosome 5 (5q11–5q35) 3, 4, 5. A recent study by Knight et al. indicated that a gene in the region named the WW and C2 domain containing 1 (WWC1) gene could inhibit breast cancer progression. The gene encodes a protein called WWC1 (or KIBRA) which interacts with PTPN14 (protein tyrosine phosphatase nonreceptor 14) and inhibits the oncogenic activity of YAP/TAZ 6.
WW and C2 domain containing 1 consists of two WW domains in the amino‐terminus, an internal C2‐like domain, and a carboxy‐terminal glutamic acid‐rich stretch. The protein was originally found to be predominantly expressed in the kidneys and brain 7 and is suspected to be involved in memory performance and associated with age‐dependent risk of Alzheimer's disease 8, 9, 10, 11. Studies also found that the protein was involved in regulation of cell migration 12, 13, 14. Genetic screening of Drosophila suggests that this gene may function as a potential tumor suppressor by regulating the Hippo signaling pathway 15, 16, 17 which controls tissue growth and tumorigenesis, inhibits cell proliferation, and promotes apoptosis 18, 19, 20, 21, 22, 23. WWC1 associates with both large tumor suppressor (Lats) 1 and Lats2 to regulate the Hippo signaling in human cells 24. One study indicated that activation of the Hippo signaling pathway by WWC1 in breast cancer cells could suppress epithelial‐to‐mesenchymal transition (EMT) 25. Another showed that WWC1 regulated epithelial cell polarity by suppressing apical exocytosis through direct inhibition of aPKC kinase activity in the PAR3/PAR6/aPKC complex 26. Loss of epithelial cell polarity is intricately related to tumor progression and invasion 27. A previous study also found that WWC1 formed a complex with dynein light chain 1, an estrogen receptor‐α (ER)‐interacting protein, and played a role in ER transactivation in breast cancer cells 24.
Despite the growing evidence on WWC1's involvement in breast cancer progression, no study has evaluated its relationship with tumor characteristics and patient survival. To address these issues, we conducted a clinical study to examine WWC1 expression in association with patient clinical and pathological features as well as disease outcomes.
Materials and methods
Patients and tumor samples
Breast tumor samples were collected from patients during surgery in a university hospital between January 1998 and July 1999 and in Mauriziano Hospital between October 1996 and August 2012. Both hospitals are affiliated with University of Turin. The patients were followed regularly after surgery for postoperative treatment and checkup. The average age of patients at surgery was 58 years (range: 23–89). The median relapse‐free survival (RFS) and overall survival (OS) were 82.2 months (range: 1.7–196.4) and 83.6 months (range: 4.2–196.4), respectively. Information on disease stage, tumor grade, hormone receptor status, follow‐up time, and survival outcomes was extracted from patient medical records and follow‐up contacts. A detailed description of the information has been described elsewhere 28. The study was approved by the ethical review committees in the hospitals, and informed written consents were obtained from the patients. The study methodologies conformed to the standards set by the Declaration of Helsinki.
RNA extraction
Total RNA was extracted from 470 fresh‐frozen breast tumor samples using the AllPrep DNA/RNA Kit (Qiagen, Hilden, Germany). The concentrations and qualities of RNA were measured with the NanoDrop 1000 spectrophotometer, and the specimens were stored at −80 °C until analysis. The RNA integrity was assessed on the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Quantitative reverse transcriptase PCR
WWC1 expression was analyzed in 470 tumor samples using quantitative reverse transcriptase PCR (qRT‐PCR). PCR primers were designed and synthesized by IDT (San Diego, CA, USA). Total RNA (1 μg) was reverse‐transcribed using the cDNA Reverse Transcription Kit (LifeTech, Carlsbad, CA, USA); qRT‐PCR was performed with the SYBR Select Master Mix (LifeTech) in a 384‐well plate using the Roche LightCycler 480 real‐time PCR detection system (Roche Diagnostics Ltd., Rotkreuz, Switzerland). In each PCR (10 μL), cDNA template (0.5 μL) was mixed with 200 nm primers and 5 μL SYBR PCR master mix. Cycling conditions included the following: 50 °C for 2 min to activate UDG, 95 °C for 2 min to activate Taq polymerase, 40 cycles of 95 °C for 15 s and 60 °C for 60 s, and 60 °C for 1 min. The cycle threshold (Ct) was determined for each sample in triplicate. WWC1 expression was normalized to GAPDH, and the relative value was calculated as expression index (EI), using the formula 1000 × 2(−ΔCt), where ΔCt = Ct(WWC1) − Ct(GAPDH). PCR primers were 5′‐TCCGCAGTCCTGGAAACATT‐3′ (forward) and 5′‐GTGGATTCCCAATGAGCCGA‐3′ (reverse) for WWC1, and 5′‐GTCAAGGCTGAGAACGGGAA‐3′ (forward) and 5′‐AAATGAGCCCCAGCCTTCTC‐3′ (reverse) for GAPDH.
