Abstract
Aberrant expression of microRNAs (miRNAs) may contribute to the initiation and development of multiple types of human cancer. Several miRNAs have been found to be strongly correlated with the diagnosis, progression, and prognosis of colorectal carcinoma (CRC), but the role of miR‐125a in CRC remains unclear. In the present study, the function of miR‐125a on the expression of Smad ubiquitin regulatory factor 1 (Smurf1) was investigated in vitro and in vivo. We verified that Smurf1 is a downstream target gene of miR‐125a and is involved in miR‐125a‐mediated regulation of CT26 cell (colon cancer cell) proliferation and migration. Overexpression of miR‐125a suppresses CT26 cell growth by inhibiting cell proliferation. Additionally, wound healing assays were performed to show that overexpression of miR‐125a significantly reduced CT26 cell migration, which was reversed by overexpression of Smurf1. In vivo, miR‐125a overexpression downregulated the expression of Ki67 and Smurf1, thus leading to a marked reduction in tumor growth. These results revealed that miR‐125a plays a critical role in CRC by directly targeting Smurf1, a finding that may facilitate the development of improved diagnostic and therapeutic techniques for CRC.
Keywords: colorectal carcinoma, miR‐125a, Smurf1, tumorigenesis
Abbreviations
- CRC
colorectal carcinoma
- DMEM
Dulbecco's modified Eagle's medium
- miRNA
microRNA
- Smurf1
Smad ubiquitin regulatory factor 1
- TGFβ
transforming growth factor beta
Colorectal carcinoma (CRC) is the third most common type of cancer worldwide, causing > 500 000 mortalities annually 1, 2. Several risk factors have been shown to be implicated in the progression of CRC, including lifestyle, environmental factors, and genetic polymorphisms 3. CRC follows the sequential process from adenoma to carcinoma, with significant morbidity and mortality. It has been reported, however, that the introduction of CRC screening programs has reduced the incidence of CRC 4, 5, 6. However, the optimal screening strategy has not yet been identified. It is therefore critical to develop new screening tools as potent biomarkers for CRC.
Recently, certain studies have indicated that microRNAs (miRNAs) are strongly associated with the diagnosis, progression, and prognosis of CRC 7, 8. miRNAs, a class of short noncoding RNAs, serve as important regulators of human gene expression 9. There are no < 1500 human miRNAs in the miRBase database, playing a critical role in post‐transcriptional modification of gene expression via targeting the 3′UTR of specific mRNA 10, thereby affecting certain cellular processes in embryonic development and disease conditions 11, 12, 13. miR‐143 and miR‐145 were the first miRNAs found to be downregulated in precancerous and neoplastic colorectal tissue 14. miR‐143 could act as inhibitor in CRC cell proliferation by targeting K‐ras 15. Schepeler et al. 16 showed that the overexpression of miR‐145 could inhibit the three different cells lines of CRC, indicating that it may function as a tumor suppressor. Furthermore, certain miRNAs, such as miR‐192 and miR‐215, have been found to play an important role in carcinogenesis by modulating proliferation, invasion, tube formation, the cell cycle, and angiogenesis 17, 18.
In humans, there are two existing isoforms of miR‐125: miR‐125a and miR‐125b. Several studies have shown that miR‐125b was expressed in prostate, breast, and pancreatic cancers 19, 20. miR‐125b appears to directly bind to the 3′UTR of the P53 gene and contributes to the development and progression of human cancers 21. Nishida et al. 22 reported that miR‐125b served as a key prognostic biomarker in colorectal cancer and revealed the underlying mechanism between miR‐125b and CRC.
Smad ubiquitin regulatory factor 1 (Smurf1), belonging to the neural precursor cell expressed developmentally downregulated 4 subfamily of homologous to the E6‐AP C terminus type E3 ubiquitin protein ligases, is encoded by a distinct gene located at chromosome 7, which was originally identified as a negative regulator of the bone morphogenic protein/transforming growth factor beta (TGFβ) signaling pathway in mammals. It has been shown to play a crucial role in embryogenesis and adult tissue homeostasis and has a very high homology and amino acid identity (95%), both murine and human. This suggests that mouse models can be used to appropriately investigate the biological roles of smurfs in humans 23. In the present study, Smurf1 was also found to be the potential target gene of miR‐125a in CRC.
However, to date, the role of miR‐125a in CRC tumorigenesis remains poorly understood and the potential target of miR‐125a in CRC has not yet been fully characterized. The aim of this study was to investigate the effect of miR‐125a on CRC and explore the underlying downstream targets involved.
Materials and methods
Cell culture
Mouse colon cancer CT26 and human colon cancer SW620 cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) under the condition of 5% CO2 at 37 °C.
Bioinformatics analysis
targetscan software (http://www.targetscan.org/) was used to predict the target sites of miR‐125a.
