Abstract
microRNAs (miRs) dysregulation have emerged as a crucial step in tumorigenesis, being related with cancer development, progression and response to treatment. In chronic myeloid leukaemia (CML), the resistance to tyrosine kinase inhibitors (TKI) is responsible for treatment failure and could be linked to changes in miRs expression. This work aimed to correlate the expression levels of 3 miRs, miR-21, miR-26b and miR-451, with response to TKI treatment in CML patients. miR-451 levels at diagnosis were significantly higher in patients with optimal response after 6 and 12 months of therapy. Conversely, patients without optimal response had highest levels of miR-21. miR-21 and miR-451 appear to be good biomarkers of response, able to predict optimal TKI responders (p < 0.05). Using the combined profile of both miRs, we create a predictive model of optimal response after one year of treatment. This study highlights the role of miR-21 and miR-451 expression levels at diagnosis in predicting which patients achieve the optimal response.
Subject terms: Chronic myeloid leukaemia, Tumour biomarkers
Introduction
MicroRNAs (miRs) are a group of small single-strained non-coding RNAs (≈22 nucleotides of length) that act as epigenetic regulators of specific targets by modulating gene expression via translational repression or mRNA cleavage1. Although protein inhibition is the expected result of miR function, this is not always an inhibitory process. miRs can indirectly activate the expression of some mRNAs by degradation of their natural inhibitors1,2. These small RNAs regulate around 80% of the transcriptome and play a key role in multiple cellular processes, like proliferation, development, differentiation and apoptosis2.
The dysregulation of miRs expression patterns can have numerous implications including the promotion of tumorigenesis3. miRs expression pattern differs according to tissues, cell types and developmental stages, and could act as oncomiRs, promoting processes that favour cancer cells, or as tumour suppressor miRs, regulating the expression of oncogenes4,5. Aberrant expression of miRs has been reported in solid tumours and haematological neoplasia, as chronic myeloid leukaemia (CML)6,7. The presence of BCR-ABL1 is the distinctive molecular characteristic of CML and the target for treatment with tyrosine kinase inhibitors (TKI)8. Resistance to imatinib and other TKIs has been recognised as the major challenge for CML treatment and monitoring, since some of the imatinib-resistant patients had no mutations on BCR-ABL1 oncogene9. Since miRs are potent regulators, they may be implicated in the acquisition of drug resistance because they could regulate not only BCR-ABL1 but also modulate the expression of genes involved in drug transporter or activation of essential signalling pathways6,10. In CML the most deregulated miRs studied include miR-10a, miR-130b, miR-150 and miR-203, but several others miRs emerge in the complex network of miR regulation11,12.
Overexpression of the oncomiR miR-21 has been associated with the development of neoplasias where it targets many tumour suppressor genes related to proliferation, apoptosis and survival13. One example of miR-21 targets is PTEN, a negative regulator of the PI3K/AKT pathway, which is often described as deregulated in cancer. In addition, levels of miR-21 were correlated with acquisition of resistance to multiple drugs, as gemcitabine and TKIs13,14. PTEN is also a target of miR-26b, but the role of this miR in cancer is still controversial, being sometimes described as oncomiR and others as a tumour suppressor miR15,16. Particularly relevant in CML are the miRs that target ABL and consequently BCR-ABL1, being some of the most deregulated ones. miR-451, miR-203 and miR-320a are described as downregulated in CML, since they directly target ABL and BCR-ABL1 acting as tumour suppressors, by reducing BCR-ABL oncoprotein expression12,17.
Furthermore, miRs expression is a dynamic process which reflects changes at cellular levels not only at the time of neoplasia development but also in progression or response to treatment. These characteristics made the expression levels of miR a potential biomarker for diagnosis, prognosis and treatment response18,19.
In this work, we analysed the expression levels of miR-21, miR-26b and miR-451 in CML patients and in vitro models, and correlated them with TKI response levels. The potential of each miR as a predictive biomarker was explored, with a particular emphasis for the combinatorial miR expression profile.
Results
Differential miRs expression levels at Imatinib-resistant models
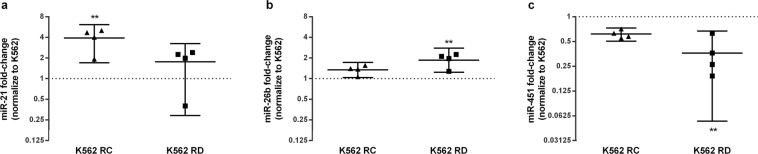
To check if the expression levels of miR-21, miR-26b and miR-451 may be related with Imatinib resistance, we evaluated the levels of each miR in three CML cell lines (one sensitive and two resistant to TKI, the K562-RC and the K562-RD cells respectively). The Imatinib-resistant cells presented different expression levels of miRs compared to the sensitive cell line (Fig. 1). The oncomiR miR-21 was up-regulated in both resistant models, with 4.3-fold higher expression in K562-RC cells (p = 0.0038; Fig. 1a). Similarly, miR-26b was increased in resistant cells, with 2.0-fold higher expression in K562-RD cells (p = 0.0023; Fig. 1b). By opposition, the tumour suppressor miR-451 was down-regulated in K562-RC and K562-RD cells, with 3.8-fold less expression in the discontinuation model (K562-RD) (p = 0.0048; Fig. 1c).