Breast cancer microarray analysis
Of the 470 tumor samples analyzed by qRT‐PCR for WWC1 expression, 204 samples were also used for microarray analysis with the Illumina Expression BeadChip (HumanRef‐8 v3) (Illumina, San Diego, CA, USA) according to the manufacturer's protocol, which was described elsewhere 29, 30. In brief, microarray data were preprocessed by the BeadStudio software and analyzed using the R statistical software and Bioconductor 31. The Lumi R package was used to normalize the data 32. The genefilter package was applied for nonspecific filtering; gene expression levels with interquartile range < 0.5 were discarded.
Statistical methods
Values of WWC1 EI were categorized into three groups according to the tertile distribution: low, medium, and high. Chi‐square test was performed to evaluate the association between WWC1 expression and clinicopathologic characteristics of breast cancer patients. The association of WWC1 expression with breast cancer survival was analyzed using the Kaplan–Meier method and log‐rank test. Univariate and multivariate Cox proportional hazards regression models were used to determine the hazard ratio (HR) and 95% confidence interval (CI) between the risk of relapse or death and WWC1 expression. In multivariate analysis, the regression models were adjusted for age at surgery, tumor grade, disease stage, and ER and progesterone receptor (PR) status. To validate the association of WWC1 expression with breast cancer survival, an online tool named Kaplan–Meier Plotter (http://kmplot.com/analysis/) was used to analyze the relationship between breast cancer survival and WWC1 expression based on the datasets available online 33. Spearman's rank correlation analysis was performed to identify genes whose expression was correlated with WWC1 using our microarray data. These WWC1‐correlated genes were interrogated by the bioinformatics software Ingenuity Pathway Analysis (IPA), which predicted potential signaling pathways influenced by WWC1. WWC1 methylation data generated by the Illumina HumanMethylation450 BeadChip (HM450) in normal tissue samples in The Cancer Genome Atlas (TCGA) Breast Invasive Carcinoma Provisional Database were downloaded from FireBrowse (http://firebrowse.org). WWC1 gene methylation and mRNA expression (RNA Seq V2 RSEM) data in TCGA were retrieved from the Web‐based tool cBioPortal (http://www.cbioportal.org/index.do) which only contains methylation data from the probes that had strong negative correlations with methylation signals 34, 35. Correlations between DNA methylation and gene expression of WWC1 were analyzed using the Spearman correlation coefficient within graphpad prism 7 (GraphPad Software Inc., San Diego, CA, USA). All the statistical tests were performed with the Statistical Analysis System (sas) software, version 9.2 (SAS Institute Inc., Cary, NC, USA). P‐values were two‐sided, and those < 0.05 were considered statistically significant.
Results
Relationship between WWC1 expression and patient characteristics
Table 1 shows the clinicopathologic characteristics of 470 breast cancer patients in association with WWC1 expression measured by qRT‐PCR. Low expression of WWC1 was significantly associated with advanced‐stage diseases (P = 0.018) and high‐grade tumors (P < 0.0001). Patients with ER‐negative tumors had lower expression of WWC1 compared with those with ER‐positive tumors (P = 0.0017). A similar relationship was also observed for PR (P = 0.00029). A correlation analysis was performed on 204 samples which had WWC1 expression data generated by both qRT‐PCR and microarray. A significant correlation (Spearman r = 0.46, P < 0.0001) was found between the two methods. In a TCGA dataset, WWC1 expression was lower in breast tumors compared to matched adjacent normal tissues (P = 0.043; Fig. 3D).
Table 1.