Western blot analysis
In brief, the total proteins of cells and tissues were extracted using the RIPA buffer (Solarbio, R0010, Shanghai, China) with inhibitor cocktail (Sigma, P8340‐5ML, St. Louis, MO, USA) and the determination of protein concentration by BCA protein assay (SBJ‐1001; Beyotime, SENBEIJIA Bio, NanJing, China). Loading buffer was added to adjust the concentration. After denaturation, add 20 μg to each sample hole. Twelve percent SDS/PAGE was used to separate protein samples, and then, a PVDF membrane was employed to transfer the corresponding proteins, after electrophoresis and transmembrane, sealed for 2 h by skimmed milk. The membrane was incubated with anti‐Smurf1 (PB0937, 1 : 1000; Wuhan Boster Biological Technology, Ltd., Wuhan, China) and anti‐β‐tubulin (2146, 1 : 1000; Cell Signaling Technology, Inc., Danvers, MA, USA) primary antibodies. β‐Tubulin was used as the loading control. Overnight incubation at 4 °C. Washed the PVDF membrane for 3 h, add horseradish peroxidase‐labeled anti‐rabbit antibody (Anti‐Rabbit IgG, HRP‐Linked Antibody 6402‐05, 1 : 5000; Amylet Scientific Inc, Wuhan, China), and incubated at 37 °C for 1–2 h. The bands were visualized using the ECL chemiluminescence colorant (Haigene, M2301, Harbin, China). The grayscale ratio of target protein β‐tubulin was analyzed. It indicates the relative content of the target protein.
Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR)
Total RNA was extracted with traditional TRIzol method. GoScriptTM Reverse Transcription System (Promega, Madison, WI, USA) was used for synthesis of cDNA. mRNA concentration was assessed by photometric analyses by NanoDrop ND‐1000 UV‐Vis Spectrophotometer (Thermo Scientific, Wilmington, DE, USA).Quantitative RT‐qPCR experiments were performed on a 7300 device (Applied Biosystems, Foster City, CA, USA). At an initial denaturation at 95 °C for 10 min, amplification was performed during 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. The primers, control, and quantification method used in qPCR were as those described in reference 24.
Dual‐luciferase assays
The CT26 cells were seeded into a 24‐well plate and cotransfected with a reporter gene [pMIR‐Smurf1‐wild‐type (WT) or pMIR‐Smurf1‐mutant (Mut)] together with either miR‐125a mimics, control, or miR‐125a inhibitor using Lipofectamine® 2000 (Gibco; Thermo Fisher Scientific, Inc.). The following sequences were used: miR‐125a mimics: 5′‐UCCCUGAGACCCUUUAACCUGUGA‐3′; miR‐125a inhibitor: 5′‐UCACAAGUUAGGGUCUCAGGGA‐3′; miR‐NC: 5′‐UUCUCCGAACGUGUCACGUTT‐3′.
At 24 h post‐transfection, luciferase assays were conducted using the Dual‐Luciferase Assay Kit (Promega Corporation), following the manufacturer's instructions.
Cell proliferation and wound healing assay
For the cell proliferation assay, cells were plated in a 96‐well plate and a CCK‐8 kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was employed to detect cell proliferation, following the manufacturer's instructions. Absorbance was measured at 450 nm using a microplate reader. For the migration assay, cells (5 × 105) were added to a six‐well plate and incubated for 24 h. A pipette tip was used to scrap the cell monolayer to cause a wound the width of which was photographed 12 h postwounding using a microscope to detect cell migration.
Mouse xenograft model
CT26 cells treated with miR‐125a agomir or the control were cultured in the fresh DMEM under the normal conditions. Male nude mice (age, 5–6 weeks; weight, 16–19 g; Jackson Laboratory, Bar Harbor, ME, USA) were randomly divided into two groups (n = 6 per group): the control (inoculated with control CT26 cells) and the observation group (inoculated with miR‐125a agomir CT26 cells). These nude mice were under the conditions of 22 °C constant temperature, 35–75% humidity, free access to food and water, and 12‐h light/dark cycle. Briefly, cells were collected and suspended in PBS at a concentration of 5 × 106 cells per ml, and the nude mice were then treated with a subcutaneous injection of cells (5 × 105 cells, 100 μL). After 6 days, tumor growth was observed every 3 days. The tumor volume (V) was calculated using the following formula: V = (L × W 2) × 0.5, where L and W stand for length and width, respectively. Euthanasia was carried out by cervical dislocation after rendering nude mice consciousless with CO2, and tumors were dissected and weighed. Subsequently, tumor tissue was preserved in 4% paraformaldehyde for further analysis. The present study was approved by the Ethics Committee of The Second Hospital of Hebei Medical University.
