Abstract
Metabolomics provides valuable tools for the study of drug effects, unraveling the mechanism of action and variation in response due to treatment. In this study we used electrochemistry-based targeted metabolomics to gain insights into the mechanisms of action of escitalopram/citalopram focusing on a set of 31 metabolites from neurotransmitter-related pathways. Overall, 290 unipolar patients with major depressive disorder were profiled at baseline, after 4 and 8 weeks of drug treatment. The 17-item Hamilton Depression Rating Scale (HRSD17) scores gauged depressive symptom severity. More significant metabolic changes were found after 8 weeks than 4 weeks post baseline. Within the tryptophan pathway, we noted significant reductions in serotonin (5HT) and increases in indoles that are known to be influenced by human gut microbial cometabolism. 5HT, 5-hydroxyindoleacetate (5HIAA), and the ratio of 5HIAA/5HT showed significant correlations to temporal changes in HRSD17 scores. In the tyrosine pathway, changes were observed in the end products of the catecholamines, 3-methoxy-4-hydroxyphenylethyleneglycol and vinylmandelic acid. Furthermore, two phenolic acids, 4-hydroxyphenylacetic acid and 4-hydroxybenzoic acid, produced through noncanconical pathways, were increased with drug exposure. In the purine pathway, significant reductions in hypoxanthine and xanthine levels were observed. Examination of metabolite interactions through differential partial correlation networks revealed changes in guanosine–homogentisic acid and methionine–tyrosine interactions associated with HRSD17. Genetic association studies using the ratios of these interacting pairs of metabolites highlighted two genetic loci harboring genes previously linked to depression, neurotransmission, or neurodegeneration. Overall, exposure to escitalopram/citalopram results in shifts in metabolism through noncanonical pathways, which suggest possible roles for the gut microbiome, oxidative stress, and inflammation-related mechanisms.
Subject terms: Depression, Scientific community
Introduction
Major depressive disorder (MDD) is a common, often disabling condition affecting over 300 million individuals worldwide1. Selective serotonin reuptake inhibitors (SSRIs) are common first-line treatments for MDD2,3. They are believed to increase the extracellular availability of the neurotransmitter serotonin by limiting its reabsorption into the presynaptic cell, so that serotonin levels are increased in the synaptic cleft and available for binding to postsynaptic receptors. Responses to antidepressant medications are modest. Only about half the patients respond to the first medication; only one in three achieves symptom remission, which is the virtual absence of symptoms and the aim of treatment4. Some patients do well on a single medication, while others require medication combinations or alternative interventions. Clinical symptoms are insufficient to guide appropriate treatment selection5 and, presently, treatments are therefore selected empirically relying on a “trial and error” approach6,7.
Metabolomics, a promising new approach to understanding depression and other neuropsychiatric disorders8–11, could help inform treatment selection12,13. Metabolomic profiles provide informative readouts on pathways and biological networks implicated in various diseases or their treatments. Metabolomic signatures have been identified for several psychiatric disorders, such as MDD14, bipolar disorder15,16, and schizophrenia17–19. Most studies of mood disorders have implicated tryptophan (TRP), tyrosine, and purine metabolism, since historically, neurotransmission and serotonergic signaling were key focus areas of investigation14. The TRP pathway along with its three branches of metabolism to serotonin/melatonin/5-hydroxyindoleacetate, kynurenine (KYN), and indole derivatives, seems to be affected in the depressed state20–28. The purine pathway, whose regulation seems to be connected to TRP metabolism, has also been implicated in depression and other psychiatric disorders29. Among patients in remission from a major depressive episode, a metabolomic signature that included methionine, glutathione along with metabolites in the purine and TRP pathways, has been identified30.
Pharmacometabolomics has also revealed that patients’ metabolomic profiles (metabotypes), both prior to and early during treatment, can inform treatment outcomes10,31. This approach has been applied to antihypertensive32 and antiplatelet33 therapies. We have used this approach to predict treatment outcomes and to identify specific metabolomic pathways that were changed in response to sertraline34,35 and to ketamine36, a promising agent for treatment-resistant depression. We have also employed a “pharmacometabolomics-informed pharmacogenomics” research strategy11 to investigate the role of genetics in response to citalopram or escitalopram37,38, thereby advancing the goal of precision medicine for depression31. However, the acute and longer-term effects of treatment with citalopram or escitalopram on pathways, critical to the pathobiology or pharmacotherapy of depression, and the relationship to clinical outcomes have not been reported.
This report used metabolomic analyses with selected metabolites in the tryptophan, tyrosine, purine, tocopherol, and the related pathways in a sample of nonpsychotic depressed outpatients who were treated for 8 weeks with citalopram or escitalopram to address the following questions:
The metabolomic signature of exposure to escitalopram/citalopram: which metabolite changes occurred from baseline to week 4, and from baseline to week 8 of treatment?
The metabolomic signature of response: which metabolomic changes were related to changes in depressive symptoms (HRSD-17), longitudinally, in the overall population and also in responders versus nonresponders?
The interrelationships between metabolites: what are the relationships among metabolites, both within and between pathways, before and after treatment with the drug?
Methods
Study design and participants
We used samples from the Mayo Clinic NIH-Pharmacogenomics Research Network-Antidepressant Pharmacogenomics Medication Study (PGRN-AMPS) which recruited a total of 803 MDD patients39. Patient selection, symptomatic evaluation, and blood sample collection for the PGRN-AMPS clinical trial have been described elsewhere24,38–40. Briefly, MDD patients were required to have a baseline HRSD17 score ≥ 14, and all patients who completed 8 weeks of treatment (n = 290) were treated with one of the two SSRIs, citalopram or escitalopram. Depressive symptoms were assessed with HRSD17 at baseline, week 4, and week 8 of SSRI treatment. Blood samples were collected at these same time points.
The HRSD17 was used to ascribe “response”—defined as at least 50% reduction in the total score from baseline to exit; “remission” —an exit HRSD17 score of 7 or less; and “complete-non-response”—less than 30% reduction in the HRSD17 total score from baseline to exit39. Genome-wide association studies for plasma concentrations of the SSRIs and metabolite levels40 and for response41 in this trial have been published previously. The trial was designed as a parallel to the large National Institute of Mental Health—funded “the Sequenced Treatment Alternatives to Relieve Depression” (STAR*D) clinical trial42 for the purpose of replication of the identified genetic markers.