Associations of WWC1 expression with clinical and pathological factors of breast cancer.
| Variables |
Total No. (N = 470) |
WWC1 expression | P‐value | ||
|---|---|---|---|---|---|
|
Low No. (%) |
Mid No. (%) |
High No. (%) |
|||
| Disease stage | |||||
| Stage 1 | 151 (33.93) | 39 (25.83) | 50 (33.11) | 62 (41.06) | 0.018 |
| Stage 2 | 222 (49.89) | 74 (33.33) | 80 (36.04) | 68 (30.63) | |
| Stages 3 and 4 | 72 (16.18) | 31 (43.06) | 18 (25.00) | 23 (31.94) | |
| Tumor grade | |||||
| Grade 1 | 48 (10.55) | 13 (27.08) | 10 (20.84) | 25 (52.08) | < 0.0001 |
| Grade 2 | 178 (39.12) | 42 (23.60) | 71 (39.89) | 65 (36.52) | |
| Grade 3 | 229 (50.33) | 95 (41.48) | 73 (31.88) | 61 (26.64) | |
| ER status | |||||
| Positive | 313 (68.19) | 85 (27.16) | 114 (36.42) | 114 (36.42) | 0.0017 |
| Negative | 146 (31.81) | 64 (43.84) | 42 (28.77) | 40 (27.40) | |
| PR status | |||||
| Positive | 171 (51.82) | 43 (25.15) | 65 (38.01) | 63 (36.84) | 0.00029 |
| Negative | 159 (48.18) | 67 (42.14) | 46 (28.93) | 46 (28.93) | |
WWC1 expression and breast cancer survival
Associations of breast cancer survival with WWC1 expression are shown in Table 2. Compared to those with high expression, patients with low expression of WWC1 had higher risk of relapse (HR = 1.49, 95% CI: 1.15–1.94, P for trend = 0.0028) and higher risk of death (HR = 1.66, 95% CI: 1.23–2.24, P for trend = 0.00089). After adjusting for age at surgery, tumor grade, disease stage, and ER and PR status, the association with relapse remained significant (HR = 1.33, 95% CI: 1.003–1.75, P for trend = 0.047). Since the associations with survival outcomes were not substantially different between low and medium WWC1 expression, we grouped the two groups together and compared their disease outcomes with those of high WWC1 expression. Survival analysis showed that patients with low and mid expression (low and mid tertile) had significantly increased risk of relapse and death (HR = 2.06, P = 0.0038, and HR = 2.76, P = 0.00095, respectively; Table 2, Fig. 1A,B) compared to those with high expression (high tertile). The association remained significant for relapse after adjusting for clinical and pathological variables (HR = 1.83, P = 0.024; Table 2).
Table 2.
Associations of WWC1 expression with breast cancer survival.
| Variables | RFS | P‐value | OS | P‐value | RFSa | P‐value | OSa | P‐value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | HR | 95% CI | |||||
| High | 1 | 1 | 1 | 1 | ||||||||
| Mid | 1.88 | 1.09–3.24 | 0.024 | 2.53 | 1.31–4.87 | 0.0055 | 1.83 | 1.02–3.27 | 0.042 | 1.78 | 0.87–3.62 | 0.11 |
| Low | 2.30 | 1.33–3.99 | 0.0030 | 3.05 | 1.58–5.89 | 0.00093 | 1.83 | 1.02–3.29 | 0.044 | 1.94 | 0.97–3.89 | 0.061 |
| Continuous | 1.49 | 1.15–1.94 | 0.0028 | 1.66 | 1.23–2.24 | 0.00089 | 1.33 | 1.003–1.75 | 0.047 | 1.36 | 0.98–1.89 | 0.068 |
| High (upper tertile) | 1 | 1 | 1 | 1 | ||||||||
| Low (lower two tertiles) | 2.06 | 1.26–3.37 | 0.0038 | 2.76 | 1.51–5.03 | 0.00095 | 1.83 | 1.08–3.08 | 0.024 | 1.86 | 0.99–3.52 | 0.055 |
The significant statistical results are indicated in bold.
Adjusted for age at surgery, tumor grade, disease stage, ER, and PR.
Figure 1.