Immunohistochemical analysis
Tumor tissues from mouse xenograft models were fixed, dehydrated, embedded in paraffin, and sectioned. Tumor tissue slices (4.5 μm) were incubated with anti‐Ki67 (1 : 100; A01K0054; Wuhan Boster Biological Technology, Ltd.) for 2 h at 37 °C. Following washing with PBS, the sections were treated with secondary antibodies (1 : 500; 10600‐P07E‐H; Wuhan Boster Biological Technology, Ltd.) for 1 h at room temperature. Normal colon tissue was used as negative control. Finally, the tumor slices were counterstained with hematoxylin and eosin (H&E) staining and photographed using light microscopy.
Statistical analysis
The statistical analysis was performed using spss software (SPSS for windows 17.0; SPSS, Inc., Chicago, IL, USA). Data are presented as mean ± SD. Student's t‐test was used to compare the mean values between the two groups; one‐way ANOVA was used to compare the mean values among three or more groups, in which the post hoc test was Bonferroni. P < 0.05 was considered to indicate a statistically significant difference.
Results
miR‐125 Regulates Smurf1 expression by targeting the 3′‐UTR of Smurf1
First, the 3′‐UTR of Smurf1 was screened using the TargetScan (http://www.targetscan.org/) to find a potential miRNA binding site; a putative binding site was identified at the 3′‐UTR of Smurf1 (Fig. 1A).
Figure 1.
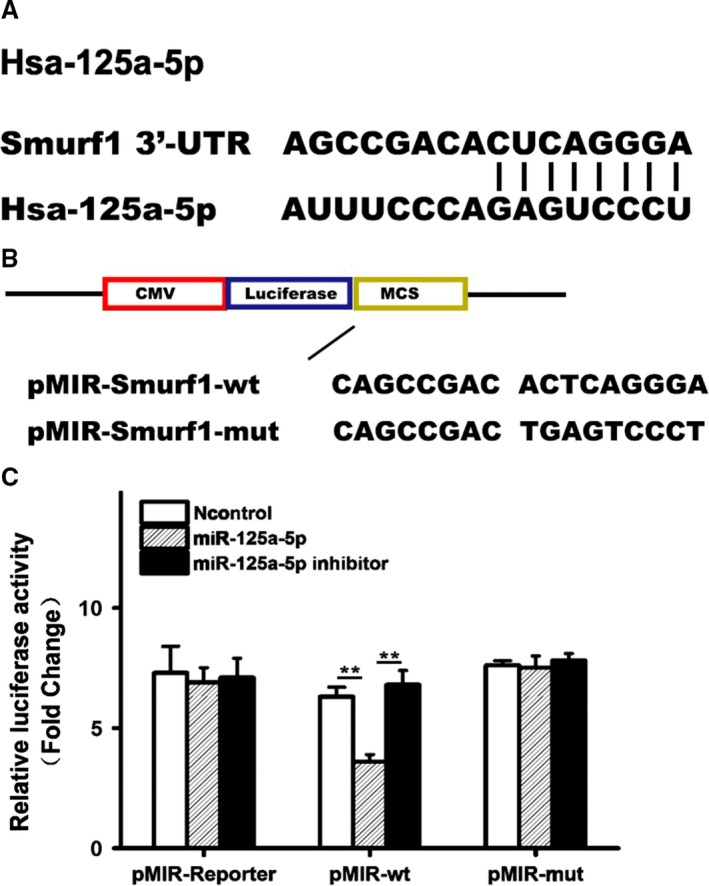
In vitro miR‐125a regulates Smurf1 expression by binding to the 3′‐UTR of Smurf1. (A) Schematic representation of the Smurf1 3′‐UTR as a direct target for miR‐125a. (B) WT or Mut miR‐125a target sssss of the Smurf1 3′‐UTR. (C) Luciferase activity of cells cotransfected with the WT or Mut CDKN1A 3′‐UTR reporter genes or negative control miRNA mimics (pMIR‐reporter). Three experiment replicates were performed. Data are presented as mean ± SD. One‐way ANOVA. ** P < 0.01.
In order to gain insights into whether miR‐125 successfully targeted the Smurf1 gene, the WT reporter gene was mutated to obtain a reporter vector containing Mut miR‐125 target sequences of Smurf1 (Fig. 1B). Next, CT26 cells were cotransfected with the pMIR‐Smurf1‐WT or pMIR‐Smurf1‐Mut reporter together with either miR‐125a mimics, the control, or miR‐125a inhibitor, and luciferase assays were performed. The transfection of miR‐125a mimics led to a significant reduction in the luciferase activity of the WT 3′UTR reporter gene, compared with the controls, whereas the use of an miR‐125a inhibitor significantly reversed this response (Fig. 1C). However, miR‐125a mimics and the miR‐125a inhibitor failed to affect the luciferase activity of the Mut reporter gene (Fig. 1C).
To examine the role of miR‐125a in regulating the expression of Smurf1, we examined the protein and the mRNA expression of Smurf1 using western blot analysis and RT‐qPCR in CT26 cells transfected with miR‐125a mimics or the controls. As expected, the treatment with miR‐125a mimics significantly reduced the mRNA and protein expression of Smurf1, compared with the controls (Fig. 2A,B). These results suggested that miR‐125a may repress the expression of Smurf1 by directly targeting the 3′UTR of Smurf1.