Metabolomic profiling
A targeted, liquid chromatography–electrochemical coulometric array (LCECA) metabolomics platform43 was used to assay metabolites in plasma samples from the three time points, baseline, 4 weeks, and 8 weeks. This platform was used to identify and quantify 31 neurotransmitter-related metabolites (against standards) primarily from the TRP, tyrosine, and tocopherol pathways, including serotonin. A list of the metabolites that were quantitatively measured using this platform is presented in Table 1.
Table 1.
List of metabolites and pathways analyzed in the study
| Metabolite by pathways | Abbreviation | Metabolite by pathways | Abbreviation |
|---|---|---|---|
| Tryptophan | Phenylalanine/tyrosine | ||
| 3-Hydroxykynurenine | 3OHKY | 4-hydroxybenzoic acid | 4HBAC |
| 5-Hydroxyindoleacetic acid | 5HIAA | ||
| 5-Hydroxytryptophan | 5HTP | Purine | |
| Indole-3-acetic acid | I3AA | Guanine | G |
| Kynurenine | KYN | Guanosine | GR |
| Serotonin | 5HT | Hypoxanthine | HX |
| Tryptophan | TRP | Uric acid | URIC |
| Xanthine | XAN | ||
| Tyrosine | Paraxanthine | PXAN | |
| 4-Hydroxyphenylacetic acid | 4HPAC | Xanthosine | XANTH |
| 4-Hydroxyphenyllacetic acid | 4HPLA | ||
| Homogentisic acid | HGA | 1 Carbon + GSH | |
| Homovanillic acid | HVA | Methionine | MET |
| Methoxy-hydroxyphenyl glycol | MHPG | Cysteine | CYS |
| Tyrosine | TYR | ||
| Vanillylmandelic acid | VMA | Other | |
| Salicylate | SA | ||
| Tocopherol | Alpha-Methyltryptophan | AMTRP | |
| Tocopherol-alpha | ATOCO | Indole-3-propionic acid | I3PA |
| Tocopherol-delta | DTOCO | Theophylline | Theophylline |
| Tocopherol-gamma | GTOCO | ||
Analysis method
The long-gradient LCECA method used for this analysis can resolve compounds at picogram levels through electrochemical detection (resulting from oxidation or reduction reactions) including multiple markers of oxidative stress and protection. This method utilizes a 120-min gradient from (0%) organic modifier with an ion-pairing agent (i.e., pentane sulfonic acid) to a highly organic mobile phase with methanol (80%)/isopropanol (10%)/acetonitrile (10%). An array of 16 serial coulometric electrochemical detectors is set at incremental potentials from 0 to 900 mV, responding to oxidizable compounds such as tocopherol in lower potential sensors and higher oxidation potential compounds such as hypoxanthine in the higher potential channels.
Analysis sequence and data output
At the time of preparation, a pool was created from small aliquots of each sample in the study, which was then treated identically to a sample. All of these assays were executed in sequences that included mixed standard, five samples, pool, five samples, mixed standard, and so on and so forth. In this study, all sample run orders were randomized. The sequences decreased possible analytical artifacts during further data processing. Data were time normalized to a pool at the midpoint of the study, aligning major peaks to 0.5 s and minor peaks to 0.5–2 s. Details on the LCECA methods are described in previously published work35,41,44–50.
Data analysis
All data preprocessing and analysis were performed with R (version 3.4.2) and Bioconductor (version 3.3) statistical packages.
Preprocessing
This study’s data extraction protocol followed the STORBE guidelines46. All metabolite data were first checked for missing values (none were detected at >20% missing abundances) and were subjected to imputation by the k-nearest neighbor algorithm51. Data were then log2 transformed and scaled to unit variance prior to statistical analyses.
Univariate analysis
To define the effect of drug exposure over 4 weeks and 8 weeks of treatment, linear mixed effects models (using the R package nlme52) were fitted on each metabolite adjusting for age, gender, and HRSD17 scores at baseline with subjects as random variable. Analyses were conducted separately for 4 and 8 weeks. Linear mixed effects models were also used to determine associations between the changes in metabolites and changes in HRSD17 over time, with age and gender as covariates, and using subjects as random variable. All p-values were used to calculate the false discovery rates by Benjamini–Hochberg method53, and a cutoff point of 10% was used. A two-step regression strategy was used to find metabolites with significant temporal changes and significant differences between responders and nonresponders using the maSigPro library in R54. First, a least-squared technique was employed to identify differential metabolites in a global regression model, using dummy variables for experimental groups. Second, stepwise regression was applied to select variables that differed between the experimental groups and find significantly different metabolite profiles between the groups.
Partial correlation networks with cluster subgraph analysis
The relationship between metabolites in a complex disease setting can be represented in terms of partial correlation networks, where each node represents a metabolite and each edge between two metabolites represents that two variables are not independent after conditioning on all variables in the dataset. These edges have a weight, edge weights, which are the partial correlation coefficients. Here, we estimated the partial correlation matrix for all of the metabolites using the least absolute shrinkage and selection parameter (LASSO) to obtain the sparse inverse covariance matrix to avoid overfitting and spurious correlations. Thus, it can be reasonably expected that the regularized partial correlation networks will provide accurate estimates of the underlying relationships between the metabolites in metabolic pathways and reactions. The LASSO regularization parameter was set via EBIC or Extended Bayesian Information criterion50. Finally, the walktrap algorithm, which is based on random walks to capture cluster structures in a network, is used to identify clusters of strongly interacting metabolites45. The final network with cluster subgraphs is formed by the median pairwise partial correlations over 1000 bootstrap estimations and plotted using the Fruchterman–Reingold layout. We further included the HRSD17 scores in our partial correlation network models to perform differential network analysis. The overall statistical impact of HRSD17 scores on the metabolite interactions was calculated based on measuring structure invariance between two networks, high HRSD17 and low HRSD17 networks, constructed using a median split of the variable. Permutation tests were used to determine the significance of structure and edge invariances between the two networks55. The metabolite–metabolite partial correlations that were of differential strength between networks of high and low HRSD17 networks were further validated for significant interaction effects through linear regression analysis.