Figure 1.
miRs expression profile of imatinib-resistant CML cell lines. Sensitive cell line, K562 cells, was used as reference to determine the fold-change of miR-21 (a), miR-26b (b) and miR-451 (c) of resistant cell lines, K562-RC and K562-RD cells. miR-21 was higher in K562-RC, while miR-26b was higher in K562-RD. Moreover, miR-451 was significantly down-regulated in K562-RD. The results are presented in mean with 95% CI of four independent samples and the expression of K562 cell line is represented by dot line. **p < 0.001 compared with K562 cells.
CML patients miRs expression levels at diagnosis
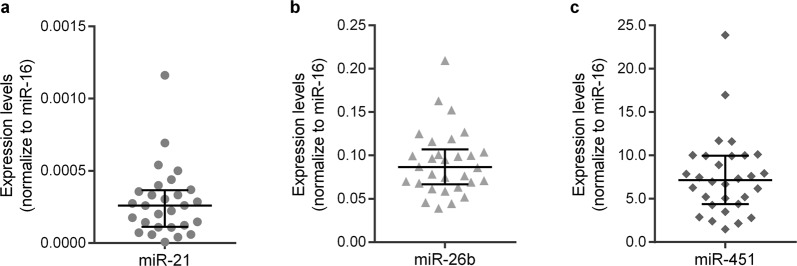
We subsequently assessed the expression of the same miRs in CML patients, samples at diagnosis (Fig. 2). Clinical and biological characteristics of our cohort of patients were summarised in Table 1. miR-26b and miR-451 were expressed in all patients, the tumour suppressor miR-451 exhibiting the highest expression at diagnosis [7.147 (95% confidence interval (CI): 5.201–8.912; Fig. 2c)], and the miR-26b the second one with a median of 0.087 (95% CI: 0.070–0.099; Fig. 2b). miR-21 was expressed in 93.3% of patients (28 out of 30 patients) with a median of 0.003 (95% CI: 0.00014–0.00033; Fig. 2a). The expression of this miR was inversely correlated with the expression of miR-451 (R = −0.655; p = 0.0002). miR-26b expression was positively correlated with miR-21 expression (R = 0.611; p = 0.001) and inversely associated with miR-451 (R = −0.462; p = 0.010).
Figure 2.
miRs expression levels in CML patients at diagnosis. The expression levels of miR-21 (a), miR-26b (b) and miR-451 (c) were normalised to miR-16 in CML patients at diagnosis time. At diagnosis, miR-451 was the miR with the highest expression and miR-21 with the lowest. The results are represented as median with interquartile range.
Table 1.
Biodemographic and clinical characteristics of CML patients.
| Characteristics | Diagnosis group (n = 30) | Follow-up group (n = 27) | ||
|---|---|---|---|---|
| Demographic features | ||||
| Gender (%) | ||||
| Male | 16 | (53.3) | 14 | (51.9) |
| Female | 14 | (46.7) | 13 | (48.1) |
| Clinical features | ||||
| Age at diagnosis (years) | ||||
| Median | 54 | 42 | ||
| Range | 18–78 | 24–78 | ||
| Disease Phase | ||||
| Chronic Phase (%) | 25 | (83.3) | ||
| Accelerate Phase or Blast Crisis (%) | 5 | (16.7) | ||
| Treatment | at analysis time point | |||
| TKI (%) | 27 | (90.0) | 22 | (81.5) |
| Imatinib (%) | 26 | (96.3) | 14 | (63.6) |
| 2nd TKI (%) | 1 | (3.7) | 8 | (36.4) |
| TKI plus INF-α (%) | 3 | (10.0) | 5 | (18.5) |
| Imatinib (%) | 2 | (66.7) | 4 | (80.0) |
| 2nd TKI (%) | 1 | (33.3) | 1 | (20.0) |
| Response | at 6 Months | at analysis time point | ||
| Optimal Response (%) | 17 | (56.7) | 24 | (88.9) |
| Without Optimal Response (%) | 9 | (30.0) | 3 | (11.1) |
| No determined (%) | 4 | (13.3) | ||
| at 12 Months | ||||
| Optimal Response (%) | 15 | (50.0) | ||
| Without Optimal Response (%) | 14 | (46.7) | ||
| No determined (%) | 1 | (3.3) | ||
TKI: Tyrosine kinase inhibitors; INF-α: interferon-alpha.
The observed miR expression levels may be related with the disease phase at diagnosis, since miR-451 shown a tendency to be downregulated at accelerated phase or blast crisis [median of 4.28, Interquartile Range (IR) 1.82–8.92, n = 5)] in comparison with patients in chronic phase (median of 7.31, IR 5.13–10.02, n = 25). However, the low number of patients in advanced phases did not allow statistically significant differences to be detected. For that it would be necessary at least 68 patients in each group (G power analysis with 80% of power).