Figure 2.
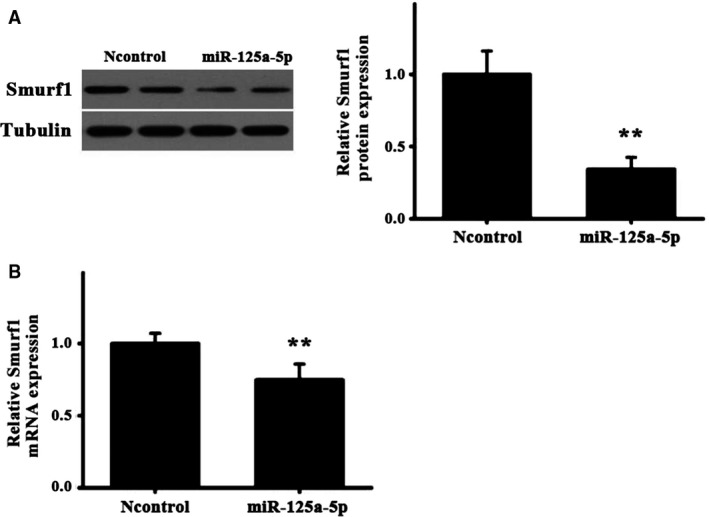
Expression of Smurf1 in cells transfected with miR‐125a. (A) The Smurf1 protein expression and quantification in CT26 cells transfected with miR‐125a or control was determined by western blotting. ** P < 0.01. (B) Quantification of Smurf1 mRNA levels was determined by reverse transcription‐quantitative polymerase chain reaction. Data are presented as mean ± SD. Student's t‐test. ** P < 0.01.
miR‐125a inhibits cell proliferation and migration in CT26 and SW620 cells
Considering that miR‐125a negatively influenced Smurf1 expression, it was hypothesized that miR‐125a could inhibit tumor cell proliferation and migration. To assess the impact of miR‐125a on the aspect of cell proliferation, CT26 and human colon cancer cell SW620 cells were transfected with miR‐125a mimics or the control, respectively. Cell proliferation assays with the CCK‐8 kit indicated that the overexpression of miR‐125a significantly inhibited cell proliferation in CT26 and SW620 cells (Figs 3A and 4A). The wound healing assays suggested that the overexpression of miR‐125a significantly decreased the migration of CT26 and SW620 cells, which could be reversed by Smurf1 overexpression (Figs 3B and 4B).
Figure 3.
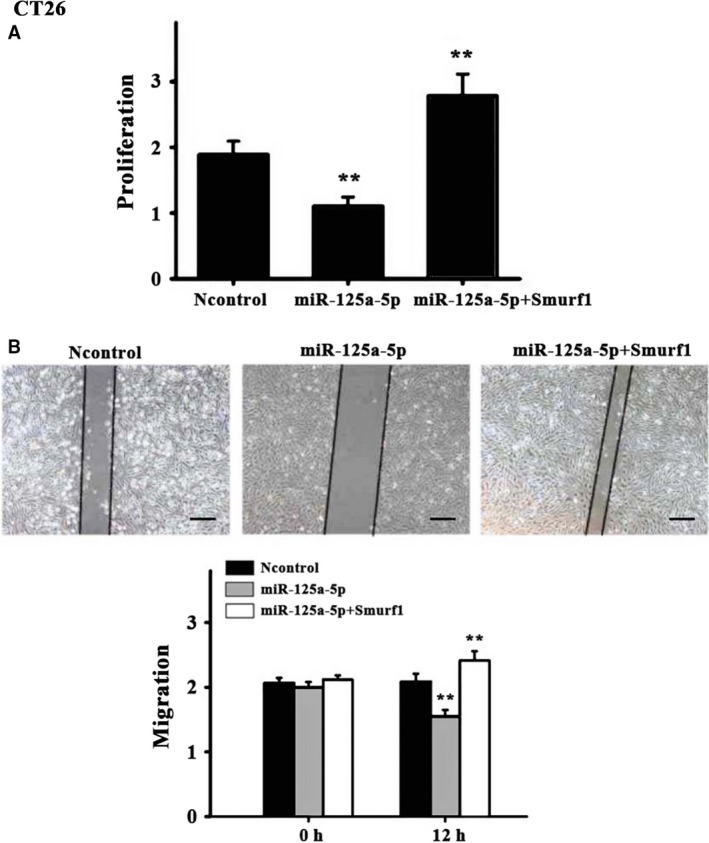
Effects of miR‐125a in CT26 cells. (A) Cell proliferation assays were performed in CT26 cells transfected with Smurf1 and/or miR‐125a‐5p using the CCK‐8 kit. (B) Representative images and quantification of the migration of CT26 cells transfected with miR‐125a, control, or miR‐125a + Smurf1. Scale bar = 25 μm; N = 6 per group. Three experiment replicates were performed. Data are presented as mean ± SD. One‐way ANOVA. ** P < 0.01.