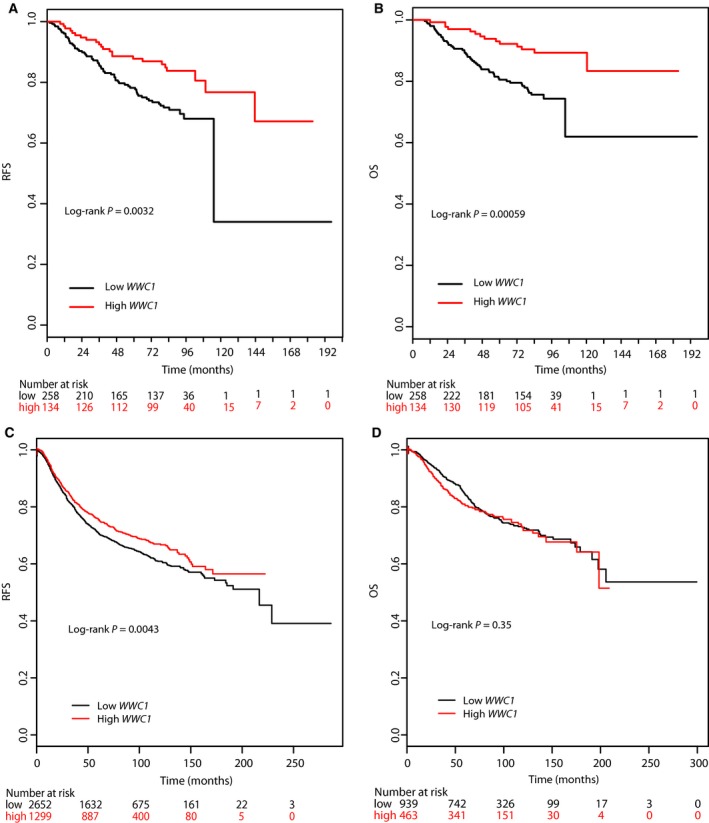
Kaplan–Meier survival curves in patients with high and low WWC1 expression. (A) RFS curves in patients of our study with high (upper tertile) and low (two lower tertiles) expression of WWC1. (B) OS curves in patients of our study with high (upper tertile) and low (two lower tertiles) expression of WWC1. (C) RFS curves in patients from online datasets with high (upper tertile) and low (two lower tertiles) expression of WWC1. (D) OS in patients from online datasets with high (upper tertile) and low (two lower tertiles) expression of WWC1.
The relationship between breast cancer survival and WWC1 expression was also analyzed using an online tool, the Kaplan–Meier Plotter, in an online database containing 5134 breast cancer samples with a mean follow‐up of 69 months from 35 publically available microarray datasets (E‐MATB‐365, E‐TABM‐43, GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE16446, GSE16716, GSE17705, GSE17907, GSE18728, GSE19615, GSE20194, GSE20271, GSE2034, GSE20685, GSE20711, GSE51653, GSE2603, GSE16971, GSE2990, GSE31448, GSE31519, GSE32646, GSE3494, GSE37946, GSE41998, GSE42568, GSE45255, GSE4611, GSE5327, GSE6532, GSE7390, and GSE9195). The microarray probe 213085_s_at was used to represent WWC1 expression which was categorized into high and low groups by upper tertile. As shown in Fig. 1C,D, high WWC1 expression was associated with RFS (log‐rank P = 0.0043), but not with OS.
WWC1‐related pathways
To explore the WWC1‐related signaling pathways, we performed co‐expression analysis on genes significantly correlated with WWC1 expression in our breast cancer microarray data. A total of 3560 out of 22 184 genes were found to be significantly co‐expressed with WWC1 (FDR q value < 0.05 and Spearman correlation coefficient > 0.2). Bioinformatics analysis of these genes by IPA suggested that the top five canonical pathways involved were EIF2 signaling, mTOR signaling, Hippo signaling, regulation of eIF4 and p70S6K signaling, and tRNA charging (Fig. 2A,B).
Figure 2.
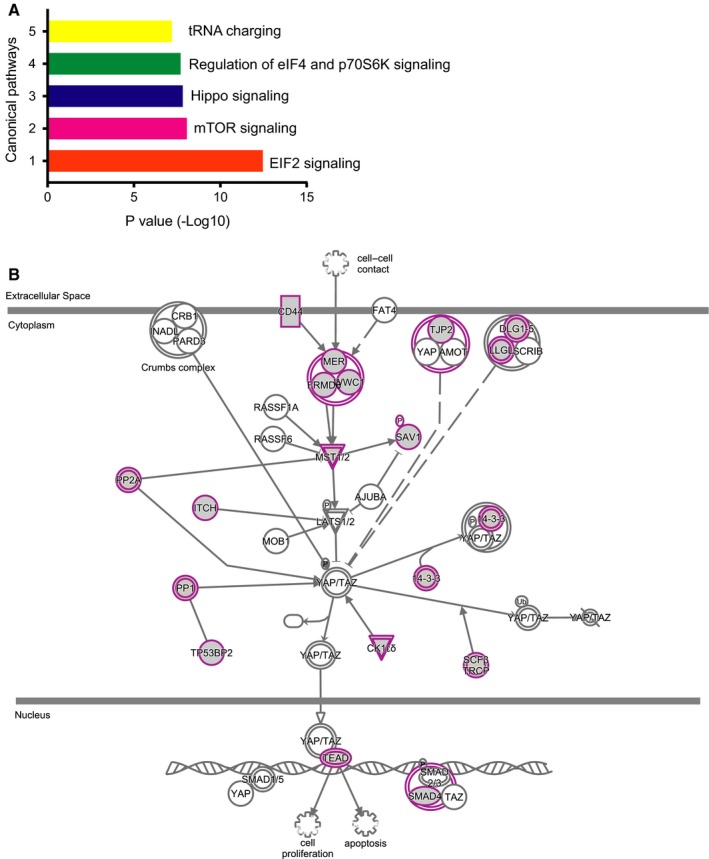
Signaling pathways identified by IPA based on the genes co‐expressed with WWC1 in our expression microarray data. (A) Top 5 canonical pathways based on P‐value; (B) WWC1 in the Hippo signaling pathway. Node shape represents the functional class of gene product. Double outline for complexes/group of different proteins. Purple for upregulation.
