Abstract
A better knowledge of the bovine milk proteome and its main drivers is a prerequisite for the modulation of bioactive proteins in milk for human nutrition, as well as for the discovery of biomarkers that are useful in husbandry and veterinary medicine. Milk composition is affected by lactation stage and reflects, in part, the energy balance of dairy cows. We aggregated the cow milk proteins reported in 20 recent proteomics publications to produce an atlas of 4654 unique proteins. A multistep assessment was applied to the milk proteome datasets according to lactation stages and milk fractions, including annotations, pathway analysis and literature mining. Fifty-nine proteins were exclusively detected in milk from early lactation. Among them, we propose six milk proteins as putative biomarkers of negative energy balance based on their implication in metabolic adaptative pathways. These proteins are PCK2, which is a gluconeogenic enzyme; ACAT1 and IVD, which are involved in ketone metabolism; SDHA and UQCRC1, which are related to mitochondrial oxidative metabolism; and LRRC59, which is linked to mammary gland cell proliferation. The cellular origin of these proteins warrants more in-depth research but may constitute part of a molecular signature for metabolic adaptations typical of early lactation.
Subject terms: Gene ontology, Data integration, Animal physiology
Introduction
Phenotyping animal traits related to performance, quality, welfare and health is often challenging but necessary to meet husbandry and societal expectations. Rapid and non-invasive tools are desirable to monitor multiple animals traits accurately and inexpensively1–3. Biological fluids such as milk are increasingly used as a source for animal trait phenotyping3. Milk composition varies depending on several factors, including the stage of lactation, the metabolic status and the health status of dairy cows3. In early lactation, energy balance and body reserve mobilization are major drivers of dairy performance, robustness and longevity of dairy cows4. Negative energy balance (NEB) occurs frequently in early lactation cows because the energy demands for milk production exceed nutrient intake5–7. Maladaptation during the periparturient period impacts milk production and increases the risk of post-partum diseases, removal from the herd and infertility8,9. Much attention has been paid to the development of milk indicators for NEB in high-producing dairy cows. Of these, some are based on proteins identified by proteomics. The abundance of ten proteins with roles in cholesterol synthesis and composition of the milk fat globule membrane (MFGM) was shown to vary with the energy balance in early lactation cows10. Furthermore, improvements in the sensitivity of proteomics techniques11, which have allowed the identification of thousands of proteins in milk12,13; have contributed to a better understanding of lactation periods10,14 and animal health15–17. The volume of publicly available proteomics data provides opportunities for in silico proteomics studies in compliance with recent guidelines18. We hypothesized that the computation of available proteomics data would allow putative biomarkers of NEB to be identified. We used the early lactation period as a proxy for NEB because modern dairy cows systematically experience some degree of negative energy balance and extensive mobilisation of body reserves during this period. We use, reuse, reprocess, and repurpose18 the cow milk proteome reported in 20 recent publications to describe the proteomic signature of milk according to lactation period and milk fraction. The aim of the current study was to identify proteins that were specifically identified in early lactation milk. These proteins could be robust biomarkers of NEB based on their presence or absence in milk or milk fractions during early lactation (all or nothing identification). Moreover, because available proteomics data were obtained from different breeds, countries and rearing practices, these proteins may constitute robust biomarkers of NEB, independently of the breed and husbandry practices. To our knowledge, this is the first attempt to reuse the publicly available milk proteome data to propose potential indicators for NEB and for dairy ruminants.
Results
Proteome atlas overview according to milk fractions
Among the 4654 proteins compiled in the atlas, 95 gene names (GN) (Fig. 1 and Supplementary information Table 1) were detected for all four milk fractions, whereas 93, 488, 15 and 3139 GN were specifically detected in skimmed milk, whey, MFGM and exosomes.
Figure 1.
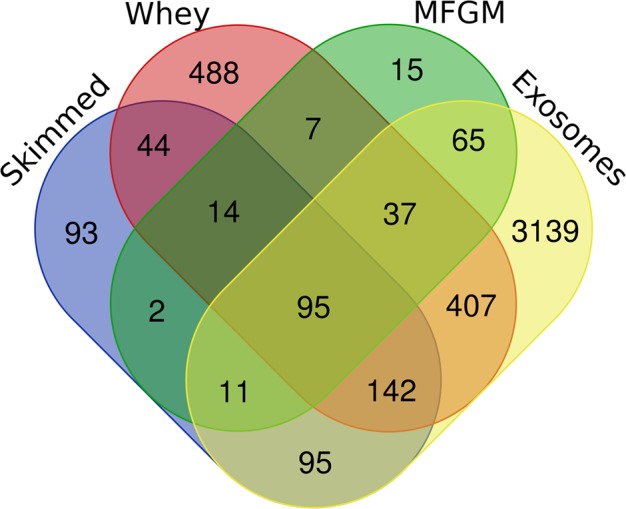
Venn diagram of common and specific gene names for proteins present in skimmed, whey, MFGM and exosome milk fractions.
The 95 GN detected in the studied milk fractions are the most abundant milk proteins. These included alpha-S1-casein (CSN1S1), alpha-S2-casein (CSN1S2), beta-casein (CSN2), α-lactalbumin (LALBA) and β-lactoglobulin (LGB), with the latter frequently detected in whey. The 8 major proteins detected in the MFGM, such as mucin 1 (MUC1), redox enzyme xanthine dehydrogenase (XDH), and butyrophilin subfamily 1 member A1 (BTN1A1), were among the list of 95 GN. Similarly, proteins classically detected in milk exosomes, such as lipopolysaccharide-binding protein (LBP), annexin (ANXA1), complement c3 (C3), protein S100-A9 (S100A9), serum amyloid A protein (SAA3) and cathelicidin-1 (CATHL1), were also among the list of 95 proteins found in all the milk fractions. Enriched Gene Ontology (GO) terms (980 GO terms) annotated 94 proteins among the 95 detected proteins in the four milk fractions. Among the top 50 enriched (P < 0.05) GO terms in the Biological Process (BP) category, 9 terms related to lipoprotein, phosphatidylcholine and cholesterol, highlighting the proteins involved in lipid metabolism. Other enriched GO terms were related to hormone and cytokine signalling, such as the growth hormone secretion that annotated a member of the RAS oncogene family protein (RAB1A). The growth factor/hormone term annotated the heat shock protein HSP 90-beta (HSP90AB1) and CSN1S1. Terms linked to inflammatory response annotated the tissue factor (F3) involved in the cytokine-mediated signalling pathway and the proteins S100A8 and S100A9 that are involved in antioxidant activity and immune response.
Proteome atlas overview according to lactation stages
Of the 4654 proteins present in the atlas, 105 GN (Fig. 2 and Supplementary information Table 1) were detected in all five lactation stages, whereas 3288, 59, 185 and 155 GN were exclusively identified during the colostrum period and early, peak and mid-lactation.
Figure 2.
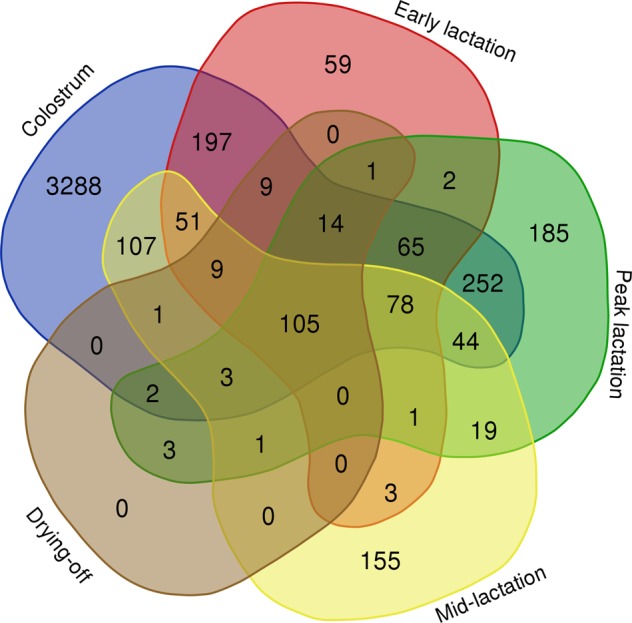
Venn diagram of common and specific gene names from the colostrum period, early lactation, peak lactation, mid-lactation and drying-off.