TKI Response and miRs expression levels
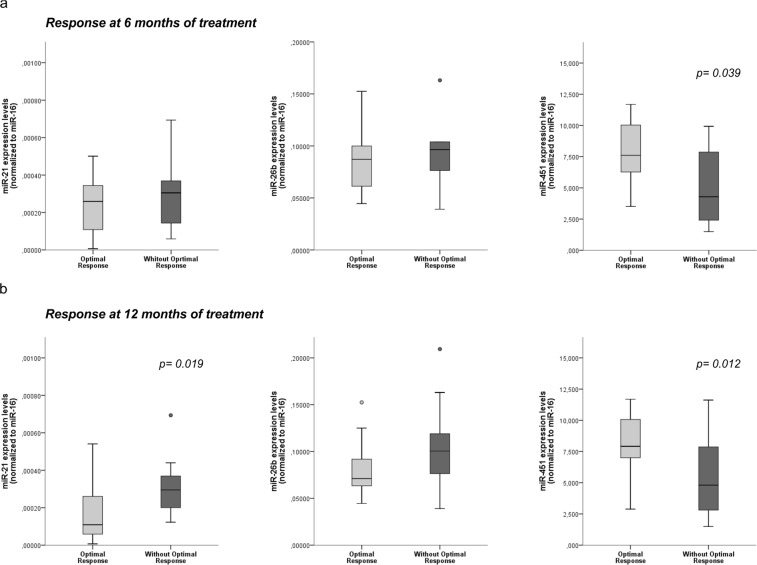
To confirm if the miRs expression at diagnosis could be related with the degree of response achieved after TKI treatment, we evaluated the expression profile according to response levels after 6 and 12 months of treatment20. Optimal response was considered when BCR-ABL1 expression levels were lower than 1% at 6 months and ≤0.1% at 12 months of treatment20. Patients with higher levels of miR-451 at diagnosis had a higher rate of optimal response after 6 months (p = 0.039; Fig. 3a). Our results reveal that the lower expression of miR-21 (p = 0.019) and higher levels of miR-451 (p = 0.012) were associated with patients with optimal response at 12 months of treatment (Fig. 3b). At both time points, the levels of miR-26b were higher in patients without optimal response, but the differences were not statistically significant.
Figure 3.
Correlation between TKI response and miRs expression levels at diagnosis. Patients were grouped according to TKI response in optimal response or without optimal response, reached after 6 and 12 months of treatment. The expression levels of miR-21, miR-26b and miR-451 at diagnosis were analysed according to the TKI response after 6 (a) and 12 (b) months of treatment. miR-451 was down-regulated in patients without optimal response after 6 and 12 months of TKI therapy, while miR-21 was up-regulated in patients without optimal response after 12 months of treatment. Statistical differences between groups are reported by the p-value.
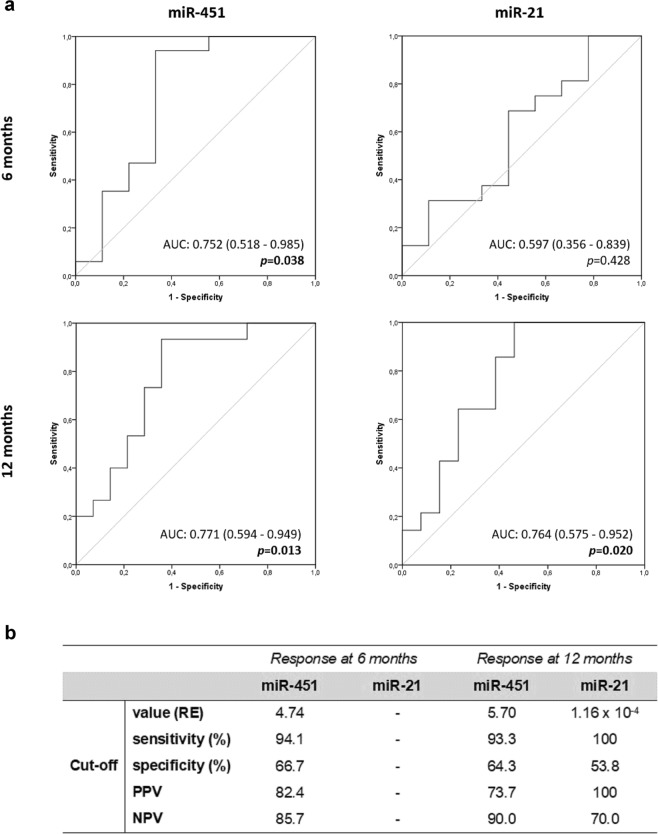
To explore the predictive power of miRs expression to predict response to TKI treatment, we performed ROC curve analysis (Fig. 4). miR-451 expression discriminated patients with optimal response from those without response after 6 months of treatment (p = 0.038; Fig. 4a). In addition, both miR-451 and miR-21 revealed significant potential as response biomarkers at 12 months of TKI (Fig. 4a). The expression of miR-451 was the most accurate parameter to predict response with an area under the curve (AUC) of 0.771 (95%CI: 0.594–0.949; p = 0.013). At diagnosis, miR-451 expression levels higher than 4.74 and 5.70 were the optimal cut-off values to discriminate optimal response after 6 and 12 months of treatment, respectively (Fig. 4b). These cut-off values present higher sensitivity, 94.1% at 6 months and 93.3% at 12 months, with a satisfactory specificity of 66.7% and 64.3% for both time points. Conversely, levels of oncomiR miR-21 below 1.16 × 10−4 at diagnosis was identified as the best cut-off value for optimal response at 12 months of treatment. This biomarker revealed the highest sensitivity (100%) and also a strong positive predictive value (PPV:100%) and a good negative predictive value (NPV: 70%) (Fig. 4b).