Candidate metabolic trait GWAS with HGA/GR and MET/TYR ratios
For 288 of the 290 subjects in this study we had genotype data for the Illumina human 610-Quad BeadChips (Illumina, San Diego, CA, USA) available, as described previously38,41. Genotype QC using PLINK and imputation followed standard protocols. Briefly, raw genotype data were filtered for variants with call rate <5%, minor allele frequency (MAF) <5%, and Hardy–Weinberg equilibrium HWE p < 1 × 10−549. The data was then subjected to prephasing using SHAPEIT2 (ver. 2.12)48, followed by imputation with IMPUTE2 (ver. 2.3.2)47 using 1000 genomes phase 3 version 556 haplotypes as a reference. Post-imputation QC included filtering variants for IMPUTE info score < 0.5, call rate and MAF < 5%, and HWE p < 1 × 10−5, resulting in a final set of 5.55 mio SNPs with 99.14% genotyping rate. To remove any potential for spurious associations due to population stratification, we used a set of about 100,000 SNPs pruned for the LD structure and retrieved the first five principal component eigenvectors (PCs). Metabolite data for the HGA/GR and Met/TYR ratios were log transformed, centered to zero mean, and scaled to unit variance. In addition, for candidate GWAS, we excluded values that were more than 4 standard deviations from the mean. We then performed GWAS for HGA/GR and MET/TYR at each time point while adjusting for age, sex, and PCs 1–5. We reran the GWAS additionally adjusting for HDRS17 scores at each time point to eliminate the effects linked to depression severity.
Results
Patient characteristics
Plasma metabolite data were available from 290 MDD patients. The average age of the patient cohort was 39.8 (±13.1) years. Females comprised of 66% of the study cohort, while males were at 34%. The response rate to the drug, based on HRSD17 scores, was 69.3% after 8 weeks, compared with 30.7% who were classified as nonresponders for this study. The depressive status of the patients, as determined by the HRSD17 scores, decreased over time with the drug treatment, from an average of 21.9 (±4.9) at baseline to 11.6 (±6.4) at week 4 and 8.6 (±5.5) at week 8. Demographic and clinical characteristics are detailed in Supplemental Table 1.
Metabolite changes at weeks 4 and 8 compared with baseline, in response to the drug
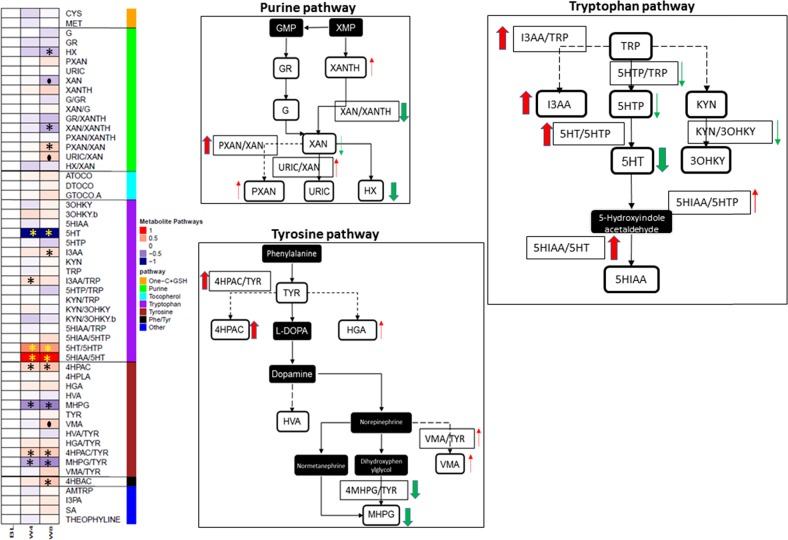
Several metabolites in the purine, tryptophan, and tyrosine pathways changed, following 4 weeks of drug therapy. However, perturbations in the metabolite levels were in general, greater and more significant after 8 weeks of treatment (Supplemental Table 2). Figure 1 illustrates changes within key pathways evaluated after 8 weeks of treatment.
Fig. 1. Metabolic signature of drug exposure.
a Shows the heatmap of metabolite changes at baseline, week 4, and week 8, normalized to baseline levels. b–d Show changes within the purine, tryptophan, and tyrosine pathways
Tryptophan pathway
Dramatic changes were observed in serotonin (5HT) and the ratio 5HIAA/5HT, both at week 4 and week 8. At both time points, 5HT showed substantial decreases and the 5HIAA/5HT ratio was significantly elevated. While TRP itself did not show a notable change, its indole-containing metabolite I3AA was significantly elevated, as was the ratio of I3AA/TRP, possibly indicating a shift away from the serotonergic pathway of TRP metabolism. Interestingly, another indole-containing compound that is known to be produced only by gut microbiota in humans, I3PA, was also increased at 8 weeks (unadjusted p-value < 0.02). No statistically significant alterations were observed in the KYN branch of TRP metabolism.
Tyrosine pathway
A similar trend of a shift to noncanonical branches of tyrosine metabolism was also observed in this pathway. MHPG, the major metabolite of the neurotransmitter norepinephrine and the ratio MHPG/TYR showed significant reductions in their blood levels at both 4 and 8 weeks while VMA, a norepinephrine end metabolite, showed significant elevations at 8 weeks compared with baseline. A phenolic acid, 4HPAC, and its ratio to TYR (4HPAC/TYR) were significantly increased at both 4 and 8 weeks. Another phenolic derivative from the phenylalanine/tyrosine pathway, 4-hydroxybenzoic acid (4HBAC), was also significantly elevated at 8 weeks.
Purine pathway
The purine metabolites HX and XAN and the ratio XAN/XANTH were decreased significantly, while the ratios PXAN/XAN and URIC/XAN were elevated at 8 weeks compared with baseline, indicating a similar decline in the canonical pathway of purine metabolism, as observed in the tryptophan and tyrosine pathways.
Other metabolites that showed significant changes, albeit at unadjusted p values <0.05, were the purine metabolites, G, PXAN, and XANTH; the TRP metabolite, 5HTP; the tyrosine metabolite, HGA; and other metabolites, such as salicylic acid (SA).
Metabolomic changes associated with changes in depressive symptoms (HRSD17)
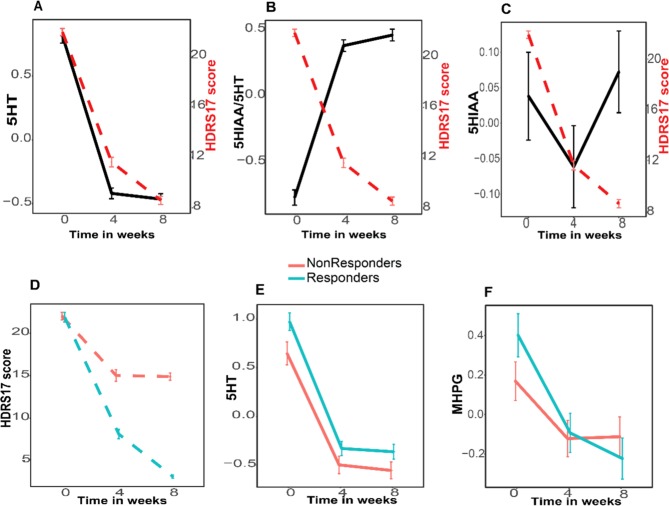
Using linear mixed models, we examined the association between temporal changes in metabolite levels (across three time points, baseline, 4 weeks, and 8 weeks) and the temporal changes in patients’ HRSD17 scores over that period of time (see Fig. 2a–c and Supplemental Table 3). In the overall population, metabolites from the TRP pathway were associated with changes in HRSD17 scores. 5HT, 5HIAA and the serotonin turnover marker 5HIAA/5HT showed significant positive and negative associations, respectively, with decreases over time in HRSD17 scores (FDR-adjusted p values <0.01).