WWC1 methylation status and expression
Based on the UCSC Genome Browser, a CpG island was identified within the WWC1 promoter region (Fig. 3A). Eight methylation probes, cg26980033, cg07243784, cg09547099, cg02865818, cg05253577, cg11614065, cg15232798, and cg14599284, were designed in the region by Illumina in the HumanMethylation450 BeadChip. A significant correlation between WWC1 methylation and expression across all the probes in the gene was observed (r = −0.29, P < 0.0001; Fig. 3B). Analyzing methylation at each specific probe in relation to gene expression in 84 breast tumor samples and matched adjacent normal tissues, we also found that methylation in most of the probes was inversely correlated with expression (Fig. 3C).
Figure 3.
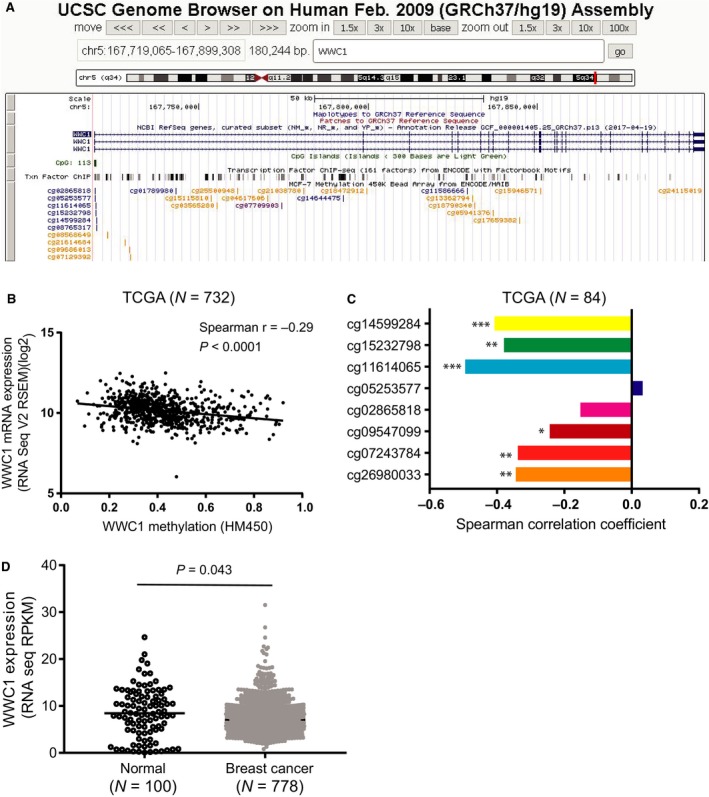
Correlation between WWC1 expression and methylation in breast cancer in the TCGA database. (A) A screenshot from the UCSC Genome Browser shows the CpG island in the WWC1 promoter and CpG site probes in the Illumina HumanMethylation450 BeadChip. (B) Scatter plot shows the Spearman correlation between WWC1 expression and methylation in all CpG sites in the WWC1 gene using the TCGA breast cancer data from cBioPortal. (C) Bar charts indicate the correlation coefficients between WWC1 expression and methylation in each of the 8 CpG sites located in the CpG island using data from the TCGA breast cancer and matched normal tissues (Spearman's rank correlation; *P < 0.05; **P < 0.001; ***P < 0.0001). (D) The WWC1 expression in normal tissue is higher than that in breast cancer using Student's t‐test.