Figure 4.
Receiver Operating Characteristic (ROC) curve analysis of miR-451 and miR-21. ROC curves (a) and the identification of optimal cut-off values to discriminate optimal response (b) point-out the ability of miR-451 and miR-21 expression at diagnosis to predict response to TKI treatment at 6 months and 12 months. For each curve was represent the Area Under the Curve (AUC) and the respective p-value. RE–Relative Expression; PPV- Positive Predictive Value; NPV- Negative Predictive Value.
miRs profile and response after a year of treatment
Since we identified both miR-21 and miR-451 as good biomarkers of response after a year of treatment with TKI, we made a profile analysis and risk association. Using the optimal cut-off values detected for each miR, we classified patients in four different profiles according to miR-21 and miR-451 expression levels: Profile 1: miR-451 > 5.69 and miR-21 < 1.16 × 10−4; Profile 2: miR-451 < 5.69 and miR-21 > 1.16 × 10−4; Profile 3: miR-451 > 5.69 and miR-21 > 1.16 × 10−4; Profile 4: miR-451 < 5.69 and miR-21 < 1.16 × 10−4. After, we correlated these miRs profiles with the probability to achieve an optimal response after 12 months of treatment. All patients included in the first profile presented an optimal response (8 optimal responses out of 8), while most of those in profile 2 do not reach optimal response (1 optimal response out of 10). Risk analysis showed that profile 2 patients have 25 times lower probability to achieve optimal response after one year of treatment (OR: 0.040; 95%CI: 0.0039–0.398; p = 0.001). On the other hand, patients with profile 1 presented 42 times higher chances to accomplish an optimal response at the same time point (OR: 42.38; 95%CI: 2.129–843.9; p = 0.0007). Profile 3 was not statistical associated with response (4 optimal responses out of 9), and no patient presented profile 4.
In this context, miR-21 and miR-451 levels at diagnosis were used to create a predictive model for an optimal response after one year of treatment. From the different regression models tested, the main effect (ME) model that takes in consideration both miRs for optimal response, was the most predictive one accounting for a total of 66% of the variance (Table 2). Applying this model, we will be able to assess the individual probability of each patient to achieve an optimal response (P) Eq. (1).
| 1 |
Table 2.
Statistical measures for predicting models for optimal response.
| Model | Varibles dichotomize | |||
|---|---|---|---|---|
| miR-21 | miR-451 | Interaction | Main Effect | |
| Raw Aggrement (%) | 79.31 | 79.31 | 79.31 | 79.31 |
| kappa (Cohen) | 0.59 | 0.58 | 0.58 | 0.58 |
| Sensitivity (%) | 100.00 | 64.29 | 64.29 | 64.29 |
| Specificity (%) | 60.00 | 93.33 | 93.33 | 93.33 |
| PPV (%) | 70.00 | 90.00 | 90.00 | 90.00 |
| NPV (%) | 100.00 | 73.68 | 73.68 | 73.68 |
| acuracy (Youden) | 60.00 | 57.62 | 57.62 | 57.62 |
| R2 (Nagelkerke) | 0.56 | 0.45 | 0.45 | 0.66 |
| AIC | 28.44 | 32.40 | 32.40 | 26.36 |
PPV: Positive predictive value; NPV: Negative predictive value; AIC: Akaike information criterion.
In Eq. (1), miR21 and miR451 represent the miR expression in a dichotomized form (miR-21 ≤ 1.16 × 10−4 = 0 and miR-21 > 1.16 × 10−4 = 1; miR-451 > 5.7 = 0 and miR-451 ≤ 5.7 = 1).
miRs expression at follow-up
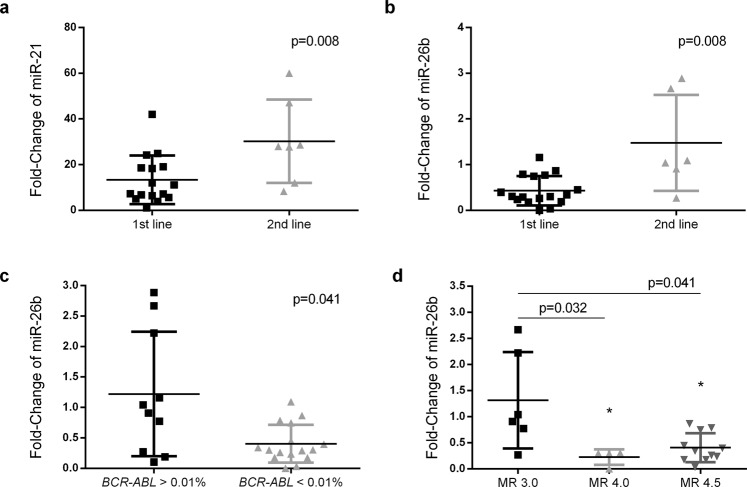
Due to lack of samples, it was not possible to determine the miRs expression at diagnosis in the follow-up group. To determine the fold-change of this group, we used the median of each miR observed in the diagnosis group as a reference, since the values did not present a normal distribution. mir-21 was up-regulated 15.0 times during the treatment, while miR-26b and miR-451 were down-regulated 2.4 and 9.2 times, respectively. Correlating the expression levels with clinical and laboratory data, we observed higher levels of miR-21 (p = 0.008; Fig. 5a) and miR-26b (p = 0.008. Figure 5b) in patients that fail 1st line of treatment and switch for other TKI. Additionally, miR-26b levels were significantly downregulated in patients with lower BCR-ABL1 quantification (p = 0.041; Fig. 5c). Furthermore, we observed that patients with a deeper molecular response (MR) presented lower levels of miR-26b than those with MR of 3.0 (MR 4.0 p = 0.032. and MR 4.5 p = 0.041) (Fig. 5d).