Fig. 2. Metabolite changes associated with HRSD17 scores.
a 5HT (Serotonin), b 5HIAA/5HT ratio, and c 5HIAA (5-hydroxyindoleacetic acid). Temporal changes in (d) HRSD17 scores, (e) 5HT, and (f) MHPG differed significantly between responders and nonresponders. The error bars represent standard error of the mean
We further subcategorized the population based on their HRSD17 scores after 8 weeks of treatment. If they had at least a 50% reduction in their HRSD17 scores, from baseline to exit, they were categorized as responders, otherwise they were nonresponders. We examined whether the temporal associations between metabolite changes and HRSD17 scores significantly differed between responders and nonresponders. The mean (±sd) HRSD17 scores in the responders and nonresponders were 21.86 (±5.17) and 22.03 (±4.28), respectively, at baseline, 10.10 (±5.77) and 15.03 (±6.58), respectively, at week 4, and 5.79 (±3.27) and 14.90 (±4.15), respectively, at week 8. 5HT temporal profiles significantly differed between the two groups, with the levels being consistently higher in the responders at baseline, week 4, and week 8, while the decline in HRSD17 scores was significantly lower at both 4 and 8 weeks compared with baseline (Fig. 2d, e). Levels of MHPG at baseline were significantly higher and the drop in MHPG levels over time was significantly greater in responders compared with nonresponders (Fig. 2f).
Relationships amongst metabolites at baseline and after 8 weeks of treatment
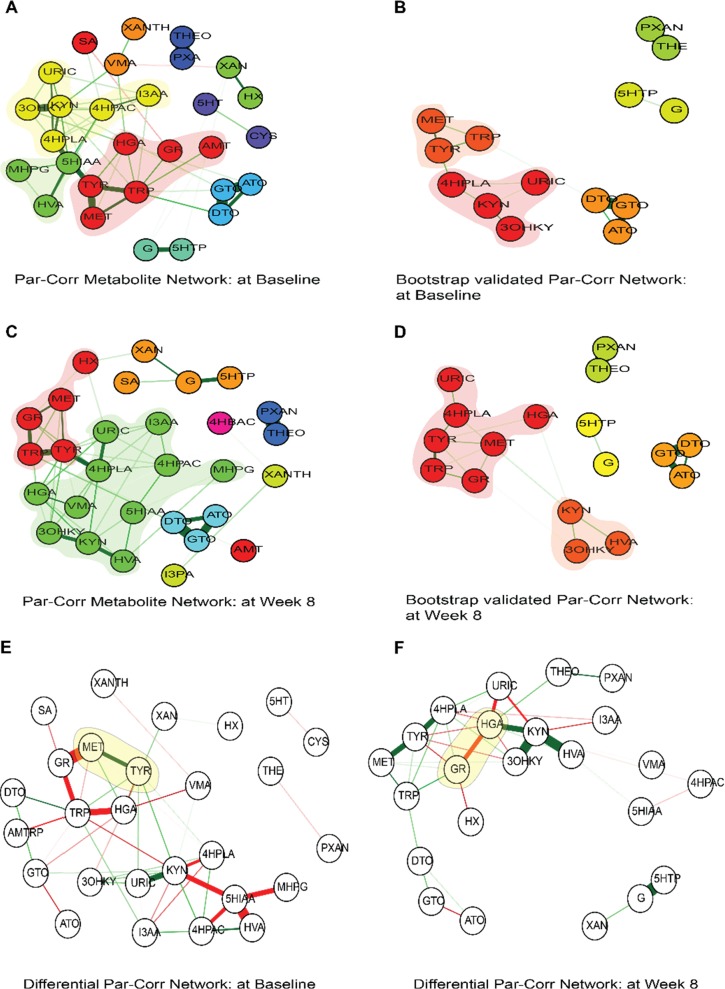
Biological systems are now increasingly viewed as complex networks of interlinked entities, topological analyses of which can reveal the underlying landscape of biological functionalities. Gaussian graphical modeling has been used to reconstruct pathway reactions in metabolomics data57. Combining a partial correlation network and genetic variation through GWAS has been shown to provide an in-depth overview of the underlying mechanistic pathways58. Here, using regularized partial correlation network analysis at baseline and also after week 8 of drug exposure (Fig. 3), we assessed the metabolite–metabolite interactions between tryptophan, tyrosine, purine, and tocopherol pathways.
Fig. 3. Partial correlation networks (PCN).
a PCN at baseline; b PCN at baseline after 1000 bootstrap estimations; c PCN at week 8, and d PCN at week 8 after 1000 bootstrap estimations. The different clusters representing communities of closely associated metabolites are shown in different colors. Differential PCN as a function of high versus low HRSD17 week 8 scores at (e) baseline and (f) week 8. The edges between metabolites most impacted by higher HRSD17 week 8 scores are bolded in green, while those by lower HRSD17 week 8 scores are bolded in red
Regularized partial correlation networks of the metabolites at baseline (Fig. 3a, b) and also at week 8 (Fig. 3c, d) showed significant correlations between several metabolites, both within and between pathways forming clusters of interacting molecules. A list of statistically significant partial correlations between metabolites at baseline and week 8 are presented in Supplemental Table 4 A, B. Important observations through cluster subgraph analysis showed that MET, TYR, and TRP formed a tight cluster both at baseline and week 8. However, the strength of interactions between MET and TYR was significantly reduced at week 8, compared with baseline (~50% reduction, permutation p value < 0.10). GR connection to this cluster was significant at week 8 through interactions with all three metabolites. HVA formed a significant correlation with KYN at week 8 that was not observed at baseline. Multiple other overlapping correlations in the two networks were observed at both baseline and week 8, suggesting that the majority of these interactions were a result of housekeeping biological interactions and were probably not entirely related to the drug effect.