Discussion
In this study, we found that WWC1 expression was low in breast tumors compared to normal tissues and that low expression was associated with aggressive tumors and poor RFS. The WWC1 gene is located in 5q34 with high expression in the brain and kidneys 7. Most studies have investigated the effect of WWC1 on cognition and memory performance. Although its function in non‐neuronal cells is less clear, emerging evidence suggests that WWC1 may play a role in cancer. Decreased WWC1 was shown to maintain the stemness of osteosarcoma cells 36. Knockdown WWC1 expression enhanced the migration and invasion of immortalized breast epithelial cells, and reduced expression of WWC1 in claudin‐low breast cancer was associated with poor prognosis 25. Loss of WWC1 function could also induce EMT and promote mammary epithelial cell transformation, whereas WWC1 overexpression was found to suppress head and neck squamous cell carcinoma cells in forming colony and oncosphere in soft agar 37. Based on the findings, WWC1 was considered a potential tumor suppressor 38. A recent study confirmed that WWC1 could inhibit breast tumor metastasis both in vitro and in vivo 6.
Consistent with the above reports, we found that low expression of WWC1 was associated with shorter RFS, suggesting that WWC1 may play a role in the suppression of tumor progression. The association between WWC1 and RFS was replicated independently in a larger dataset. Despite most studies showing WWC1 as a possible tumor suppressor, a few studies found inconsistent results. One study showed that high expression of WWC1 was associated with poor prognosis of gastric cancer patients with low expression of atypical protein kinase C 39. A recent study reported that WWC1 could enhance cell proliferation, migration, and invasion in both immortalized and cancerous prostate epithelial cells, and overexpression of WWC1 was observed in prostate cancer 14. These inconsistent findings may indicate that WWC1 has tissue‐specific and/or context‐dependent effects on cancer cells. More than 30 public microarray datasets were included in the Kaplan–Meier Plotter analysis, and these datasets had diverse patient background and treatment, which may affect OS outcome.
WW and C2 domain containing 1 was found to regulate growth suppression through the Hippo signaling pathway in Drosophila 15, 16, 17, and this finding was confirmed in mammalian cells 40. Based on our microarray data on gene expression, we confirmed that WWC1 was involved in the Hippo signaling pathway in our IPA, which focused on the genes co‐expressed with WWC1. We also found that these co‐expressed genes were enriched in the mTOR signaling pathway. Hippo and mTOR are two critical pathways that control cell growth and tissue/organ homeostasis. Previous studies have provided evidence for signaling cross talk between the mTOR and Hippo pathways. YAP, which is the main downstream target of the mammalian Hippo pathway, can regulate the mTOR pathway via its effects on miR‐29 expression 41. WWC1 could be an important signaling node linking mTOR and Hippo pathways. Promoter methylation that leads to tumor suppressor gene silencing is an important mechanism involved in tumor initiation, progression, and metastasis 42, 43. WWC1 methylation was found to be associated with unfavorable prognostic indicators in patients with chronic lymphocytic leukemia 44. Epigenetic inactivation of WWC1 was found frequently in B‐cell acute lymphocytic leukemia 45. One study reported that WWC1 expression was significantly affected by CpG island methylation in the promoter region and the promoter methylation was elevated in human clear renal cell carcinoma 46. In our study, we found a significant inverse correlation between CpG methylation and WWC1 expression, suggesting that DNA methylation may regulate WWC1 expression. We also noticed in our study that methylation regulation was not restricted to the promoter region as methylation of CpG sites in other regions of the gene could be involved (data not shown).
Genetic polymorphisms in the WWC1 gene may affect expression and contribute to disease risk. One recent study identified WWC1 as a possible genetic locus for breast cancer susceptibility among women of African ancestry 47. A genomewide association study reported that individuals carrying the T allele of rs17070145 in WWC1 had significantly better memory scores in the Buschke Selective Reminding Test compared to those carrying the C allele 9. The single nucleotide polymorphism rs17070145 resides in intron 9 of the WWC1 gene. We found that patients with genotype TT or TC in rs17070145 had better relapse‐free and OS compared to those with genotype CC, but eQTL was not detected in this locus, indicating no impact of this genotype on WWC1 expression (data not shown). The biological mechanism underlying the genotype's association with breast cancer survival remains to be elucidated.
Of note, the relatively small sample size may limit the statistical power of our study. We also did not evaluate whether our tumor samples had a deletion in chromosome 5q which may explain low expression of WWC1.