Figure 5.
Analysis of miR-21 and miR-26b fold-change in patients under TKI treatment. miR-21 (a) and miR-26b (b) fold-change were analysed according to the number of TKI that patients were exposed. The miR-26 expression levels were also studied in patients stratified according to BCR-ABL1 values (c) and to molecular response rates (d) Higher levels of miR-21 and miR-26b were detected in patients that fail 1st line of treatment, whereas miR-26b were down-regulated in patients with lower BCR-ABL1 levels and also in patients with deeper responses to treatment (MR 4.0 and MR 4.5). Statistical differences between groups are reported by the p-value.
Discussion
Most deregulated miRs act on specific signalling pathways that seem to be essential to promote and maintain cancer cell survival and growth4. This critical regulatory role, led to associations between miRs expression and response to anticancer drugs proposed by several authors10. Our results from in vitro models highlighted the relation of miR-21 and miR-26b overexpression and miR-451 downregulation with imatinib-resistant phenotype. As expected, an unbalance between oncomiRs and tumour suppressor miRs were observed in the resistant models. In line with in vitro models results, we found a correlation between miR-21 and miR-451 expression at diagnosis with the response to TKI treatment. In our cohort, we observed that the optimal response to TKI after 6 months was related to high levels of miR-451. After 12 months of treatment, patients with an optimal response presented not only high levels of miR-451 but also low levels of miR-21, compared to those without optimal response.
miR-451 acts as a tumour suppressor and has been reported to be downregulated in some neoplasias, such as gastric cancer, glioblastoma and leukaemia21–23. Particularly in CML, the expression of miR-451 was associated with a favourable prognosis, since this miR targets ABL and BCR-ABL1 directly12. According to Lopotová et al.24, the miR-451 expression is downregulated in CML at diagnosis compared with healthy controls, and this could be due to BCR-ABL activity25. The same authors suggested a reciprocal regulatory loop between this miR and BCR-ABL1 levels that could be interrupted by imatinib treatment24. This interaction could have a critical impact on response level, and levels of miR-451 constitute an excellent marker to predict response to TKI. In our results, miR-451 levels were able to predict patients that will achieve response at an early point (6 months) and after one year of treatment, with very good sensitivity and specificity. In the same line, Su et al. (2015) suggested miR-451 as a potential prognostic biomarker in gastric cancer23. The introduction of new biomarkers, as could be the case of miR-451, will improve patient monitoring and could have an impact on therapeutic selection.
With opposite functions, miR-21 is one of the most overexpressed miRs in solid and haematological tumours13. We observed that low levels of this miR at CML diagnosis were related with the achievement of optimal response to TKI therapy. miR-21 present multiple targets as PTEN, PDCD4 and TIMPs that contribute to the promotion of proliferation, survival and invasion pathways, however according to cell type, the principal mechanism of action may be different13,26. In some cells, it could be related to the up-regulation of BCL-2 to prevent apoptosis, while in others the most significant effect may be the inhibition of PTEN and consequently the activation of PI3K/AKT pathway13,27. In CML, BCR-ABL, with its constitutive tyrosine kinase activity, stimulates multiple signalling pathways such as PI3K/AKT and MAPK/ERK. The downregulation of PTEN and inhibition of PDCD4 has been described as the most crucial action of miR-21 in CML cells26,28. The correlation between miR-21 expression and resistance to anticancer agents, describe by us and other groups, highlight the possible clinical application of this miR as a target for therapy29. Several groups showed an increase in cell death and a synergistic effect with other drugs when miR-21 was inhibited by antagomiRs or some molecules that modulate miR-21, as curcumin30–32. In this work, we did not test miR-21 as a therapeutic target, but we assess the power as a tool for predicting a good response to TKI treatment. Levels of miR-21 measured at diagnosis work as a biomarker of response after one year of TKI therapy. In childhood B cell acute lymphoblastic leukaemia, the same miR is an independent prognostic marker, were high levels were associated with poor response and shorter overall survival33.