As expected, of the 105 GN, four were the major caseins: CSN1S1, CSN1S2, CSN2 and kappa-casein (CSN3). Other detected proteins related to protein and lactose synthesis, e.g., lactotransferrin (LTF), LALBA and beta-1,4-galactosyltransferase 1 (B4GALT1). Some proteins linked to lipolysis and fatty acid esterification were also detected, including perilipin-2 (PLIN2); apolipoproteins E, A-IV, A-I (APOE, APOA4, APOA1); and lipoprotein lipase (LPL), which in the mammary gland hydrolyses the triglycerides circulating in chylomicra and very low density lipoproteins, and in milk hydrolyses the triglycerides of the fat globule core. F 45 kDa calcium-binding protein (SDF4), which is linked to calcium metabolism, and leucine-rich alpha-2-glycoprotein 1 (LRG1), implicated in brown fat cell differentiation, were also detected. Six hundred seventy-eight enriched GO terms annotated 101 of the 105 proteins detected in all lactation stages. Among the top 50 (P < 0.05) of the enriched GO terms in the BP category, one term related to phosphatidylcholine annotated heart fatty acid-binding protein (FABP3) and highlighted lipid metabolism. The term lactose biosynthetic process and 3 terms related to hormones annotated LALBA, B4GALT1 and CSN1S1, which are all proteins that support lactation.
Focus on milk proteins specific for early lactation as potential biomarkers of NEB
The 59 GN exclusively detected during early lactation (i.e., 6 to 21 days in milk (DIM); Fig. 2) are listed in Table 1.
Table 1.
The 59 proteins detected in milk during early lactation (i.e., 6 to 21 DIM).
| No. | Gene Name | Protein ID | Protein name | Milk fraction localization | References |
|---|---|---|---|---|---|
| 1 | ABCC4 | A0JND8_BOVIN | ABCC4 protein | MFGM | 10 |
| 2 | ACAT1 | THIL_BOVIN | Acetyl-CoA acetyltransferase, mitochondrial | Skimmed | 13, 17 |
| 3 | ALDH3B1 | AL3B1_BOVIN | Aldehyde dehydrogenase family 3 member B1 | Skimmed | 10 |
| 4 | ALOX12 | Q6SYC4_BOVIN | Arachidonate 12-lipoxygenase | MFGM | 10 |
| 5 | ARL6IP5 | PRAF3_BOVIN | PRA1 family protein 3 | Skimmed | 13, 17 |
| 6 | ATP5F1a | AT5F1_BOVIN | ATP synthase F(0) complex subunit B1, mitochondrial | Skimmed | 13, 17 |
| 7 | ATP5Ha | ATP5H_BOVIN | ATP synthase subunit d, mitochondrial | Skimmed | 13, 17 |
| 8 | ATP5Ja | ATP5J_BOVIN | ATP synthase-coupling factor 6, mitochondrial | Skimmed | 13, 17 |
| 9 | ATP5J2a | ATPK_BOVIN | ATP synthase subunit f, mitochondrial | Skimmed | 13, 17 |
| 10 | BAT1 | Q861P7_BOVIN | HLA-B-associated transcript 1 | Skimmed | 13 |
| 11 | Bt.64131 | F1MIR4 | RAB2A, member RAS oncogene family | MFGM | 10 |
| 12 | C13H20ORF116a | Q1LZB0_BOVIN | Chromosome 20 open reading frame 116 ortholog | Skimmed | 13 |
| 13 | C1QBP | C1QBP_BOVIN | Complement component 1 Q subcomponent-binding protein, mitochondrial | Skimmed | 13, 17 |
| 14 | CAT | CATA_BOVIN | Catalase | Skimmed | 13, 17 |
| 15 | COPG | Q0V888_BOVIN | Coatomer protein complex, subunit gamma 1 | Skimmed | 13, 17 |
| 16 | COX5A | COX5A_BOVIN | Cytochrome c oxidase subunit 5A, mitochondrial | Skimmed | 13, 17 |
| 17 | COX5B | COX5B_BOVIN | Cytochrome c oxidase subunit 5B, mitochondrial | Skimmed | 13, 17 |
| 18 | COX7A2 | CX7A2_BOVIN | Cytochrome c oxidase subunit 7A2, mitochondrial | Skimmed | 13, 17 |
| 19 | CUZD1a | F1MD73 | Uncharacterized protein | MFGM | 10 |
| 20 | DDRGK1 | DDRGK_BOVIN | DDRGK domain-containing protein 1 | Skimmed | 13, 17 |
| 21 | DDX39B | DX39B_BOVIN | Spliceosome RNA helicase DDX39B | Skimmed | 17 |
| 22 | EEF1B | EF1B_BOVIN | Elongation factor 1-beta | Skimmed | 13, 17 |
| 23 | GPAM | GPAT1_BOVIN | Glycerol-3-phosphate acyltransferase 1, mitochondrial | MFGM | 10 |
| 24 | HIST1H4Aa | P62803 | Histone 4 | MFGM | 10 |
| 25 | IGa | A5D7Q2 | Uncharacterized protein | MFGM | 10 |
| 26 | IVD | IVD_BOVIN | Isovaleryl-CoA dehydrogenase, mitochondrial | Skimmed | 13, 17 |
| 27 | LF | Q95M55_BOVIN | Lactoferrin | MFGM | 10 |
| 28 | LMAN1 | Q8MJ82_BOVIN | Lectin mannose binding 1 | Skimmed | 13, 17 |
| 29 | LOC789567 | A6H7H3_BOVIN | LOC789567 protein | Skimmed | 13, 17 |
| 30 | LRPAP1 | Q148K7_BOVIN | Low density lipoprotein receptor-related protein associated protein 1 | Skimmed | 13, 17 |
| 31 | LRRC59 | LRC59_BOVIN | Leucine-rich repeat-containing protein 59 | Skimmed | 13, 17 |
| 32 | Man8 | O78186_BOVIN | MHC class I antigen | MFGM | 10 |
| 33 | MGC137099 | Q2KII3_BOVIN | Hepatitis A virus cellular receptor 1 N-terminal domain containing protein | MFGM | 10 |
| 34 | MYCBP | MYCBP_BOVIN | c-Myc-binding protein | Skimmed | 13, 17 |
| 35 | NDUFA5 | NDUA5_BOVIN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | Skimmed | 13, 17 |
| 36 | NDUFAB1 | ACPM_BOVIN | Acyl carrier protein, mitochondrial | Skimmed | 13, 17 |
| 37 | OSTC | OSTC_BOVIN | Oligosaccharyltransferase complex subunit OSTC | Skimmed | 13 |
| 38 | PAFAH1B2 | PA1B2_BOVIN | Platelet-activating factor acetylhydrolase IB subunit beta | Skimmed | 13, 17 |
| 39 | PCK2 | F1MDS3_BOVIN | Phosphoenolpyruvate carboxykinase 2, mitochondrial | Skimmed | 13, 17 |
| 40 | PLSCR2 | PLS2_BOVIN | Phospholipid scramblase 2 | MFGM | 10 |
| 41 | RPL10A | RL10A_BOVIN | 60S ribosomal protein L10a | Skimmed | 13, 17 |
| 42 | RPL18 | RL18_BOVIN | 60S ribosomal protein L18 | Skimmed, MFGM | 10, 13, 17 |
| 43 | RPL6 | RL6_BOVIN | 60S ribosomal protein L6 | Skimmed | 13, 17 |
| 44 | RPL7 | RL7_BOVIN | 60S ribosomal protein L7 | Skimmed | 13, 17 |
| 45 | RPL7A | RL7A_BOVIN | 60S ribosomal protein L7a | Skimmed | 13, 17 |
| 46 | RPS13 | RS13_BOVIN | 40S ribosomal protein S13 | Skimmed | 13, 17 |
| 47 | SAA | P35541 | Serum amyloid A protein | MFGM | 10 |
| 48 | SDHA | SDHA_BOVIN | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Skimmed | 13, 17 |
| 49 | SEC. 11C | Q2KI36_BOVIN | Signal peptidase complex catalytic subunit SEC. 11 | Skimmed | 13, 17 |