Conclusions
In summary, we found that low WWC1 expression was associated with shorter RFS of breast cancer patients compared to high expression and that the association was independent from tumor grade, disease stage, and hormone receptor status. We further replicated this association in a large independent dataset available online. The study also showed that WWC1 expression was inversely associated with tumor grade and disease stage and hormone receptor‐positive tumors had higher expression than receptor‐negative ones. These associations indicate that tumor biology and cell differentiation may influence the expression of WWC1. Bioinformatics analysis of our microarray data on gene expression confirmed that WWC1 expression was involved in the Hippo signaling pathway. Analysis of online methylation data indicated that DNA methylation could influence WWC1 expression. Further investigating the biological actions of WWC1 in breast cancer will help to elucidate the mechanisms of tumor progression.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
HY and ZW designed the study, analyzed the data, and wrote the manuscript. ZW, YS, YF, and MT performed the laboratory experiments. HY, ZW, DK, and NB interpreted the data. All authors approved the final version of the manuscript, including the authorship list.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J and Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Polyak K (2007) Breast cancer: origins and evolution. J Clin Invest 117, 3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johannsdottir HK, Jonsson G, Johannesdottir G, Agnarsson BA, Eerola H, Arason A, Heikkila P, Egilsson V, Olsson H, Johannsson OT et al (2006) Chromosome 5 imbalance mapping in breast tumors from BRCA1 and BRCA2 mutation carriers and sporadic breast tumors. Int J Cancer 119, 1052–1060. [DOI] [PubMed] [Google Scholar]
- 4. Natrajan R, Lambros MB, Rodriguez‐Pinilla SM, Moreno‐Bueno G, Tan DS, Marchio C, Vatcheva R, Rayter S, Mahler‐Araujo B, Fulford LG et al (2009) Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res 15, 2711–2722. [DOI] [PubMed] [Google Scholar]
- 5. Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A et al (2010) Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene 29, 2013–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knight JF, Sung VYC, Kuzmin E, Couzens AL, de Verteuil DA, Ratcliffe CDH, Coelho PP, Johnson RM, Samavarchi‐Tehrani P, Gruosso T et al (2018) KIBRA (WWC1) is a metastasis suppressor gene affected by chromosome 5q loss in triple‐negative breast cancer. Cell Rep 22, 3191–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kremerskothen J, Plaas C, Buther K, Finger I, Veltel S, Matanis T, Liedtke T and Barnekow A (2003) Characterization of KIBRA, a novel WW domain‐containing protein. Biochem Biophys Res Commun 300, 862–867. [DOI] [PubMed] [Google Scholar]
- 8. Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, Murray GD, Whalley LJ, Starr JM and Deary IJ (2009) Association of KIBRA and memory. Neurosci Lett 458, 140–143. [DOI] [PubMed] [Google Scholar]
- 9. Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D et al (2006) Common Kibra alleles are associated with human memory performance. Science 314, 475–478. [DOI] [PubMed] [Google Scholar]
- 10. Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu‐Lince D, Craig DW et al (2010) Evidence for an association between KIBRA and late‐onset Alzheimer's disease. Neurobiol Aging 31, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaper K, Kolsch H, Popp J, Wagner M and Jessen F (2008) KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging 29, 1123–1125. [DOI] [PubMed] [Google Scholar]
- 12. Duning K, Schurek EM, Schluter M, Bayer M, Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA et al (2008) KIBRA modulates directional migration of podocytes. J Am Soc Nephrol 19, 1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH and Parker PJ (2009) An aPKC‐exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol 7, e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stauffer S, Chen X, Zhang L, Chen Y and Dong J (2016) KIBRA promotes prostate cancer cell proliferation and motility. FEBS J 283, 1800–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baumgartner R, Poernbacher I, Buser N, Hafen E and Stocker H (2010) The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell 18, 309–316. [DOI] [PubMed] [Google Scholar]
- 16. Genevet A, Wehr MC, Brain R, Thompson BJ and Tapon N (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu J, Zheng Y, Dong J, Klusza S, Deng WM and Pan D (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar BA (2006) From cell structure to transcription: Hippo forges a new path. Cell 124, 267–273. [DOI] [PubMed] [Google Scholar]
- 19. Harvey K and Tapon N (2007) The Salvador‐Warts‐Hippo pathway ‐ an emerging tumour‐suppressor network. Nat Rev Cancer 7, 182–191. [DOI] [PubMed] [Google Scholar]
- 20. Harvey KF, Pfleger CM and Hariharan IK (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467. [DOI] [PubMed] [Google Scholar]