Figure 4.
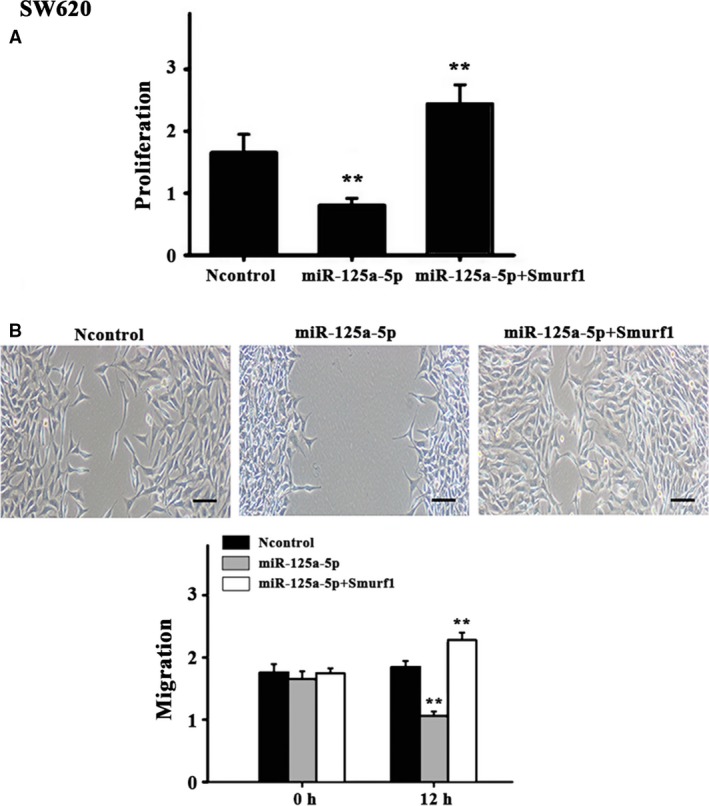
Effects of miR‐125a in SW620 cells. (A) Cell proliferation assays were performed in SW620 cells transfected with Smurf1 and/or miR‐125a‐5p using the CCK‐8 kit. (B) Representative images and quantification of the migration of SW620 cells transfected with miR‐125a, control, or miR‐125a + Smurf1. Scale bar = 25 μm; N = 6 per group. Three experiment replicates were performed. Data are presented as mean ± SD. One‐way ANOVA. ** P < 0.01.
miR‐125a inhibits tumor growth in nude mice
CT26 cells were used to establish a mouse xenograft tumor model, cells were transfected with miR‐125a agomir or the control, in order to elucidate the functional implications of miR‐125a in vivo. As presented in Fig. 5A–D, the treatment of miR‐125a strongly halted tumor growth (tumor size and weight), compared with the controls. Next, immunohistochemical staining was employed to examine the positive expression of Ki67 and Smurf1 proteins, with the results showing that the density of positive Ki67 and smurf‐positive cells was significantly lower in tumors treated with miR‐125a agomir than in those treated with the controls (Fig. 6A). Furthermore, western blot analysis indicated that the Smurf1 expression was lower in the observation group than that in the controls (Fig. 6B). These results confirmed that miR‐125a downregulates Smurf1, leading to a marked reduction in tumor growth.
Figure 5.
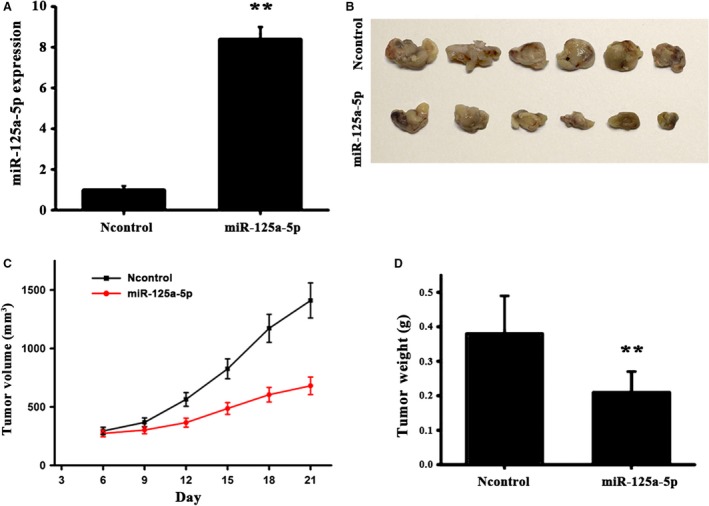
MiR‐125a inhibits tumor growth in vivo. (A) The expression level of miR‐125a in the mouse xenograft tumor mode was determined by reverse transcription‐quantitative polymerase chain reaction. (B) Tumor size of nude mice following subcutaneous injection with CT26 cells pretransfected with AgomiR‐125a or AgomiR‐NC. (C) Tumor volume growth curves. (D) Tumor weight. N = 6 per group. Scale bar = 1 cm. NC, negative control. Data are presented as mean ± SD. Student's t‐test. ** P < 0.01.