Differential partial correlation networks associated with HRSD17 scores at week 8
HRSD17 scores at week 8 indicated the depression status of the patients post drug treatment. We compared two partial correlation networks constructed with lower and higher values of HRSD17 scores at week 8 (the outcome status), using a median split, as a node. Our aim was to examine if the associations between metabolites were different between patients who responded to the drug better than those who responded poorly. Several metabolite–metabolite associations across the tyrosine, tryptophan, and purine pathways were found to be changed as a function of higher or lower outcome status (Supplemental Table 5 A, B). At baseline, GR–MET, TYR–MET, and KYN–URIC partial correlations were most impacted, while at week 8, KYN–HVA, KYN–3OHKY, 5HTP-G, and HGA–GR values were most impacted by HRSD17 week 8 status (Fig. 3e, f). Two sets of metabolite–metabolite interactions associated with the outcome status, HGA–GR interactions at week 8 and MET–TYR interactions at baseline, were further found to be statistically significant in linear regression models (highlighted in yellow in Fig. 3, e, f). The interaction plots based on linear regression models are presented in Supplemental Figs. 1 and 2. An interesting observation from the differential analysis of networks at baseline was that the partial correlations between metabolites that were differential between the low versus high HRSD17 networks involved several gut-microbe-related metabolites such as HGA, I3AA, 5HIAA, 4HPLA, and 4HPAC amongst others (Fig. 3e).
Genetic influences on ratios of interacting metabolite pairs change during SSRI treatment
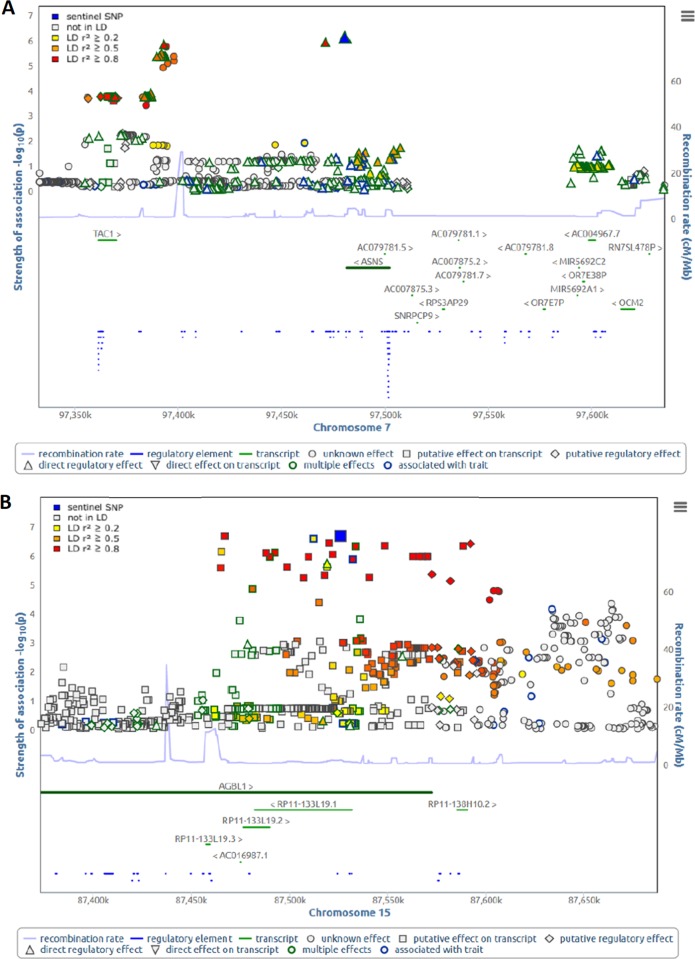
To identify potential modulators of significant metabolite–metabolite interactions and their differential interactions over time, we performed genome-wide association studies with the pairwise ratios of HGA/GR and MET/TYR in 288 subjects at each time point. To this end, we computed additive genetic associations of the two ratios with 5.55 mio autosomal SNPs at each time point, while adjusting for age, sex, time point-specific HRSD17 score, the first five PCs to account for population stratification. The strongest signal for the HGA/GR ratio was for rs55933921 on chromosome 7 (baseline: P = 8.59 × 10−7; week 4: P = 3.05 × 10−3; week 8: P = 1.14 × 10−3) in a locus spanning two genes, TAC1 (protachykinin-1) and ASNS (asparagine synthetase [glutamine-hydrolyzing]). The strongest signal for the MET/TYR ratio was for rs2701431 on chromosome 15 (baseline: P = 5.57 × 10−3; week 4: P = 2.00 × 10−4; week 8: P = 8.48 × 10−8) in the AGBL1 (ATP/GTP-binding protein like 1) locus (Fig. 4). Of note, genetic associations between these loci and metabolite ratios were the strongest at the time point that showed insignificant metabolite–metabolite interactions on the HRSD17 score.
Fig. 4.
Plots showing regional association plots generated with SNiPA61 for: a the HGA/GR ratio at baseline and b the MET/TYR ratio at week 8
Discussion
We have applied a “targeted” electrochemistry-based metabolomics platform to quantitate the metabolomic profiles in MDD patients before and after SSRI treatment. Specifically, we assayed 31 neurotransmission-related metabolites that might have some relevance to MDD pathophysiology, based on published literature, including compounds from the tryptophan, tyrosine, and purine pathways, in plasma samples from 290 MDD patients at baseline, 4-, and 8 weeks of escitalopram/citalopram treatment. Metabolomic profiles were correlated with treatment response, defined as 50% reduction from baseline HRSD17 scores. We found that plasma 5HT concentration was the most significantly decreased metabolite among all of the 31 metabolites after the drug treatment. Higher baseline 5HT levels were associated with better response to SSRI treatment. The 5HT levels in responders remained higher than that those in nonresponders all through the treatment period. Compared with the baseline metabolic state, significant shifts of metabolomic profiles to noncanonical branches of the three major pathways after drug exposure were noted, such as increase in the production of indoles in TRP metabolism, phenolic acids in phenylalanine/tyrosine metabolism, and the PXAN/XAN ratio in the purine pathway. In addition, changes in the catecholamine branch of tyrosine metabolism like increase in VMA and decrease in MHPG, and changes in the end products of purine pathway, such as decreases in XAN and HX, were identified in the MDD patients after drug exposure. Patients who had high MHPG at baseline responded better to SSRI treatment and their MHPG levels decreased more significantly than nonresponders. Temporal change in serotonin and 5HIAA significantly correlated with changes in HRSD17 scores over time. Partial correlation analysis between metabolites revealed that MET, TYR, and TRP formed a tight cluster of interacting molecules in these MDD patients. However, the strength of interactions (partial correlations) varied significantly pre- and post treatment. GR association with this cluster was significant at week 8. GWAS for the HGA/GR ratio identified two genetic loci that mapped to the TAC1 and ASNS genes, which are known to be involved in depression and neurotransmission, while the ratio MET/TYR identified the gene AGBL1 previously linked to neurodegeneration in mice.