Using a single measure to predict prognosis, progression, response and many other aspects seem very simple and sometimes difficult to explain the cell complexity. Based on this, we look for the combinatorial miR expression profile since two miRs showed biomarkers characteristics. Based on cut-off values identified for miR-21 and miR-451, we created 4 different profiles to assess the risk/ probability to achieve optimal response after 12 months of TKI treatment. Patients with a high level of miR-451 and low levels of the oncomiR miR-21 at diagnosis (profile1) presented 42 times more probability of achieving an optimal response. According to miRs functions, profile 1 appears as the best combination possible supporting a more controlled environment against cancer cells growth. In opposition, profile 2, which comprehend higher oncomiRs expression, lead to a very low probability of these patients reaching optimal response.
By identifying patients at high risk of relapse or progression under treatment, we can prevent several complications and disease progression. Improving the disease monitoring or introducing changes in therapeutic options for these subgroups of patients could be the best approach to achieve better results. These predictive models might play an important role in identifying subgroups of patients based on clinical and biological features. Sokal, EURO, EUTOS and ELTS are scores used in CML for discriminating overall survival (Sokal and EURO), predicting complete cytogenetic remission (CCgR) 18 months after the start of therapy, and predict the probability of CML related death, respectively34–37. These scores are based on age, spleen size and haematological parameters at diagnosis, such as basophils, blast cells and platelets. However, none of them were designed to predict optimal response to TKI treatment. Using our miRs results, we create a predictive model (Eq. 1) to assess the probability of each patient achieving and optimal response to TKI treatment after one year of therapy. The identification of new biomarkers of response/resistance to TKI is in constant progress, and several molecules present good discriminating power, namely specific gene signatures. Nevertheless, independent validation of all new biomarkers and scores would be very helpful to implement in the clinic the most accurate one to predict the optimal response.
In order to increase the number of patients studied and due to the impossibility of obtaining samples at diagnosis from all patients, we use the medians of miRs expression at diagnosis to estimate the fold-change in miR expression induced by TKI treatment. Although not statistically significant, the expression levels of miR-26b at diagnosis was higher in patients that did not reach optimal response with TKI. However, this miR presented some relevance at follow-up measures in a group of treated patients. The majority of studies describe miR-26b as a tumour suppressor miR that can inhibit cell proliferation in breast cancer, hepatocellular carcinoma and cervical cancers16,38,39. However, some contradictory results have suggested that this miR could act as oncomiR. Palumbo et al.15 revealed that the regulation of PTEN in pituitary tumour cells is mediated by miR-26b action15. miR-26b by targeting PTEN, as also describe to miR-21, leave the PI3K/AKT signalling pathway without regulation, promoting tumorigenesis. In agreement with these authors, our results suggest that lower levels of miR-26b were associated with a good response to the first line of treatment and with more profound responses. These results suggest that miR-26b might be an indicator of good responses levels.
The link between miR-21, miR-26b and miR-451 and TKI response levels need to be validated in a bigger cohort of patients before moving into clinical practice. On the other hand, understanding the crucial role of these miRs in disease progression and treatment response might be very useful for exploring these miRs as a new therapeutic target in CML. In conclusion, expression levels of miR-21 and miR-451 at diagnosis play a crucial role in CML response to TKI treatment and may constitute a new potential biomarkers of response and for the guidance of therapeutic options.
Material and Methods
Cell lines
K562 cells sensitive to Imatinib and two Imatinib-resistant models (K562-RC and K562-RD cells) were used as in vitro models of CML. The sensitive cell line was obtained from ATCC, and the Imatinib-resistant cell lines were developed in our laboratory based on two strategies: a continuous exposure (K562-RC) and a discontinuous exposure to Imatinib (K562-RD), as described in Alves et al.40.
Study population
Fifty-seven CML patients were enrolled in the present study and recruited at Clinical Hematology Department of Centro Hospitalar Universitário de Coimbra (CHUC), Portugal. Patients were grouped according to different time points: at diagnosis (diagnosis group, n = 30) and follow-up (follow-up group, n = 27). Clinical and biological characteristics were summarised in Table 1. Treatment response criteria were defined according to the European Leukemia Net (ELN) guidelines20, where optimal response corresponds to BCR-ABL levels lower than 1% at 6 months and ≤0.1% at 12 months. Patients that presented levels higher than the established cut-off were incorporated in “without optimal response” group. The study was conducted according to the Helsinki Declaration, and all participants provided informed consent for participation before enrolment. The Ethics Committee of the Faculty of Medicine (University of Coimbra, Portugal) approved all research procedures (ref. CE-014/2014).