Figure 6.
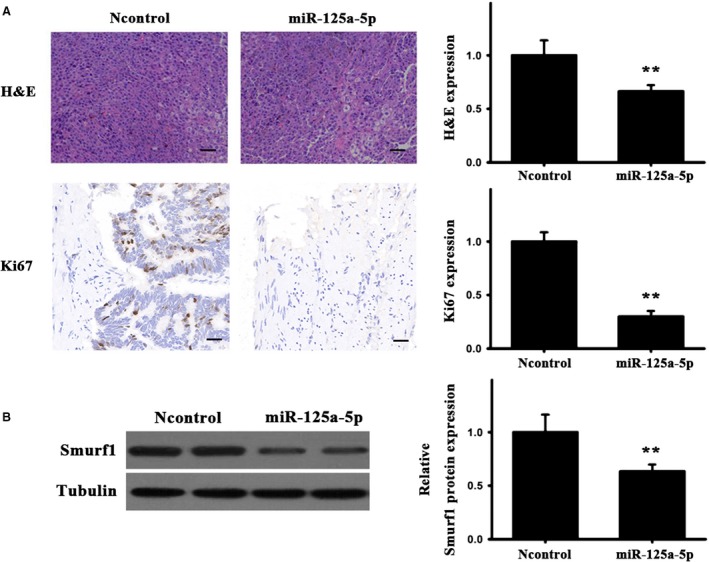
Effects of miR‐125a on Ki67 and Smurf1 expression. (A) Ki67 protein detection and quantification by H&E and immunohistochemical staining. (B) Smurf1 protein expression and quantification in tumors, as determined by western blotting. In the mouse xenograft tumor model, the expression of smruf1 protein was less than normal group. N = 3 per group. Three experiment replicates were performed. Scale bar = 50 μm. Data are presented as mean ± SD. Student's t‐test. ** P < 0.01.
Discussion
MicroRNAs are critical regulators involved in numerous cellular processes, such as proliferation, migration, apoptosis, and differentiation. In addition, miRNAs have been closely associated with the initiation and progression of malignant tumors 25, 26. Although this study is not the first one to explore the implication of miR‐125a in cancer, the present study presented some novel findings. To begin with, Smurf1 was first shown here in to be a potential target of miR‐125a in CRC. Next, in vitro assays indicated that the overexpression of miR‐125a exerted antiproliferative and antimigratory effects in CT26 and SW620 cells. Finally, consistent with the in vitro results, it was identified that miR‐125a downregulated Smurf1 in a mouse tumor xenograft model. In combination, these results suggested that miR‐125a suppresses tumorigenesis in CRC by directly targeting Smurf1.
The abnormal expression of miRNAs has been demonstrated in a variety of human cancer types 27, 28, 29, 30, 31, 32. miR‐143 and miR‐145 have been shown to be downregulated in neoplastic colorectal tissue 33. Schepeler et al. 16 reported that the growth of three types of CRC cell lines (LS174T, DLD1, HCT116) was significantly inhibited when transfected with miR‐145, suggesting that miR‐145 may serve as a tumor inhibitor. miRNA‐143 could exert a critical effect in CRC cell proliferation by targeting the K‐ras gene, which is involved in various signaling pathways mediating cellular biological processes 34, 35. Recent studies have shown that miR‐125a functions as a tumor inhibitor of the malignant phenotype of CRC cells by binding to signal transducer and activator of transcription 3 36. In addition, miR–125a has been reported to exhibit an important function in inhibiting osteosarcoma cell migration and invasion by binding to matrix metalloproteinase‐11 (MMP‐11) 37. Furthermore, the miR–125a overexpression significantly inhibited the proliferation and metastasis of hepatocellular carcinoma by targeting MMP‐11 and vascular endothelial growth factor 38. Together, these studies suggested that miR‐125a plays a crucial role in the tumorigenesis of different types of cancer by targeting different genes. However, the association of miR‐125a with the pathogenesis and progression of CRC remains unclear. The present study was the first to confirm that miR‐125a markedly affected CT26, SW620 cell lines, and tumor tissue. The overexpression of miR‐125a significantly suppressed CT26, SW620 cell proliferation, and migration in vitro and inhibited tumor growth by downregulating Ki67 expression in vivo. However, the disadvantage of this study is that although miR‐125a has been reported to be downregulated in human colorectal cancer tissues, it has not been verified again in the paper. A follow‐up clinical study will be conducted to clarify the correlation between miR‐125a expression in human colon cancer and the prognosis of CRC treatment. In combination, these findings indicated that the modulation of cell proliferation and migration by miR‐125a contributed to the initiation and progression of CRC.