Overall, significant perturbations within and between the tryptophan, tyrosine, and purine pathways due to the drug exposure, were noted. These findings are consistent with our previous metabolomic study of sertraline, another SSRI, in depressed patients35, where perturbations in TRP, in particular, changes in methoxyindole pathway and the ratio of KYN/TRP were correlated with treatment outcomes. Interestingly, plasma concentrations of the indoles synthesized from TRP, I3AA, and I3PA, were found to be significantly increased in the MDD patients in this study after the SSRI treatment. Plasma concentrations for I3AA and I3PA are known to be influenced by gut microbiota. Both I3AA and I3PA are aryl hydrocarbon receptor (AHR) agonists59, which could activate AHR transcriptional activity and modulate inflammation in the gut60–63 and brain64,65.
Indole-3-propionic acid is a potent hydroxyl radical scavenger produced exclusively by the commensal gut bacteria Clostridium sporogenes66 and normally found in the plasma and cerebrospinal fluid. I3AA, on the other hand, has been found to correlate significantly with both anxiety and depressive symptoms in chronic kidney disease patients (CKD)67. I3AA can be produced from indole by gut microflora68 in the intestines, or metabolized in tissues from tryptamine69 and other TRP derivatives. At uremic concentrations, I3AA has been linked to oxidative stress via AHR in CKD patients70. However, neither I3AA nor I3PA changes were correlated to changes in HRSD17 scores in our findings.
The most notable change in metabolic profiles after SSRI exposure occurred in the TRP metabolite, 5HT concentrations, which was expected from the mechanism of action of SSRI38. Plasma 5HT originates from the enterochromaffin cells in the gut and gets actively absorbed and stored by blood platelets, which highly express the 5HT transporter SLC6A4. SSRIs target SLC6A4 and inhibit 5HT uptake by platelets in blood. Therefore, a dramatic decrease in plasma 5HT concentration after the SSRI treatment can be expected in these patients. A higher concentration of plasma 5HT, which was stored in platelets, may reflect an elevated activity of the 5HT transporter, and, as a result, greater sensitivity to SSRIs in those patients. This hypothetical situation might explain why patients with higher plasma 5HT concentrations responded better to SSRIs.
Altered metabolic activity of the purine cycle has been linked with several MDD-related systemic responses, such as increased proinflammatory and oxidative processes35. The end products of purine metabolism, uric acid, a potent antioxidant, have been reported to be found in decreased levels in MDD71–73, while lower cerebro-spinal fluid (CSF) levels of hypoxanthine and xanthine, the two metabolites preceding uric acid, have previously been linked with depression74. Ali-Sisto et al., on the contrary, reported increased levels of xanthine to be associated with MDD75. In our study, we observed higher baseline levels of xanthine and hypoxanthine that decreased with the drug treatment. We did not detect increases in uric acid, but we did observe significant increases in the ratios of paraxanthine/xanthine and xanthosine/xanthine due to the drug exposure. This may indicate a potential beneficial effect of the drug through reducing oxidative stress by direct or indirect inhibition of the xanthine oxidase enzyme system. XO is known to generate vascular oxidative stress through reactive oxygen species production by catalyzing the hypoxanthine → xanthine → urate synthesis76. On the other hand, we observed increased associations between uric acid with 4HPLA and also with HGA at week 8 compared with baseline. Uric acid is known to function as an antioxidant (primarily in plasma) and pro-oxidant (primarily within the cell)77. Paraxanthine showed strong correlations to theophylline at all time points. It may also be possible that we were not able to detect significant increases in the levels of the antioxidant, uric acid, and the known psychostimulant paraxanthine78 due to their increased turnover rates brought about by the drug treatment.
In the tyrosine/phenylalanine pathway, 4HPAC and 4HBAC, the phenolic acid metabolites, were found to be significantly increased in MDD patients after SSRI treatment. Although these can potentially come from diets rich in plant-based foods, evidence suggests that these compounds can be produced through microbial fermentation of aromatic amino acids (AAAs) in the colon79. Although changes in the concentration of those metabolites were not associated with SSRI response in MDD patients, those changes possibly indicate alterations in gut microbiome or gut metabolism after citalopram/escitalopram treatment. 4HBAC is known for its antioxidant properties, as effective scavengers of free radicals and reactive nitrogen species, such as peroxynitrite80. 4HPAC is also known for its antioxidant, antiinflammatory, and anticancer activities79.
The strong interactions between MET–TYR–TRP, observed through partial correlation networks at baseline confirms the connection between folate-mediated methionine formation, leading to methyl donation reactions that form the monoamine neurotransmitters serotonin, dopamine, and epinephrine81,82. In depression, this balance is known to be perturbed83. With the drug exposure, we see further alterations in this balance at week 8. In addition, we see that GR significantly correlates with MET and TRP at week 8 post treatment. This may be indicative of changes in methylation status of the serotonin transporter84 through epigenetic mechanisms, in response to the SSRI treatment in these depression patients. At week 8, KYN–3OHKY association decreased significantly with concomitant increases in associations between KYN and the dopamine degradation product, HVA, and this association was comparitively stronger in patients who did not respond well to the treatment.
Using the ratios of metabolites significantly interacting as intermediate phenotypes leads us to rediscover loci known to be involved in neurotransmission/depression and neurodegeneration. The strongest association signals for baseline GR/HGA were within a locus on chromosome 7 containing two central genes: TAC1 (protachykinin-1) that has been linked to depression and anxiety85 and ASNS (asparagine synthetase [glutamine-hydrolyzing]) that is an important enzyme, the deficiency of which leads to substantial neurodevelopmental deficits86. Interestingly, patients with this deficiency (it is an inborn error of metabolism) also show modest changes in neurotransmitters. The strongest signal for MET/TYR was within a locus on chromosome 15 containing the gene AGBL1 (ATP/GTP binding protein-like 1) that has a role in controlling the length of the polyglutamate side chains on tubulin. This process is critical for neuronal survival, and the lack of such control has been reported to result in neurodegeneration in mice87. These findings underscores the utility of our “Pharmacometabolomics-Informs- Pharmacogenomics” approach33 to identify candidate genes for further functional studies. Using this strategy, we have previously identified SNP signals in the DEFB1 and AHR genes that were associated with severity of depressive symptoms in these MDD patients28. DEFB1 is an antimicrobial peptide which is highly expressed and active in the gut88, playing a potentially important role in maintaining gut–microbiome homeostasis89. These results fit within the broadening body of information in support of important roles for the “microbiota–gut–brain axis” and inflammation in MDD pathophysiology.