| 50 | SEC. 61A1 | S61A1_BOVIN | Protein transport protein Sec. 61 subunit alpha isoform 1 | Skimmed | 13, 17 |
| 51 | SLC15A2 | B8Y6C2_BOVIN | Solute carrier family 15 member 2 | MFGM | 10 |
| 52 | SSR1 | SSRA_BOVIN | Translocon-associated protein subunit alpha | Skimmed | 13, 17 |
| 53 | SURF4 | SURF4_BOVIN | Surfeit locus protein 4 | Skimmed | 13, 17 |
| 54 | SYPL1 | A8PVV5_BOVIN | SYPL1 protein | Skimmed | 13, 17 |
| 55 | TMED9 | TMED9_BOVIN | Transmembrane emp24 domain-containing protein 9 | Skimmed | 13, 17 |
| 56 | TMEM43 | A6QQR5_BOVIN | TMEM43 protein | Skimmed | 13, 17 |
| 57 | TREM1 | TREM1_BOVIN | Triggering receptor expressed on myeloid cells 1 | Skimmed | 13, 17 |
| 58 | TXNDC4a | TXND4_BOVIN | Thioredoxin domain-containing protein 4 | Skimmed | 13 |
| 59 | UQCRC1 | QCR1_BOVIN | Cytochrome b-c1 complex subunit 1, mitochondrial | Skimmed | 13, 17 |
aUpdated since the period of data mining. The Gene name and ID were converted before November 2018 by use of the Retrieve/ID Mapping tool of the Uniprot database.
Of the 59 GN, 52 were annotated by 326 enriched GO terms. Among the most significantly (P < 0.001) enriched GO terms in the BP category (Supplementary information Fig. 1), 3 terms are related to the oxidation-reduction process and oxidative metabolism. These terms annotated proteins of the respiratory chain such as the mitochondrial cytochrome b-c1 complex subunit 1 protein (UQCRC1); mitochondrial acyl carrier protein (NDUFAB1); and two mitochondrial cytochrome c oxidase subunits, 5B and 5A (COX5B, COX5A), which are terminal oxidases in mitochondrial electron transport. We listed the mitochondrial succinate dehydrogenase flavoprotein subunit (SDHA), which is involved in the complex II of the mitochondrial electron transport chain. We identified the aldehyde dehydrogenase family 3 member B1 (ALDH3B1), which oxidizes medium- and long-chain saturated and unsaturated aldehydes, and the mitochondrial isovaleryl-CoA dehydrogenase (IVD) that is involved in the synthesis of 3-hydroxy-3-methylglutaryl-CoA from 3-isovaleryl-CoA as it enters the β-oxidation step. The terms related to oxidative metabolism also annotated catalase (CAT), which protects cells from the toxic effects of hydrogen peroxide, and the 12 S type arachidonate 12-lipoxygenase (ALOX12), which participates in lipid hydroperoxidation. Seven terms are related to translation and transport. One term, ribosome biogenesis, annotated the mitochondrial complement component 1 Q subcomponent-binding protein (C1QBP). Three terms are related to ketone metabolism and annotated the mitochondrial acetyl-CoA acetyltransferase or acetoacetyl-CoA thiolase (ACAT1). Other milk proteins were not annotated and may contribute to the oxidative metabolism, such as the mitochondrial phosphoenolpyruvate carboxykinase 2 (PCK2), the mitochondrial glycerol-3-phosphate acyltransferase 1 (GPAM), the triggering receptor expressed on myeloid cells 1 (TREM1) and the signal peptidase complex catalytic subunit (SEC. 11C). The Leucine-rich repeat-containing protein 59 (LRRC59) is also annotated in early lactation milk. This protein is required for nuclear import of the fibroblast growth factor 1 (FGF1).
Algorithms implemented in ProteINSIDE proposed 15 out of the 59 proteins (25% of the early milk proteins list) as mitochondrial proteins. The mitochondrial proteins are COX7A2, NDUFA5, GPAM, ATP5F1, ATP5H, ATP5J, ATP5J2, C1QBP, COX5B, COX5A, NDUFAB1, SDHA, UQCRC1, IVD and ACAT1. Finally, several proteins belong to protein complexes of the ATP synthase (4 proteins), cytochrome c oxidase (3 proteins) and 60S ribosomal protein (5 proteins).
Of these 59 proteins, only the RPL18 was detected in both the MFGM and skimmed milk fractions. Fourteen proteins were detected exclusively in the MFGM fraction: SLC15A2, Man8, Bt.64131, the 12S type arachidonate 12-lipoxygenase (ALOX12), GPAM, HIST1H4A, PLSCR2, LF, ABCC4, CUZD1, IG, MGC137099, ALDH3B1 and SAA. The 44 other proteins were detected exclusively in the skimmed milk (Table 1). Obviously, these proteins are also present in whole milk. However, the ability to detect and quantify minor proteins will strongly depend on their concentration; therefore, their technical enrichment in these fractions before mass spectrometry identification could be valuable.
Discussion
We merged available proteomic datasets to produce an atlas of 4654 nonredundant proteins. We identified 59 proteins specifically found in early lactation milk, which we proposed as putative biomarkers of NEB. Of these, we focused on six milk proteins because of their link with ketogenesis, gluconeogenesis and oxidative metabolism, which are well known metabolic pathways enhanced in dairy cows during early lactation.
Early lactation is a classical situation of physiological undernutrition and NEB because feed intake increases at a slower pace than the requirements for milk production. The prioritization of nutrient partitioning to the mammary gland and milk synthesis leads to mobilization of body fat, glycogen, proteins and minerals. Dairy cows may mobilize up to 90 kg of fat and 24 kg of protein7. Intense lipomobilisation leads to the release of adipose free fatty acids (FFA) into plasma. At the same time, lipogenesis, FA esterification and glucose utilization decrease in adipose tissue7. Much of the mobilized protein appears to be derived primarily from skeletal muscle through the downregulation of tissue protein synthesis and increased proteolysis19. The liver coordinates and interconverts nutrients to support pregnancy and lactation by increasing gluconeogenesis and ketogenesis6,7. The NEB of dairy cows leads to these metabolic adaptations during early lactation20,21. We mined the 59 proteins exclusively detected in milk sampled between 6 and 21 DIM from cows likely in NEB, relative to these well-known homeorhetic adaptations.