RNA extraction, cDNA synthesis, and real-time PCR
Total RNA was extracted from CML patients’ samples and cell lines using the mirVana miRNA Isolation Kit (Ambion), according to the manufacturer’s protocol. RNA quantity and quality were measured using a NanoDrop ND-1000 spectrophotometer. TaqMan® Advanced miRNA cDNA Synthesis Kit (Applied Biosystems) was used to convert 10 ng of total RNA, according to the manufacturer’s protocol. Briefly, this kit included several steps: the Poly(A)-tailing reaction, the ligation reaction, reverse transcriptase reaction followed by a miR-Amp reaction. cDNA was diluted to 1:10 with DNase/RNase free water before use. The levels of miR-21, miR-451, miR-26b and miR-16 were determined in each sample. The quantitative real-time polymerase chain reaction (qPCR) was performed using TaqMan Advanced miRNA Assays (Applied Biosystems) in a QuantStudio 3 system (Applied Biosystems). For each miR, we used one TaqMan Advanced miRNA Assays with specific probes and primers. miR-16 exhibited the most constant expression levels between the samples and was identified as endogenous control by relative quantification software (Thermo Fisher Scientifc). Based on this and supported with statistical analysis (Supplementary Figure S1) and miR-16 was used as endogenous control. Relative expression levels of the studied miRNAs were calculated using the relative quantification 2−ΔCT method and normalised with miR-16 (ΔCT = CT of target–CT of miR-16). The fold change was calculated by 2−ΔΔCT method using K562 as a reference for cell lines and the expression at diagnosis for patients follow-up.
Statistical analysis
The statistical analyses were performed using IBM.SPSS®, version 22, except when otherwise indicated. Normal distribution was tested and the following analyses were performed according to these results. Differences between two groups were assessed by nonparametric Mann-Whitney U test and for three or more groups by nonparametric Kruskal-Wallis test followed by Dunn’s multiple comparison tests. The receiver operating characteristic (ROC) curves analysis was performed to evaluate the variables accuracy as biomarkers. For each ROC curve, an optimal cut-off point was determined by the maximum Youden’s J Index, which corresponds to the value of the parameter that maximised the sum of specificity and sensitivity. The positive predictive value (PPV) and negative predictive value (NPV) were calculated for each identified biomarker. The association between profiles and the optimal response was analysed by calculating the odds ratio (OR) and its 95% confidence interval (CI), using Fisher’s exact test with GraphPad Prism version 6.0. G power analysis was performed Gpower software (version 3.1.9.2). The formulas for predicting an optimal response using both miR-21 and miR-451 expression levels were achieved using R software and its general linear model (glm) function with binomial family and logit link, either using continuous or binary miR profile, considering the single effect, main effect, interaction or full factorial models. The Akaike information criterion (AIC’s) minimal value identified the best model. All the statistical tests were considered significant when p < 0.05.
Supplementary information
Acknowledgements
R.A. was supported by the Portuguese Foundation for Science and Technology (FCT) with a PhD grant (SFRH/BD/51994/2012). The work was supported by CIMAGO (Project 10/14), by Faculty of Medicine of the University of Coimbra and Santander Totta Bank with the grant (FMUC-BST-2016-214), by funds from FEDER through the Operational Program Competitiveness Factors (COMPETE) and by FCT under the strategic projects from FCT/MCTES/PIDDAC (CNC.IBILI. Centre Reference: UID/NEU/04539/2013).
Author Contributions
R.A., A.M.A. and A.B.S.R. conceived of the study. P.F.T., A.B.R., D.L. and G.M. recruited and provided the clinical information of the participants. R.A., A.C.G. and J.J. conducted the experiments. R.A., A.C.G. and B.O. performed the statistical analysis. R.A., A.C.G. and J.J. wrote the paper. A.M.A. and A.B.S.R. revised the manuscript. All authors read the final version and approved of its submission.
Data Availability
All data generated or analysed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46132-9.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh S, Azizi SG, Soleimani M, Farshi Y, Kashani Khatib Z. The Role of MicroRNAs in Myeloproliferative Neoplasia. International Journal of Hematology-Oncology and Stem Cell Research. 2016;10:172–185. [PMC free article] [PubMed] [Google Scholar]
- 3.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Current Opinion in Cell Biology. 2009;21:470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Ichimura A, Ruike Y, Terasawa K, Tsujimoto G. miRNAs and regulation of cell signaling. The FEBS Journal. 2011;278:1610–1618. doi: 10.1111/j.1742-4658.2011.08087.x. [DOI] [PubMed] [Google Scholar]
- 5.Berindan-Neagoe I, Monroig PDC, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA: A Cancer Journal for Clinicians. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litwińska Z, Machaliński B. miRNAs in chronic myeloid leukemia: small molecules, essential function. Leukemia & Lymphoma. 2017;58:1297–1305. doi: 10.1080/10428194.2016.1243676. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes Q. MicroRNA: Defining a new niche in Leukemia. Blood Reviews. 2017;31:129–138. doi: 10.1016/j.blre.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Apperley JF. Chronic myeloid leukaemia. The Lancet. 2015;385:1447–1459. doi: 10.1016/S0140-6736(13)62120-0. [DOI] [PubMed] [Google Scholar]
- 9.Volpe G, Panuzzo C, Ulisciani S, Cilloni D. Imatinib resistance in CML. Cancer Letters. 2009;274:1–9. doi: 10.1016/j.canlet.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 10.An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharmaceutica Sinica B. 2017;7:38–51. doi: 10.1016/j.apsb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon JEA, Wong JJL, Rasko JEJ. MicroRNAs in myeloid malignancies. British Journal of Haematology. 2013;162:162–176. doi: 10.1111/bjh.12364. [DOI] [PubMed] [Google Scholar]