Several limitations of this study warrant consideration. Compared with other MDD patients recruited in the PGRN-AMPS trial, study participants were “selected” because they were able to complete all three visits (i.e., baseline, 4, and 8 weeks) and provide blood samples, which would reduce the number of patients in the final sample who did poorly. In addition, only Caucasians were included in this study, and thus, a given inherent limitation was developed from analyzing a subset of MDD patients. Furthermore, the LCECA platform captures information on only redox-active compounds in the tyrosine, tryptophan, purine, and sulfur amino acid pathways and several markers of vitamin status and oxidative processes. The integration of data from lipidomics and mass spectrometry-based metabolomics platforms in future studies, as well as inclusion of several confounding variables, such as body mass index, diet, and lifestyle factors, would definitely help to better unravel the mechanistic aspect of the drug response.
In conclusion, we analyzed the metabolomic profile in 290 MDD patients before and after citalopram/escitalopram treatment. Noncanonical metabolic pathways related to TRP, tyrosine, and purine metabolism were found to be activated after the drug exposure. There was crosstalk among these pathways at baseline depression levels, which was significantly impacted by the drug exposure. Significant increases in gut–microbiota-related metabolites, such as the indoles and the phenolic acids, were observed in the overall population. Patients who responded to the drug compared to those who did not, had significant differences in baseline levels as well as in the trajectories of several metabolites, including several gut–microbiota related metabolites, suggesting that the drug exposure might be impacting gut-microbial ecology differently in the two groups. Overall, amelioration of oxidative stress and increases in anti-inflammatory processes seem to be part of the mechanism involved in response to citalopram/escitalopram treatment.
Supplementary information
Acknowledgements
The authors are grateful for the support of NIH, to Lisa Howerton for her administrative support, and to the study participants and their families of the Mayo Pharmacogenomics Research Network-Antidepressant Pharmacogenomics Medication Study (PGRN-AMPS). The research and the authors are supported by funding from the NIH. This work was funded by grant support to Rima Kaddurah-Daouk through NIH grants R01MH108348, R01AG046171 & U01AG061359, RF1AG051550. Sudeepa Bhattacharyya was supported by 5R01MH108348, 5R01AG046171-03S1. R.M.W. was supported by NIH grants RO1 GM28157, U19 GM61388, U54 GM114838, and NSF1624615.
Conflict of interest
R.M.W. is a cofounder and stockholder in OneOme, LLC, a pharmacogenomic clinical decision support company. A.J.R. has received consulting fees from Akili, Brain Resource Inc., Compass Inc., Curbstone Consultant LLC., Emmes Corp., Johnson and Johnson (Janssen), Liva-Nova, Mind Linc, Sunovion, and Taj Medical; speaking fees from Liva-Nova; and royalties from Guilford Press and the University of Texas Southwestern Medical Center, Dallas, TX (for the Inventory of Depressive Symptoms and its derivatives). He is also named the coinventor on two patents: US Patent No. 7,795,033: Methods to Predict the Outcome of Treatment with Antidepressant Medication and US Patent No. 7,906,283: Methods to Identify Patients at Risk of Developing Adverse Events during Treatment with Antidepressant Medication. M.A.F. has received grant support from AssureRx Health Inc, Myriad, Pfizer Inc, NIMH (R01 MH079261), the National Institute on Alcohol Abuse and Alcoholism (P20AA017830) in the National Institutes of Health at the US Department of Health and Human Services, and the Mayo Foundation. He has been a consultant (for Mayo) to Janssen Global Services, LLC; Mitsubishi Tanabe Pharma Corp; Myriad Genetics, Inc; Sunovion Pharmaceuticals, Inc; and Teva Pharmaceutical Industries Ltd. He has received continuing medical education, travel, and presentation support from the American Physician Institute and CME Outfitters. L.W. was supported by NIH grants U19 GM61388, U54 GM114838, and NSF164615. She is a cofounder and stockholder in OneOme. R.K.-D. is an inventor on key patents in the field of metabolomics. D.N.’s stipend has been supported in part by NIH T32 GM072474 and the Mayo Graduate School. A.T.A.’s research was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number T32 GM008685. M.A. was supported by the National Institute on Aging [R01AG057452, RF1AG051550, and R01AG046171], National Institute of Mental Health [R01MH108348], and Qatar National Research Fund [NPRP8-061-3-011]. The funders listed above had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the paper; and decision to submit the manuscript for publication.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ahmed T. Ahmed, Matthias Arnold
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0507-5).
References
- 1.World Health Organization W. Depression and Other Common Mental Disorders: Global Health Estimates. Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Crismon ML, et al. The Texas Medication Algorithm Project: report of the Texas Consensus Conference Panel on medication treatment of major depressive disorder. J. Clin. Psychiatry. 1999;60:142–156. doi: 10.4088/JCP.v60n0302. [DOI] [PubMed] [Google Scholar]
- 3.Anderson IM, et al. Evidence-based guidelines for treating depressive disorders with antidepressants: a revision of the 2000 British Association for Psychopharmacology guidelines. J. Psychopharmacol. 2008;22:343–396. doi: 10.1177/0269881107088441. [DOI] [PubMed] [Google Scholar]
- 4.Rush AJ, et al. Report by the ACNP Task Force on response and remission in major depressive disorder. Neuropsychopharmacol.: Off. Publ. Am. Coll. Neuropsychopharmacol. 2006;31:1841–1853. doi: 10.1038/sj.npp.1301131. [DOI] [PubMed] [Google Scholar]
- 5.Rush AJ, et al. Selecting among second-step antidepressant medication monotherapies: predictive value of clinical, demographic, or first-step treatment features. Arch. Gen. Psychiatry. 2008;65:870–880. doi: 10.1001/archpsyc.65.8.870. [DOI] [PubMed] [Google Scholar]
- 6.US Food and Drug Administration Ws. Antidepressant Use in Children, Adolescents, and Adults. (2007).