All 59 proteins may be putative biomarkers of NEB. None of them were previously listed in a study reporting the variation in abundance of milk protein relatively to the energy status of dairy cows10 or in a comparison between colostrum and 7 DIM22. Despite being present in our atlas, 11 proteins (stomatin (STOM), ectonucleotide pyrophosphatase/phosphodiesterase family member 3 (ENPP3), acyl-CoA synthetase long-chain family member 1 (ACSL1), NADH-cytochrome b5 reductase 3 (CYB5R3), Isocitrate dehydrogenase [NADP] cytoplasmic (IDH1), lactoperoxidase (LPO), serum albumin (ALB), LGB, LALBA, LBP and cell death-inducing DFFA-like effector a (CIDEA)) were not specific to early lactation and therefore were not included in our present/absent list. We believed that the binary approach that we utilized has produced a robust list of biomarkers because these proteins were present exclusively in milk from cows in early lactation. This suggests that their abundance strongly increases during early lactation, which may simplify their detection and quantification to qualify these biomarkers relative to the energy status of cows in further studies. Moreover, the reliability of the 59 putative biomarkers of NEB that we propose is strengthened because most of them were previously identified as affected by lactation stage23 and heat stress24,25 during early lactation. Indeed, out of the 59 proteins detected in early lactation milk, 43 were previously identified in the adipose tissue25, liver24 or mammary gland23 of lactating cows experiencing different EB (Fig. 3). Of these, 12 proteins belong to 3 protein complexes (ATP synthase, cytochrome c oxidase and 60S ribosomal protein), which reduced the diversity of proteins on our list. However, for future studies aimed to evaluate these putative biomarkers of NEB in compliance with the biomarker discovery pipeline26, we need to focus on a restricted number of proteins. Therefore, we selected 6 proteins we believe are pertinent because of their biological functions relative to the homeorhetic adaptation of early lactation cows. These proteins, which are the mitochondrial phosphoenolpyruvate carboxykinase 2 (PCK2), ACAT1, IVD, SDHA, UQCRC1 and LRRC59, are potential indicators of NEB because of their roles in metabolic adaptations to early lactation.
Figure 3.
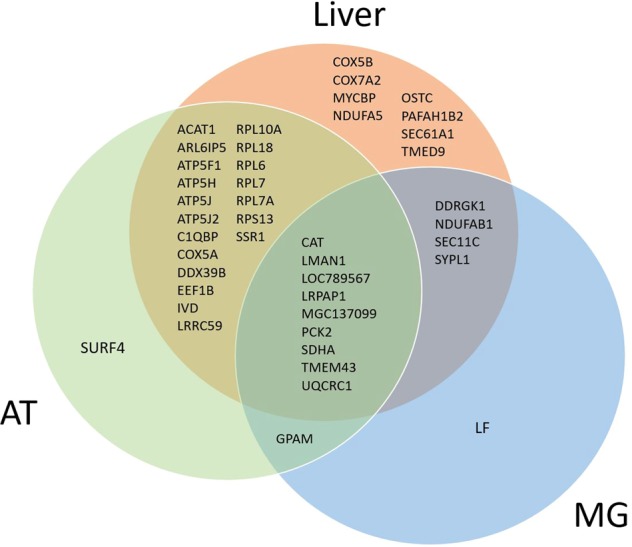
Venn diagram of the 43 gene names from early milk proteins that were previously identified in the liver24, adipose tissue25 and mammary gland23 of dairy cows.
We propose LRRC59, a protein originating from the cytosol, membrane and/or endoplasmic reticulum, as a putative biomarker of NEB. This protein was identified in the AT and liver proteomes24,25, but not in mammary gland. The LRRC59 is required for the nuclear import of the FGF127, a growth factor that participates in the regulation of proliferation and differentiation of the mammary gland cells. Immunoreactive FGF1 was found in considerable concentration in the epithelium of the mammary gland in heifers during mammogenesis and lactation28. The LRRC59 is a membrane‐anchored protein located in the endoplasmic reticulum that may be secreted in milk via exocytosis.
PCK2 is the mitochondrial isoform of the hepatic gluconeogenic enzyme that was detected in early lactation milk. The massive increase in glucose requirements around calving is partially met by endogenous glucose synthesis via liver gluconeogenesis in dairy cows. The gene expression of PCK2 was increased in the liver of cows in early lactation29, and the processes of milk secretion may transfer the PCK2 protein from the plasma into the milk. Indeed, in humans, the mapping of the human tissue proteome based on an integrated omics analysis has indicated that the liver is the major secretory tissue30. However, milk PCK2 could also arise from a mammary synthesis, since the abundance of PCK2 mRNA in the mammary tissue, as in liver and in skeletal muscle, varied depending on the genetic merit and lactation performances of the dairy cows31. Moreover, PCK2 activity was reported to increase 43-fold during the transition from pregnancy into lactation in the guinea pig mammary gland32. The biological function of PCK2 expression by the liver or other organs may be to provide phosphoenolpyruvate for gluconeogenesis. Due to the similarity between PCK2 and PCK1, PCK2 was also proposed to contribute to the synthesis of glycerol-3-phosphate, a precursor for fatty acid esterification into triglycerides, especially in tissues deprived of glucose-6-phosphatase such as mammary gland31.
We proposed ACAT1 as a potential biomarker of NEB because it plays a major role in ketone metabolism. Early lactating cows mobilize lipids stored in adipose tissue and exhibit an increased plasma FFA concentration. Plasma FFA are taken up by the liver and partially oxidized to ketone bodies. Milk β-hydroxybutyrate, a ketone body, is commonly used to detect subclinical ketosis33,34, which is consistent with the detection of ACAT1 in early lactation milk.
We proposed IVD as a potential NEB biomarker because it is also related to ketogenesis. IVD is involved in the L-leucine degradation pathway and in the synthesis of 3-hydroxy-3-methylglutaryl-CoA, an intermediate in ketogenesis. IVD was detected in bovine liver35, AT proteomes25, and in mammary gland transcriptome36. The origin and biological significance of milk ACAT1 and IVD remain unclear. Nevertheless, these proteins may constitute molecular signatures of a ketogenic state typical of early lactation.
We proposed SDHA and UQCRC1, two mitochondrial proteins involved in the oxidative phosphorylation and ATP production, as putative biomarkers of NEB. SDHA is involved in the complex II of the mitochondrial electron transport chain and has a role in the tricarboxylic acid cycle pathway. The SDHA was reported in milk somatic cells37. During lactation, mammary fatty acid and cholesterol synthesis require large amounts of energy and reduction equivalents in the form of NADPH. In the ruminant mammary gland, most NADPH is synthesised from the decarboxylation of isocitrate5. Therefore, the presence of SDHA in milk may reflect NADPH requirements and the oxidative metabolic activity of the mammary gland. The UQCRC1 is involved in complex III of the respiratory chain, electron transfer coupled to proton pumping and NADPH synthesis and was reported in bovine mammary gland23,36. Mitochondrial NADPH production implies the transfer of electrons and the translocation of protons. We speculate that UQCRC1 in milk reflects the oxidative metabolic activity of the mammary gland. The presence of these five mitochondrial proteins (PCK2, ACAT1, IVD, SDHA and UQCRC1) in early lactation milk reflects the upregulation of metabolic pathways (ketogenesis, respiratory chain, tricarboxylic acid cycle and β-oxidation cycle) in the mammary gland and other key tissues of early lactation cows experiencing NEB and a glucose deficit. The identification of several mitochondrial proteins is consistent with the increase in the number of mitochondria present in bovine epithelial cells from parturition until the peak of lactation, which emphasizes the pertinence of mitochondrial proteins as putative biomarkers of NEB38.
Our in silico proteomics approach allowed us to produce an atlas of milk proteins, of which we listed 59 milk proteins as being present exclusively in early lactation milk (post-colostrum period), a period characterized by NEB, lipomobilisation and metabolic imbalance in dairy cows. Among this list of putative biomarkers of NEB, we selected six proteins—LRRC59, PCK2, ACAT1, IVD, SDHA and UQCRC1—based on their roles in multiple pathways of energy metabolism, and therefore potentially associated with states of metabolic imbalance. All six proteins were detected in skimmed milk; consequently, technological enrichment of these proteins may be possible in further assays. Notably, given the power of proteomics, minor proteins can be detected despite their low concentration. Whether these proteins arise from tissues that are central in the homeorhetic adaptation of early lactation warrants further study. Further research is also needed to qualify these biomarkers when a relationship between the abundance of these proteins in milk and the energy status of early lactating cows is implied. The results obtained in this study proves the usefulness of mining the present atlas to understand and phenotype some traits of dairy cows.