- 21. Huang J, Wu S, Barrera J, Matthews K and Pan D (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434. [DOI] [PubMed] [Google Scholar]
- 22. Pan D (2007) Hippo signaling in organ size control. Genes Dev 21, 886–897. [DOI] [PubMed] [Google Scholar]
- 23. Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D and Hariharan IK (2002) salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478. [DOI] [PubMed] [Google Scholar]
- 24. Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J and Kumar R (2006) Essential role of KIBRA in co‐activator function of dynein light chain 1 in mammalian cells. J Biol Chem 281, 19092–19099. [DOI] [PubMed] [Google Scholar]
- 25. Moleirinho S, Chang N, Sims AH, Tilston‐Lunel AM, Angus L, Steele A, Boswell V, Barnett SC, Ormandy C, Faratian D et al (2013) KIBRA exhibits MST‐independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 32, 1821–1830. [DOI] [PubMed] [Google Scholar]
- 26. Yoshihama Y, Sasaki K, Horikoshi Y, Suzuki A, Ohtsuka T, Hakuno F, Takahashi S, Ohno S and Chida K (2011) KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr Biol 21, 705–711. [DOI] [PubMed] [Google Scholar]
- 27. Martin‐Belmonte F and Perez‐Moreno M (2012) Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer 12, 23–38. [DOI] [PubMed] [Google Scholar]
- 28. Wang Z, Katsaros D, Biglia N, Shen Y, Fu Y, Loo LWM, Jia W, Obata Y and Yu H (2018) High expression of long non‐coding RNA MALAT1 in breast cancer is associated with poor relapse‐free survival. Breast Cancer Res Treat 171, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mu L, Tuck D, Katsaros D, Lu L, Schulz V, Perincheri S, Menato G, Scarampi L, Harris L and Yu H (2012) Favorable outcome associated with an IGF‐1 ligand signature in breast cancer. Breast Cancer Res Treat 133, 321–331. [DOI] [PubMed] [Google Scholar]
- 30. Wang Z, Katsaros D, Shen Y, Fu Y, Canuto EM, Benedetto C, Lu L, Chu WM, Risch HA and Yu H (2015) Biological and clinical significance of MAD2L1 and BUB1, genes frequently appearing in expression signatures for breast cancer prognosis. PLoS ONE 10, e0136246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin SM, Du P, Huber W and Kibbe WA (2008) Model‐based variance‐stabilizing transformation for Illumina microarray data. Nucleic Acids Res 36, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q and Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123, 725–731. [DOI] [PubMed] [Google Scholar]
- 34. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Basu‐Roy U, Bayin NS, Rattanakorn K, Han E, Placantonakis DG, Mansukhani A and Basilico C (2015) Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Commun 6, 6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, Ramaswamy S and Ellisen LW (2017) ACTL6A Is Co‐Amplified with p63 in Squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell 31, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mussell AL, Denson KE, Shen H, Chen Y, Yang N, Frangou C and Zhang J (2018) Loss of KIBRA function activates EGFR signaling by inducing AREG. Oncotarget 9, 29975–29984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoshihama Y, Izumisawa Y, Akimoto K, Satoh Y, Mizushima T, Satoh K, Chida K, Takagawa R, Akiyama H, Ichikawa Y et al (2013) High expression of KIBRA in low atypical protein kinase C‐expressing gastric cancer correlates with lymphatic invasion and poor prognosis. Cancer Sci 104, 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao L, Chen Y, Ji M and Dong J (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem 286, 7788–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD and Guan KL (2012) YAP mediates crosstalk between the Hippo and PI(3)K‐TOR pathways by suppressing PTEN via miR‐29. Nat Cell Biol 14, 1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaissiere T, Sawan C and Herceg Z (2008) Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 659, 40–48. [DOI] [PubMed] [Google Scholar]
- 43. Jaenisch R and Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 (Suppl), 245–254. [DOI] [PubMed] [Google Scholar]
- 44. Shinawi T, Hill V, Dagklis A, Baliakas P, Stamatopoulos K, Agathanggelou A, Stankovic T, Maher ER, Ghia P and Latif F (2012) KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics 7, 211–215. [DOI] [PubMed] [Google Scholar]
- 45. Hill VK, Dunwell TL, Catchpoole D, Krex D, Brini AT, Griffiths M, Craddock C, Maher ER and Latif F (2011) Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B‐cell acute lymphocytic leukemia. Epigenetics 6, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schelleckes K, Schmitz B, Ciarimboli G, Lenders M, Pavenstadt HJ, Herrmann E, Brand SM and Brand E (2017) Promoter methylation inhibits expression of tumor suppressor KIBRA in human clear cell renal cell carcinoma. Clin Epigenetics 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S, Huo D, Ogundiran TO, Ojengbede O, Zheng W, Nathanson KL, Nemesure B, Ambs S, Olopade OI and Zheng Y (2018) Genetic variation in the Hippo pathway and breast cancer risk in women of African ancestry. Mol Carcinog 57, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]