To further identify the underlying mechanisms of miR‐125a inhibiting cell growth, proliferation, and migration in CRC, it was demonstrated that Smurf1 is a potential target of miR‐125a. Smurf1 was the first strong oncogene candidate, since it has been known to be associated with the TGFβ signaling pathway. In general, TGFβ targets its receptors resulting in the phosphorylation of SMAD2/SMAD3, which then, in complex with SMAD4, regulates transcription. Hruban et al. 39 indicated that the TGFβ signaling pathway is normally disrupted via a mutation of SMAD4 and inactivation of TGFβR I and II in pancreatic cancer. Kwei et al. 40 reported Smurf1 as an amplified oncogene promoting cell invasion in pancreatic cancer, suggesting that Smurf1 may be a tractable drug target. Other studies have also revealed that the overexpression of Smurf1 in pluripotent mesenchymal cells and pre‐osteoblasts could repress osteoblast differentiation and reported that the Smurf‐transgenic mice presented with low bone mass due to reduced bone formation 41, 42. The present study was the first to reveal that miR‐125a can decrease the Smurf1 mRNA and protein expression levels in colon cancer cells by directly targeting the 3′‐UTR of Smurf1, which suggests that Smurf1 is a potent target of miR‐125a for CRC prevention and treatment.
Conclusions
In conclusion, the present results demonstrated that the overexpression of miR‐125a can repress the malignant phenotype of CRC cells in vitro and inhibit tumor growth in xenograft mouse models in vivo, with Smurf1 as a potential and functional target. Collectively, the results of this study provide sufficient evidence that miR‐125a is a potential therapeutic target for CRC and that the miR‐125a/Smurf1 pathway may contribute to the development of new therapeutic strategies for CRC. Although this study is not the first one to explore the implication of miR‐125a in cancers, we provided the evidence that Smurf1 was one potential target of miR‐125a in CRC, which is some novel finding in this study.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
DL, XX, and JC conceived and designed the project, DL and JM acquired the data, XX, JM, and JC analyzed and interpreted the data, and all authors wrote the paper.
Acknowledgements
We thank other laboratory members for support.
References
- 1. Gellad ZF and Provenzale D (2010) Colorectal cancer: national and international perspective on the burden of disease and public health impact. Gastroenterology 138, 2177–2190. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D and Jemal A (2013) Cancer statistics. CA Cancer J Clin 63, 11–30. [DOI] [PubMed] [Google Scholar]
- 3. Aran V, Victorino AP, Thuler LC and Ferreira CG (2016) Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer 15, 195–203. [DOI] [PubMed] [Google Scholar]
- 4. Hewitson P, Glasziou P, Watson E, Towle B and Trwig L (2008) Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (Hemoccult): an update. Am J Gastroenterol 103, 1541. [DOI] [PubMed] [Google Scholar]
- 5. Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, Bresalier R, Andriole GL, Buys SS, Crawford ED, et al (2012) Colorectal‐cancer incidence and mortality with screening flexible sigmoidoscopy. New Engl J Med 366, 2345–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp‐Vogelaar I, Van BM, Hankey BF, Bengamin F, Shi W, Bond GH, Schapiro M, et al (2012) Colonoscopic polypectomy and long‐term prevention of colorectal cancer deaths. N Engl J Med 366, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava K and Srivastava A (2012) Comprehensive review of genetic association studies and meta‐analyses on miRNA polymorphisms and cancer risk. PLoS ONE 7, e50966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tokarz P and Blasiak J (2012) The role of MiRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol 59, 467–474. [PubMed] [Google Scholar]
- 9. Tili E, Michaille JJ and Calin AGA (2008) Expression and function of micro RNAs in immune cells during normal or disease state. Int J Med Sci 5, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin He and Gregory J (2004) Hannon. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 11. Kantharidis P, Wang B, Carew RM and Lan HY (2011) Diabetes complications: the microRNA perspective. Diabetes 60, 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trionfini P and Benigni A (2017) MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol 28, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato M and Natarajan R (2015) MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci 30, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michael MZ, Connor SMO, Pellekaan NGVH, Young GP and James RJ (2003) Reduced accumulation of specific micrornas in colorectal neoplasia 1note: Susan M. O'Connor and Nicholas G. van Holst Pellekaan contributed equally to this work. Mol Cancer Res 1, 882–891. [PubMed] [Google Scholar]
- 15. Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G and Ba Y (2009) Role of miR‐143 targeting KRAS in colorectal tumorigenesis. Oncogene 28, 1385–1392. [DOI] [PubMed] [Google Scholar]
- 16. Schepeler T, Jørgen T Reinert, Ostenfeld MS, Christensen LL and Andersen CL (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68, 6416–6424. [DOI] [PubMed] [Google Scholar]