Methods
Construction of the milk proteome atlas
A computational workflow was used to aggregate data from publications reporting cow milk proteome to create an atlas of all proteins present, independently of their abundance and without statistical analysis. Briefly, we collected publications on bovine milk by a literature search on Bos taurus using PubMed.gov (NCBI) and the Web of Science (Clarivate Analytics) search engines up to February 2018. The search provided 87 milk proteome publications that were reviewed and curated based on the availability of information, such as the days in milk and the health status of the cows, as well as accessible supplementary materials. Milk protein data could come from tank samples or individual milking but only from cows without mammary infection according to the somatic cell counts or as declared by the authors. Twenty publications complied with the selection criteria:10,13,15,17,22,39–53. The main objective of this computational data aggregation from 20 publications, with 35 datasets (Table 2), is to obtain an overview of milk proteins independently of breed, age, country and methodologies of protein isolation and identification. Methods for protein isolation were density gradient ultracentrifugation15,39, centrifugation and washing10,22,40, (ultra)centrifugation with acidification48 and with major proteins depletion42,45,46, or (ultra)centrifugation without acidification13,17,41,43,44,47,49–53.
Table 2.
Characteristics of the 35 datasets selected for data extraction.
| Datasets | Variation factor studied by authors | Factor selected for data extraction | Milk fraction class | Lactation stage class | Country | Breed | Sampling | Protein isolation | Protein identificationa | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | lactation stage | 12 and 24 h post-partum | Exosomes | Colostrum | Australia | Holstein-Friesian | Fresh | Density gradient ultracentrifugation | nanoLC-MS/MS | 15 |
| 2 | lactation stage | 48 h post-partum | Exosomes | Colostrum | Australia | Holstein-Friesian | Fresh | Density gradient ultracentrifugation | nanoLC-MS/MS | 15 |
| 3 | lactation stage | 72 h post-partum | Exosomes | Colostrum | Australia | Holstein-Friesian | Fresh | Density gradient ultracentrifugation | nanoLC-MS/MS | 15 |
| 4 | milk maturation | 0 to 5 d post-partum | Exosomes | Colostrum | China | Holstein | Fresh | Density gradient ultracentrifugation | iTRAQ labelling, SCX, nano-LC-MS/MS | 39 |
| 5 | lactation stage | 1 d post-partum | MFGM | Colostrum | USA | Holstein | Fresh | Centrifugation, washing | iTRAQ labelling, SCX, nano-LC-MS/MS | 22 |
| 6 | milk characterization | 7 d post-partum | MFGM | Early lactation | USA | Holstein | Fresh | Centrifugation, washing | iTRAQ labelling, SCX, nano-LC-MS/MS | 22 |
| 7 | lactation stage | dry period length | MFGM | Early lactation | not reported | Holstein-Friesian | Snap-frozen (−20 °C) | Centrifugation, washing | FASP, dimethyl labelling, LC-LTQ-Orbitrap/MS | 10 |
| 8 | breed | Holstein | MFGM | Early lactation | China | Holstein | Fresh | Centrifugation, washing | LC-MS/MS | 40 |
| 9 | milk maturation | 48 h post-partum | Skimmed milk | Colostrum | Brazil | Holstein | Fresh | Acidification, centrifugation, major protein depletion | 2D Maldi-TOF/TOF-MS | 42 |
| 10 | milk maturation | 72 h post-partum | Skimmed milk | Colostrum | Brazil | Holstein | Fresh | Acidification, centrifugation, major protein depletion | 2D Maldi-TOF/TOF-MS | 42 |
| 11 | milk characterization | 1 d post-partum | Skimmed milk | Colostrum | Denmark | Holstein-Friesian | Snap-frozen (−80 °C) | Centrifugation | 2D LC-MS/MS | 41 |
| 12 | lactation stage | 1 d post-partum | Skimmed milk | Colostrum | Denmark | Holstein-Friesian | Fresh | Centrifugation | 2D LC-MS/MS | 17 |
| 13 | lactation stage | 10 d post-partum | Skimmed milk | Early lactation | Denmark | Holstein-Friesian | Fresh | Centrifugation | 2D LC-MS/MS | 17 |
| 14 | milk characterization | milk fractionation | Skimmed milk | Early lactation | Denmark | Holstein-Friesian | Fresh | Centrifugation | 2D LC-MS/MS | 13 |
| 15 | diet | low RDP:RUPb ratio | Skimmed milk | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, sonication | HPLC | 45 |
| 16 | diet | high RDP:RUPb ratio | Skimmed milk | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, sonication | HPLC | 45 |
| 17 | type, processing of diet | corn-grain based diet | Skimmed milk | Mid-lactation | China | Holstein | Fresh | Centrifugation | 2D Maldi-TOF/TOF-MS | 50 |
| 18 | mastitis | 3 h post-challenge by Escherichia coli | Skimmed milk | Mid-lactation | Denmark | Holstein-Friesian | Filtered and snap frozen (−20 °C) | Acidification, centrifugation | iTRAQ labelling, SCX, nano-LC-MS/MS | 48 |
| 19 | breed | Holstein-Friesian | Skimmed milk | Mid-lactation | Australia | Holstein-Friesian | Bulk milk and snap frozen (−80 °C) | Centrifugation | Nano LC-ESI-MS/MS | 51 |
| 20 | breed | Jersey | Skimmed milk | Mid-lactation | Australia | Jersey | Bulk milk and snap frozen (−80 °C) | Centrifugation | Nano LC-ESI-MS/MS | 51 |
| 21 | mastitis | healthy | Skimmed milk | Mid-lactation | Spain | Holstein-Friesian | Snap frozen (−80 °C) | Centrifugation | Maldi-MS/MS | 49 |
| 22 | mastitis | healthy | Skimmed milk | Mid-lactation | USA | Holstein | Fresh | Centrifugation | 2D Maldi-TOF/TOF-PSD | 47 |
| 23 | lactation stage | 1 d post-partum | Whey | Colostrum | Denmark | Holstein-Friesian | Snap-frozen (−80 °C) | Centrifugation | 2D LC-MS/MS | 41 |
| 24 | lactation stage | 0 to 5 d post-partum | Whey | Colostrum | China | Holstein | Fresh | Centrifugation | iTRAQ labelling, SCX, LC-MS/MS | 43 |
| 25 | lactation stage | calving day | Whey | Colostrum | Belgium | Holstein-Friesian | Snap frozen (−20 °C) | Ultracentrifugation | FASP, dimethyl labelling, LC-LTQ-Orbitrap/MS | 44 |
| 26 | mammary gland involution | 3 d post drying-off | Whey | Drying-off | New Zealand | Holstein-Friesian | Fresh | Centrifugation | Gel electrophoresis LC-MS/MS | 52 |
| 27 | mammary gland involution | 8 d post drying-off | Whey | Drying-off | New Zealand | Holstein-Friesian | Fresh | Centrifugation | Gel electrophoresis LC-MS/MS | 52 |
| 28 | milk characterization | milk fractionation | Whey | Early lactation | Denmark | Holstein-Friesian | Fresh | Centrifugation | 2D LC-MS/MS | 13 |
| 29 | lactation stage | 9 d post-partum | Whey | Early lactation | Belgium | Holstein-Friesian | Snap frozen (−20 °C) | Ultracentrifugation | FASP, dimethyl labelling, LC-LTQ-Orbitrap/MS | 44 |
| 30 | breed | Holstein | Whey | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, acidification, ultracentrifugation, major proteins depletion | LC-MS/MS | 46 |
| 31 | breed | Jersey | Whey | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, acidification, ultracentrifugation, major proteins depletion | LC-MS/MS | 46 |
| 32 | diet | low RDP:RUPb ratio | Whey | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, acidification, ultracentrifugation, major proteins depletion | LC-MS/MS | 45 |
| 33 | diet | high RDP:RUPb ratio | Whey | Peak lactation | USA | Holstein | Fresh, bronopol and natamycin addition, snap frozen (−80 °C) | Centrifugation, acidification, ultracentrifugation, major proteins depletion | LC-MS/MS | 45 |
| 34 | mammary gland involution | full lactation | Whey | Mid-lactation | New Zealand | Holstein-Friesian | Fresh | Centrifugation | Gel electrophoresis LC-MS/MS | 52 |
| 35 | mastitis | 0 h post-challenge by Streptococcus uberis | Whey | Mid-lactation | United Kingdom | not reported | Frozen | Centrifugation | LC-MS/MS | 53 |
aLC-MS/MS: Liquid chromatography coupled with tandem mass spectrometry
iTRAQ: Isobaric tags for relative and absolute quantitation
SCX column: strong cation exchange column
FASP: Filter-aided sample preparation
LTQ-Orbitrap/MS: linear ion trap quadrupole-Orbitrap-mass spectrometry
Maldi-TOF/TOF: matrix-assisted laser desorption/ionization (Maldi) time-of-flight/time-of-flight (TOF/TOF)
PSD: Post-source decay
bRDP: dietary rumen degradable protein; RUP: rumen undegradable protein.