- 12.Yeh C-H, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Molecular Cancer. 2016;15:37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y-H, Tsao C-J. Emerging role of microRNA-21 in cancer. Biomedical Reports. 2016;5:395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai H, Xu R, Cao Z, Wei D, Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Letters. 2010;585:402–408. doi: 10.1016/j.febslet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 15.Palumbo T, et al. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN–AKT pathway. Oncogene. 2012;32:1651. doi: 10.1038/onc.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, Shen D, Wang W, Xian J. Aberrant expression of microRNA-26b and its prognostic potential in human cervical cancer. International Journal of Clinical and Experimental Pathology. 2015;8:5542–5548. [PMC free article] [PubMed] [Google Scholar]
- 17.Xishan Z, Ziying L, Jing D, Gang L. MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in chronic myeloid leukemia. Scientific Reports. 2015;5:12460. doi: 10.1038/srep12460. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Kotagama K, Chang Y, Mangone M. miRNAs as Biomarkers in Chronic Myelogenous Leukemia. Drug Development Research. 2015;76:278–285. doi: 10.1002/ddr.21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avigad S, et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes, Chromosomes and Cancer. 2015;55:328–339. doi: 10.1002/gcc.22334. [DOI] [PubMed] [Google Scholar]
- 20.Baccarani M, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–884. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nan Y, et al. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Research. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 22.Soltani I, et al. Downregulation of miR-451 in Tunisian chronic myeloid leukemia patients: potential implication in imatinib resistance. Hematology. 2017;22:201–207. doi: 10.1080/10245332.2016.1252020. [DOI] [PubMed] [Google Scholar]
- 23.Su Z, Zhao J, Rong Z, Geng W, Wang Z. MiR-451, a potential prognostic biomarker and tumor suppressor for gastric cancer. International Journal of Clinical and Experimental Pathology. 2015;8:9154–9160. [PMC free article] [PubMed] [Google Scholar]
- 24.Lopotová T, Žáčková M, Klamová H, Moravcová J. MicroRNA-451 in chronic myeloid leukemia: miR-451–BCR-ABL regulatory loop? Leukemia Research. 2011;35:974–977. doi: 10.1016/j.leukres.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Scholl V, Hassan R, Zalcberg I. R. miRNA-451: A putative predictor marker of Imatinib therapy response in chronic myeloid leukemia. Leukemia Research. 2012;36:119–121. doi: 10.1016/j.leukres.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Panagal M, et al. MicroRNA21 and the various types of myeloid leukemia. Cancer Gene Therapy. 2018 doi: 10.1038/s41417-018-0025-2. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira AF, et al. ApoptomiRs expression modulated by BCR–ABL is linked to CML progression and imatinib resistance. Blood Cells, Molecules, and Diseases. 2014;53:47–55. doi: 10.1016/j.bcmd.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Xu T, Chen C. The critical roles of miR-21 in anti-cancer effects of curcumin. Annals of Translational Medicine. 2015;3:330. doi: 10.3978/j.issn.2305-5839.2015.09.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abba ML, et al. MicroRNAs as novel targets and tools in cancer therapy. Cancer Letters. 2017;387:84–94. doi: 10.1016/j.canlet.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 30.Taverna S, et al. Curcumin inhibits in vitro and in vivo chronic myelogenous leukemia cells growth: a possible role for exosomal disposal of miR-21. Oncotarget. 2015;6:21918–21933. doi: 10.18632/oncotarget.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Anti-miR-21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Science. 2010;101:948–954. doi: 10.1111/j.1349-7006.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W-Z, et al. Targeting miR-21 sensitizes Ph+ ALL Sup-b15 cells to imatinib-induced apoptosis through upregulation of PTEN. Biochemical and Biophysical Research Communications. 2014;454:423–428. doi: 10.1016/j.bbrc.2014.10.107. [DOI] [PubMed] [Google Scholar]
- 33.Labib HA, Elantouny NG, Ibrahim NF, Alnagar AA. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22:392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 34.Sokal JE, et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood. 1984;63:789. [PubMed] [Google Scholar]
- 35.Hasford J, et al. A New Prognostic Score for Survival of Patients With Chronic Myeloid Leukemia Treated With Interferon AlfaWriting Committee for the Collaborative CML Prognostic Factors Project Group. JNCI: Journal of the National Cancer Institute. 1998;90:850–859. doi: 10.1093/jnci/90.11.850. [DOI] [PubMed] [Google Scholar]
- 36.Hasford J, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: the EUTOS score. Blood. 2011;118:686. doi: 10.1182/blood-2010-12-319038. [DOI] [PubMed] [Google Scholar]
- 37.Pfirrmann M, et al. Prognosis of long-term survival considering disease-specific death in patients with chronic myeloid leukemia. Leukemia. 2016;30:48–56. doi: 10.1038/leu.2015.261. [DOI] [PubMed] [Google Scholar]
- 38.Jin F, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death & Disease. 2017;8:e2540. doi: 10.1038/cddis.2016.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, et al. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell International. 2013;13:7. doi: 10.1186/1475-2867-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alves R, et al. Drug transporters play a key role in the complex process of Imatinib resistance in vitro. Leukemia Research. 2015;39:355–360. doi: 10.1016/j.leukres.2014.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.