- 17. Song B, Wang Y, Kudo K, Gavin EJ, Xi Y and Ju J (2008) miR‐192 regulates dihydrofolate reductase and cellular proliferation through the p53‐microRNA circuit. Clin Cancer Res 14, 8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Ørntoft TF, Andersen CL and Dobbelstein M (2008) p53‐Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68, 10094–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ and deVere White (2007) An androgen‐regulated miRNA suppresses Bak1 expression and induces androgen‐independent growth of prostate cancer cells. Proc Natl Acad Sci USA 104, 19983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomson JM (2006) Extensive post‐transcriptional regulation of microRNAs and its implications for cancer. Gene Dev 20, 2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le MT, Teh C, Shyh‐Chang N, Xie H, Zhou B, Korzh V, Lodish HF and Lim B (2009) MicroRNA‐125b is a novel negative regulator of p53. Gene Dev 23, 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishida N, Yokobori T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M (2011) MicroRNA miR‐125b is a prognostic marker in human colorectal cancer. Int J Oncol 38, 1437–1443. [DOI] [PubMed] [Google Scholar]
- 23. Praveen K, Gal LC and Michael B (2018) Smurfs in protein homeostasis, signaling, and cancer. Front Oncol 8, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y and Abraham E (2009) miR‐147, a microRNA that is induced upon Toll‐like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci USA 106, 15819–15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Calin GA and Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866. [DOI] [PubMed] [Google Scholar]
- 26. Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G Klein‐Szanto and A, (2008) The microRNAs miR‐373 and miR‐520c promote tumour invasion and metastasis. Nat Cell Biol 10, 202–210. [DOI] [PubMed] [Google Scholar]
- 27. Gramantieri L, Ferracin M, Fornari F, Vernoese A, Sabbioni S, Liu CG, Giovannini C, Ferrazzi E and Grazi GL (2007) Cyclin G1 is a target of miR‐122a, a MicroRNA frequently down‐regulated in human hepatocellular carcinoma. Cancer Res 67, 6092–6099. [DOI] [PubMed] [Google Scholar]
- 28. Chan JA, Krichevsky AM and Kosik KS (2005) MicroRNA‐21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65, 6029–6033. [DOI] [PubMed] [Google Scholar]
- 29. Yan LX, Huang XF, Shao Q, Huang M, Deng L, Wu QL, Zeng YX and Shao JY (2008) MicroRNA miR‐21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65, 7065–7070. [DOI] [PubMed] [Google Scholar]
- 31. Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima‐Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, et al (2008) Downregulation of miR‐138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 99, 280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicoloso MS and Calin GA (2008) MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol 18, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li JM, Zhao RH, Li ST, Xie CX, Jiang HH, Ding WJ, Du P, Chen W, Yang M and Cui L (2012) Down‐regulation of fecal miR‐143 and miR‐145 as potential markers for colorectal cancer. Saudi Med J 33, 24–29. [PubMed] [Google Scholar]
- 34. Omerovic J, Laude A and Prior I (2007) Ras proteins: paradigms for compartmentalised and isoform specific signalling. Cell Mol Life Sci 64, 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonfrate L, Altomare DF, Di Lena M, Travaglio E, Rotelli MT, De Luca A and Portincasa P (2013) MicroRNA in colorectal cancer: new perspectives for diagnosis, prognosis and treatment. J Gastrointestin Liver Dis 22, 311–320. [PubMed] [Google Scholar]
- 36. Fan Z, Cui H, Xu X, Lin Z and Jiao S (2015) MiR‐125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget 6, 25266–25280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waresijiang N, Sun J, Abuduaini R, Jiang T, Zhou W and Yuan H (2016) The downregulation of miR–125a–5p functions as a tumor suppressor by directly targeting MMP–11 in osteosarcoma. Mol Med Rep 13, 4859–4864. [DOI] [PubMed] [Google Scholar]
- 38. Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z and Xu G (2012) Ectopic expression of MiR‐125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS ONE 7, e40169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hruban RH, Maitra A, Schulick R, Laheru D, Herman J, Kern SE and Goggins M (2008) Emerging molecular biology of pancreatic cancer. Gastrointest Cancer Res 2 (4 Suppl), S10. [PMC free article] [PubMed] [Google Scholar]
- 40. Kwei KA, Hunter SA, Ryan B, Kelli M, Karikari CA, Matt VDR, Manuel H, Anriban M, Bashyam MD and Pollack JR (2011) SMURF1 amplification promotes invasiveness in pancreatic cancer. PLoS ONE 6, e23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ying SX, Hussain ZJ and Zhang YE (2003) Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein‐2‐induced osteoblast conversion by targeting Smad5 for degradation. J Biol Chem 278, 39029–39036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR and Chen D (2004) Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo . J Biol Chem 279, 12854. [DOI] [PMC free article] [PubMed] [Google Scholar]