Methods for protein identification were FASP, dimethyl labelling, LC-LTQ-Orbitrap/MS44, electrophoresis gel LC-MS/MS52, iTRAQ labelling, SCX and LC-MS/MS10,22,39,43,48, Maldi-(TOF/TOF)-MS detection42,47,49,50 and from 1 to 15 repetitions of (nano)LC-MS/MS runs13,15,17,40,41,45,46,51,53. Protein identifiers (ID) reported in the publications were extracted from tables in Portable Document Format (PDF) or from supplementary data files using Tabula software (www.tabula.technology, Last update February 11, 2017). All collected IDs were aggregated in an atlas of proteins using Excel software (2016). Each protein ID was annotated according to i) milk: fraction, processing, and period of lactation; ii) animal phenotype: breed, health status, and production parameters; and iii) protein identification: number of proteins and proteomic methods used. Because publications report different protein nomenclatures, all protein IDs were changed by the corresponding gene names (GN), with the Retrieve/ID Mapping tool of the UniProt database (The UniProt54), the Protein Identifier Cross-Reference service55 and/or the ProteCONVERT tool of the ProteINSIDE web interface56 being used to homogenize data and to generate an atlas of GN.
Categorization of the milk proteome atlas
For the categorization of the database, four milk fractions were defined based on the protein isolation techniques used in laboratories. The milk fractions were i) skimmed milk, with aggregating proteins isolated by centrifugation under 100 000 g combined with or without casein depletion by acidification (14 datasets); ii) whey, with aggregating proteins isolated by centrifugation over 100 000 g (13 datasets); iii) MFGM, with aggregating proteins isolated from cream milk (4 datasets); and iv) exosomes, with aggregating proteins isolated from skimmed milk with a protocol based on a sucrose gradient12 (4 datasets). Five lactation stages were defined according to the DIM, to account for differences in the physiological status and energy balance of the dairy cow. The lactation stages were the i) colostrum period, with aggregating proteins detected in colostrum (samples collected during the first 5 days post-partum; 12 datasets); ii) early lactation (between 6 and 21 DIM, 7 datasets); iii) peak lactation (between 22 and 80 DIM, 6 datasets); iv) mid-lactation corresponding to post-peak lactation (after 81 DIM, but not during the dry-off period, 8 datasets); and v) drying-off (from milk collected at 3 and 8 days after stopping regular milking, 2 datasets). The early lactation stage was defined as 6 to 21 DIM, corresponding to classic physiological NEB6,57–59 and excluding the colostrum period60.
The categorization of the data from the 20 publications using criteria for the milk fractions and lactation stages produced 35 datasets (Table 2) from different cow breeds, experiments and countries. Among the data, the Holstein-Friesian cows were mainly represented. One-half of studies originated from USA, Australia and Denmark. Finally, we discarded those GN identified more than once in each dataset. Based on the 35 datasets, the atlas aggregated 8841 GN corresponding to proteins.
Limitations
The first limitation of the computational approach was the use of 20 of the 87 relevant publications on cows available in mid-2018. Indeed, 67 publications were excluded due to a focus on mastitis, a lack of information on the lactation period of the milk collection, incomplete milk fractionation details, insufficient description of animals and husbandry conditions, or absent protein ID. Moreover, the conversion of protein ID into GN led to some data loss, mainly of protein isoforms. Another limitation is that the data reported in the literature depended on the success of the protein identification; thus, a protein could be absent either because it was not detectable, non-identified or absent in the milk sample. Thus, the 59 proteins exclusively detected in early lactation milk may be present at other lactation stages but at very low concentration, and thus they were not identified by proteomics. Finally, the atlas aggregates the proteins that were present because they were identified in milk without regard for their abundance.
Mining of the milk proteome atlas
From the 8841 GN of the global atlas, we discarded GN identified more than once in each fraction, thereby yielding 7135 unique GN; we did the same for each lactation stage, eventually yielding 6323 GN. The protein lists were compared using Venn diagrams (Draw Venn61 tool, VIB / Ugent) to identify proteins specific of a milk fraction (whatever the lactation stage) and of a lactation stage (whatever the milk fraction).
The resulting lists were mined using the ProteINSIDE webservice56. Briefly, lists of GN were subjected to a ‘custom analysis’ to access two types of results: (1) the biological knowledge retrieval from bovine (Bos taurus) providing mainly the protein’s function, as declared in major databases, tissue-specificity, and subcellular location, and (2) functional annotations according to Gene Ontology (GO) by querying the QuickGO database. The ProteINSIDE tool relies mainly on GO enrichment tests (p value_Benjamini and Hochberg < 0.05) among the Biological Process (BP), Molecular Function (MF) and Cellular Component (CC) categories. GO terms imported were selected by evidence codes and agreed on by the curator review in the ProteINSIDE webservice. GN were annotated by GO for bovine but also for human species to benefit from the most complete GO annotation available for human genes. The top 50 enriched GO terms were considered when the significance was lower than P < 0.05.
The potential biomarkers were selected following a workflow with 4 steps. First, we determined the presence/absence of early milk proteins based on the lactation stage. Because of this binary approach (‘all or nothing’ approach), no statistical analysis was applied. Second, we compared the list of the 59 proteins exclusively detected in milk from early lactation with the lists of proteins previously (based on the literature) reported as expressed in key adaptive tissues of lactating cows. Third, we mined each protein using PubMed.gov (NCBI) and the Web of Science (Clarivate Analytics) search engines up to February 2018. Fourth, we performed data mining on protein databases (ProteINSIDE webservice, UniProtKB database) and on pathway webservices (KEGG PATHWAY database, Reactome Pathway database) to complete the analyses with the literature available up to November 2018.
Supplementary information
Acknowledgements
Authors thank I. Veissier (INRA, UMR1213, Saint-Genès-Champanelle, France) for help drafting this manuscript and M. Reichstadt (INRA, UMR1213, Saint-Genès-Champanelle, France) for technical support of the ProteINSIDE software.
Author Contributions
M.D. and M.B. drafted and wrote the manuscript. M.D., J.P., L.B., I.C.M., M.B. reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-46142-7.
References
- 1.Verma, A. & Ambatipudi, K. Challenges and opportunities of bovine milk analysis by mass spectrometry. Clinical Proteomics13, 10.1186/s12014-016-9110-4 (2016). [DOI] [PMC free article] [PubMed]
- 2.Bislev SL, et al. A Bovine PeptideAtlas of milk and mammary gland proteomes. Proteomics. 2012;12:2895–2899. doi: 10.1002/pmic.201200057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roncada P, et al. Farm animal milk proteomics. Journal of Proteomics. 2012;75:4259–4274. doi: 10.1016/j.jprot.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Zachut M, Moallem U. Consistent magnitude of postpartum body weight loss within cows across lactations and the relation to reproductive performance. J Dairy Sci. 2017;100:3143–3154. doi: 10.3168/jds.2016-11750. [DOI] [PubMed] [Google Scholar]
- 5.Bell AW, Bauman DE. Adaptations of Glucose Metabolism During Pregnancy and Lactation. Journal of Mammary Gland Biology and Neoplasia. 1997;2:265–278. doi: 10.1023/a:1026336505343. [DOI] [PubMed] [Google Scholar]
- 6.Drackley JK. Biology of dairy cows during the transition period: The final frontier? Journal of Dairy Science. 1999;82:2259–2273. doi: 10.3168/jds.S0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 7.Chilliard, Y. In Biology of lactation (eds Martinet, J., Houdebine, L.-M. & Head, H.-H.) Ch. 20, 503–552 (INRA Paris, 1999).
- 8.Roberts T, et al. Metabolic parameters in transition cows as indicators for early-lactation culling risk. Journal of Dairy Science. 2012;95:3057–3063. doi: 10.3168/jds.2011-4937. [DOI] [PubMed] [Google Scholar]
- 9.McArt JAA, Nydam DV, Oetzel GR, Overton TR, Ospina PA. Elevated non-esterified fatty acids and beta-hydroxybutyrate and their association with transition dairy cow performance. Veterinary Journal. 2013;198:560–570. doi: 10.1016/j.tvjl.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, et al. Changes in Milk Proteome and Metabolome Associated with Dry Period Length, Energy Balance, and Lactation Stage in Postparturient Dairy Cows. Journal of Proteome Research. 2013;12:3288–3296. doi: 10.1021/pr4001306. [DOI] [PubMed] [Google Scholar]
- 11.Greenwood SL, Honan MC. Symposium review: Characterization of the bovine milk protein profile using proteomic techniques. Journal of Dairy Science. 2019;102:2796–2806. doi: 10.3168/jds.2018-15266. [DOI] [PubMed] [Google Scholar]
- 12.Reinhardt TA, Lippolis JD, Nonnecke BJ, Sacco RE. Bovine milk exosome proteome. Journal of Proteomics. 2012;75:1486–1492. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Nissen A, Bendixen E, Ingvartsen KL, Rontvedt CM. Expanding the bovine milk proteome through extensive fractionation. Journal of Dairy Science. 2013;96:7854–7866. doi: 10.3168/jds.2013-7106. [DOI] [PubMed] [Google Scholar]
- 14.Zhang, L. N., van Dijk, A. D. J. & Hettinga, K. An interactomics overview of the human and bovine milk proteome over lactation. Proteome Science15, 10.1186/s12953-016-0110-0 (2017). [DOI] [PMC free article] [PubMed]
- 15.Samuel, M. et al. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Scientific Reports7, 10.1038/s41598-017-06288-8 (2017). [DOI] [PMC free article] [PubMed]
- 16.Van Altena SEC, et al. A proteomics-based identification of putative biomarkers for disease in bovine milk. Veterinary Immunology and Immunopathology. 2016;174:11–18. doi: 10.1016/j.vetimm.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Nissen A, Andersen PH, Bendixen E, Ingvartsen KL, Rontved CM. Colostrum and milk protein rankings and ratios of importance to neonatal calf health using a proteomics approach. Journal of Dairy Science. 2017;100:2711–2728. doi: 10.3168/jds.2016-11722. [DOI] [PubMed] [Google Scholar]
- 18.Vaudel M, et al. Exploring the potential of public proteomics data. Proteomics. 2016;16:214–225. doi: 10.1002/pmic.201500295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhla B, et al. Involvement of Skeletal Muscle Protein, Glycogen, and Fat Metabolism in the Adaptation on Early Lactation of Dairy Cows. Journal of Proteome Research. 2011;10:4252–4262. doi: 10.1021/pr200425h. [DOI] [PubMed] [Google Scholar]
- 20.Wankhade PR, et al. Metabolic and immunological changes in transition dairy cows: A review. Veterinary World. 2017;10:1367–1377. doi: 10.14202/vetworld.2017.1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross J, van Dorland HA, Bruckmaier RM, Schwarz FJ. Performance and metabolic profile of dairy cows during a lactational and deliberately induced negative energy balance with subsequent realimentation. Journal of Dairy Science. 2011;94:1820–1830. doi: 10.3168/jds.2010-3707. [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt TA, Lippolis JD. Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. Journal of Dairy Science. 2008;91:2307–2318. doi: 10.3168/jds.2007-0952. [DOI] [PubMed] [Google Scholar]
- 23.Zheng X, et al. Quantitative proteome analysis of bovine mammary gland reveals protein dynamic changes involved in peak and late lactation stages. Biochemical and biophysical research communications. 2017;494:292–297. doi: 10.1016/j.bbrc.2017.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Skibiel AL, Zachut M, do Amaral BC, Levin Y, Dahl GE. Liver proteomic analysis of postpartum Holstein cows exposed to heat stress or cooling conditions during the dry period. J Dairy Sci. 2018;101:705–716. doi: 10.3168/jds.2017-13258. [DOI] [PubMed] [Google Scholar]
- 25.Zachut M, et al. Proteome dataset of subcutaneous adipose tissue obtained from late pregnant dairy cows during summer heat stress and winter seasons. Data Brief. 2017;12:535–539. doi: 10.1016/j.dib.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnology. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 27.Zhen Y, et al. Nuclear import of exogenous FGF1 requires the ER-protein LRRC59 and the importins Kpnalpha1 and Kpnbeta1. Traffic (Copenhagen, Denmark) 2012;13:650–664. doi: 10.1111/j.1600-0854.2012.01341.x. [DOI] [PubMed] [Google Scholar]
- 28.Sinowatz F, Schams D, Habermann F, Berisha B, Vermehren M. Localization of fibroblast growth factor I (acid fibroblast growth factor) and its mRNA in the bovine mammary gland during mammogenesis, lactation and involution. Anatomia, histologia, embryologia. 2006;35:202–207. doi: 10.1111/j.1439-0264.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- 29.Ostrowska M, et al. Expression of PC, PCK1, PCK2, LDHB, FBP1 and G6PC genes in the liver of cows in the transition from pregnancy to lactation. Animal Science Papers and Reports. 2013;31:281–290. [Google Scholar]
- 30.Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419–1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 31.Weikard R, Goldammer T, Brunner RM, Kuehn C. Tissue-specific mRNA expression patterns reveal a coordinated metabolic response associated with genetic selection for milk production in cows. Physiological genomics. 2012;44:728–739. doi: 10.1152/physiolgenomics.00007.2012. [DOI] [PubMed] [Google Scholar]
- 32.Jones DH, Raymer DM, Schoelen SL. The activity of phosphoenolpyruvate carboxykinase throughout the lactation cycle of the guinea-pig mammary-gland. Proceedings of the Society for Experimental Biology and Medicine. 1989;192:16–22. doi: 10.3181/00379727-192-42948. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen NI, Friggens NC, Chagunda MGG, Ingvartsen KL. Predicting risk of ketosis in dairy cows using in-line measurements of beta-hydroxybutyrate: A biological model. Journal of Dairy Science. 2005;88:2441–2453. doi: 10.3168/jds.S0022-0302(05)72922-2. [DOI] [PubMed] [Google Scholar]
- 34.Oetzel GR. Monitoring and testing dairy herds for metabolic disease. Veterinary Clinics of North America-Food Animal Practice. 2004;20:651−+. doi: 10.1016/j.cvfa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Hazekawa I, Nishina Y, Sato K, Shichiri M, Shiga K. Substrate activating mechanism of short-chain acyl-coA, medium-chain acyl-coA, long-chain acyl-coA, and isovaleryl-coA deshydrogenases from bovine liver - a resonance raman-study on the 3-ketoacyl-coA complexes. Journal of Biochemistry. 1995;118:900–910. doi: 10.1093/jb/118.5.900. [DOI] [PubMed] [Google Scholar]
- 36.Farhadian, M., Rafat, S. A., Hasanpur, K., Ebrahimi, M. & Ebrahimie, E. Cross-Species Meta-Analysis of Transcriptomic Data in Combination With Supervised Machine Learning Models Identifies the Common Gene Signature of Lactation Process. Frontiers in Genetics9, 10.3389/fgene.2018.00235 (2018). [DOI] [PMC free article] [PubMed]
- 37.Verbeke J, Van Poucke M, Peelman L, De Vliegher S. Differential expression of CXCR1 and commonly used reference genes in bovine milk somatic cells following experimental intramammary challenge. BMC genetics. 2015;16:40. doi: 10.1186/s12863-015-0197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu B, Jiang Y, Zhao F, Xiao J, Li QZ. Changes of endoplasmic reticulum and mitochondria in mammary epithelial cells during mammogenesis in Chinese Holstein dairy cows. Acta Histochemica. 2012;114:448–453. doi: 10.1016/j.acthis.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Yang M, et al. Comparative proteomic analysis of milk-derived exosomes in human and bovine colostrum and mature milk samples by iTRAQ-coupled LC-MS/MS. Food Research International. 2017;92:17–25. doi: 10.1016/j.foodres.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 40.Murgiano L, et al. Comparison of Milk Fat Globule Membrane (MFGM) Proteins of Chianina and Holstein Cattle Breed Milk Samples Through Proteomics Methods. Nutrients. 2009;1:302–315. doi: 10.3390/nu1020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nissen A, Bendixen E, Ingvartsen KL, Rontved CM. In-depth analysis of low abundant proteins in bovine colostrum using different fractionation techniques. Proteomics. 2012;12:2866–2878. doi: 10.1002/pmic.201200231. [DOI] [PubMed] [Google Scholar]
- 42.Golinelli LP, Conte CA, Paschoalin VMF, Silva JT. Proteomic Analysis of Whey from Bovine Colostrum and Mature Milk. Brazilian Archives of Biology and Technology. 2011;54:761–768. doi: 10.1590/S1516-89132011000400016. [DOI] [Google Scholar]
- 43.Yang M, et al. Comparative proteomic exploration of whey proteins in human and bovine colostrum and mature milk using iTRAQ-coupled LC-MS/MS. Int. J. Food Sci. Nutr. 2017;68:671–681. doi: 10.1080/09637486.2017.1279129. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Lina, Boeren Sjef, Hageman Jos A., van Hooijdonk Toon, Vervoort Jacques, Hettinga Kasper. Bovine Milk Proteome in the First 9 Days: Protein Interactions in Maturation of the Immune and Digestive System of the Newborn. PLOS ONE. 2015;10(2):e0116710. doi: 10.1371/journal.pone.0116710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tacoma R, Fields J, Ebenstein DB, Lam YW, Greenwood SL. Ratio of dietary rumen degradable protein to rumen undegradable protein affects nitrogen partitioning but does not affect the bovine milk proteome produced by mid-lactation Holstein dairy cows. Journal of Dairy Science. 2017;100:7246–7261. doi: 10.3168/jds.2017-12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tacoma R, Fields J, Ebenstein DB, Lam Y-W, Greenwood SL. Characterization of the bovine milk proteome in early-lactation Holstein and Jersey breeds of dairy cows. Journal of Proteomics. 2016;130:200–210. doi: 10.1016/j.jprot.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boehmer JL, Bannerman DD, Shefcheck K, Ward JL. Proteomic Analysis of Differentially Expressed Proteins in Bovine Milk During Experimentally Induced Escherichia coli Mastitis. Journal of Dairy Science. 2008;91:4206–4218. doi: 10.3168/jds.2008-1297. [DOI] [PubMed] [Google Scholar]
- 48.Danielsen M, et al. Quantitative milk proteomics - Host responses to lipopolysaccharide-mediated inflammation of bovine mammary gland. Proteomics. 2010;10:2240–2249. doi: 10.1002/pmic.200900771. [DOI] [PubMed] [Google Scholar]
- 49.Alonso-Fauste I, et al. Proteomic characterization by 2-DE in bovine serum and whey from healthy and mastitis affected farm animals. Journal of Proteomics. 2012;75:3015–3030. doi: 10.1016/j.jprot.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 50.Li SS, Shen JS, Ren DX, Liu JX. Effects of the processing methods of corn grain and soybean meal on milk protein expression profiles in dairy cows. Animal. 2015;9:267–274. doi: 10.1017/s1751731114002225. [DOI] [PubMed] [Google Scholar]
- 51.Vincent, D. et al. Milk Bottom-Up Proteomics: Method Optimization. Frontiers in Genetics6, 10.3389/fphys.2015.00360 (2016). [DOI] [PMC free article] [PubMed]
- 52.Boggs I, et al. Proteomics data in support of the quantification of the changes of bovine milk proteins during mammary gland involution. Data in brief. 2016;8:52–55. doi: 10.1016/j.dib.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mudaliar M, et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Molecular Biosystems. 2016;12:2748–2761. doi: 10.1039/c6mb00290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium, T. U. UniProt: the universal protein knowledgebase, http://www.uniprot.org/ (2017).
- 55.Wein SP, et al. Improvements in the protein identifier cross-reference service. Nucleic Acids Research. 2012;40:W276–W280. doi: 10.1093/nar/gks338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaspric Nicolas, Reichstadt Matthieu, Picard Brigitte, Tournayre Jérémy, Bonnet Muriel. Protein Function Easily Investigated by Genomics Data Mining Using the ProteINSIDE Online Tool. Genomics and Computational Biology. 2015;1(1):16. doi: 10.18547/gcb.2015.vol1.iss1.e16. [DOI] [Google Scholar]
- 57.Pires JAA, Delavaud C, Faulconnier Y, Pomiès D, Chilliard Y. Effects of body condition score at calving on indicators of fat and protein mobilization of periparturient Holstein-Friesian cows. Journal of Dairy Science. 2013;96:6423–6439. doi: 10.3168/jds.2013-6801. [DOI] [PubMed] [Google Scholar]
- 58.Weber C, et al. Variation in fat mobilization during early lactation differently affects feed intake, body condition, and lipid and glucose metabolism in high-yielding dairy cows. Journal of Dairy Science. 2013;96:165–180. doi: 10.3168/jds.2012-5574. [DOI] [PubMed] [Google Scholar]
- 59.Chilliard Y. Dietary-fat and adipose-tissus metabolism in ruminants, pigs, and rodents - A review. Journal of Dairy Science. 1993;76:3897–3931. doi: 10.3168/jds.S0022-0302(93)77730-9. [DOI] [PubMed] [Google Scholar]
- 60.Eichinger, M. Etude de la qualité (immunologique et bactériologique) de colostrums de vaches laitières de la communauté de communes Chamousset en Lyonnais dans le cadre d’une valorisation du colostrum bovin., Université Claude-Bernard, Lyon 1 (2014).
- 61.Diagram, D. V. VIB/UGent, http://bioinformatics.psb.ugent.be/webtools/Venn/ (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