- 7.Rush AJ, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am. J. Psychiatry. 2006;163:1905–1917. doi: 10.1176/ajp.2006.163.11.1905. [DOI] [PubMed] [Google Scholar]
- 8.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 9.Kaddurah-Daouk R, Krishnan KR. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacol.: Off. Publ. Am. Coll. Neuropsychopharmacol. 2009;34:173–186. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- 10.Kaddurah-Daouk R, Weinshilboum RM. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin. Pharmacol. Ther. 2014;95:154–167. doi: 10.1038/clpt.2013.217. [DOI] [PubMed] [Google Scholar]
- 11.Neavin D, Kaddurah-Daouk R, Weinshilboum R. Pharmacometabolomics informs pharmacogenomics. Metab.: Off. J. Metab. Soc. 2016;12:121. doi: 10.1007/s11306-016-1066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadad BS, et al. Peripheral biomarkers of major depression and antidepressant treatment response: current knowledge and future outlooks. J. Affect. Disord. 2018;233:3–14. doi: 10.1016/j.jad.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rush AJ, Ibrahim HM. A clinician’s perspective on biomarkers. Focus. 2018;16:124–134. doi: 10.1176/appi.focus.20170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paige LA, Mitchell MW, Krishnan KR, Kaddurah-Daouk R, Steffens DC. A preliminary metabolomic analysis of older adults with and without depression. Int. J. Geriatr. Psychiatry. 2007;22:418–423. doi: 10.1002/gps.1690. [DOI] [PubMed] [Google Scholar]
- 15.Miller CL, Llenos IC, Dulay JR, Weis S. Upregulation of the initiating step of the kynurenine pathway in postmortem anterior cingulate cortex from individuals with schizophrenia and bipolar disorder. Brain Res. 2006;1073–1074:25–37. doi: 10.1016/j.brainres.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 16.Myint AM, et al. Tryptophan breakdown pathway in bipolar mania. J. Affect. Disord. 2007;102:65–72. doi: 10.1016/j.jad.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Holmes E, et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaddurah-Daouk R, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol. Psychiatry. 2007;12:934–945. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 19.Tkachev D, Mimmack ML, Huffaker SJ, Ryan M, Bahn S. Further evidence for altered myelin biosynthesis and glutamatergic dysfunction in schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:557–563. doi: 10.1017/S1461145706007334. [DOI] [PubMed] [Google Scholar]
- 20.Hayaishi O. Utilization of superoxide anion by indoleamine oxygenase-catalyzed tryptophan and indoleamine oxidation. Adv. Exp. Med. Biol. 1996;398:285–289. doi: 10.1007/978-1-4613-0381-7_45. [DOI] [PubMed] [Google Scholar]
- 21.Maes M, et al. Treatment with interferon-alpha (IFN alpha) of hepatitis C patients induces lower serum dipeptidyl peptidase IV activity, which is related to IFN alpha-induced depressive and anxiety symptoms and immune activation. Mol. Psychiatry. 2001;6:475–480. doi: 10.1038/sj.mp.4000872. [DOI] [PubMed] [Google Scholar]
- 22.Stone TW, Darlington LG. Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Discov. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 23.Wirleitner B, Neurauter G, Schrocksnadel K, Frick B, Fuchs D. Interferon-gamma-induced conversion of tryptophan: immunologic and neuropsychiatric aspects. Curr. Med. Chem. 2003;10:1581–1591. doi: 10.2174/0929867033457179. [DOI] [PubMed] [Google Scholar]
- 24.Schiepers OJG, Wichers MC, Maes M. Cytokines and major depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2005;29:201–217. doi: 10.1016/j.pnpbp.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Gabbay V, et al. The possible role of the kynurenine pathway in adolescent depression with melancholic features. J. Child Psychol. Psychiatry, Allied Discip. 2010;51:935–943. doi: 10.1111/j.1469-7610.2010.02245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley KAL, et al. The role of the kynurenine pathway in suicidality in adolescent major depressive disorder. Psychiatry Res. 2015;227:206–212. doi: 10.1016/j.psychres.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reus GZ, et al. Kynurenine pathway dysfunction in the pathophysiology and treatment of depression: evidences from animal and human studies. J. Psychiatr. Res. 2015;68:316–328. doi: 10.1016/j.jpsychires.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, et al. Beta-defensin 1, aryl hydrocarbon receptor and plasma kynurenine in major depressive disorder: metabolomics-informed genomics. Transl. Psychiatry. 2018;8:10. doi: 10.1038/s41398-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park DI, et al. Purine and pyrimidine metabolism: convergent evidence on chronic antidepressant treatment response in mice and humans. Sci. Rep. 2016;6:35317. doi: 10.1038/srep35317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaddurah-Daouk R, et al. Cerebrospinal fluid metabolome in mood disorders-remission state has a unique metabolic profile. Sci. Rep. 2012;2:667. doi: 10.1038/srep00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaddurah-Daouk R, Weinshilboum R. Metabolomic signatures for drug response phenotypes: pharmacometabolomics enables precision medicine. Clin. Pharmacol. Ther. 2015;98:71–75. doi: 10.1002/cpt.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikoff WR, et al. Pharmacometabolomics reveals racial differences in response to atenolol treatment. PLoS ONE. 2013;8:e57639. doi: 10.1371/journal.pone.0057639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yerges-Armstrong LM, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2013;94:525–532. doi: 10.1038/clpt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, et al. Pharmacometabolomics of response to sertraline and to placebo in major depressive disorder—possible role for methoxyindole pathway. PLoS ONE. 2013;8:e68283. doi: 10.1371/journal.pone.0068283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaddurah-Daouk R, et al. Pretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: a proof of concept. Transl. Psychiatry. 2011;1:e26. doi: 10.1038/tp.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotroff DM, et al. Metabolomic signatures of drug response phenotypes for ketamine and esketamine in subjects with refractory major depressive disorder: new mechanistic insights for rapid acting antidepressants. Transl. Psychiatry. 2016;6:e894. doi: 10.1038/tp.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Y, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin. Pharmacol. Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta M, et al. TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: pharmacometabolomics-informed pharmacogenomics. Mol. Psychiatry. 2016;21:1717–1725. doi: 10.1038/mp.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mrazek DA, et al. Treatment outcomes of depression: the pharmacogenomic research network antidepressant medication pharmacogenomic study. J. Clin. Psychopharmacol. 2014;34:313–317. doi: 10.1097/JCP.0000000000000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji Y, et al. Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. Br. J. Clin. Pharmacol. 2014;78:373–383. doi: 10.1111/bcp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji Y, et al. Pharmacogenomics of selective serotonin reuptake inhibitor treatment for major depressive disorder: genome-wide associations and functional genomics. Pharm. J. 2013;13:456–463. doi: 10.1038/tpj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rush AJ, et al. Bupropion-SR, sertraline, or venlafaxine-XR after failure of SSRIs for depression. N. Engl. J. Med. 2006;354:1231–1242. doi: 10.1056/NEJMoa052963. [DOI] [PubMed] [Google Scholar]
- 43.Matson WR, et al. n-Electrode three-dimensional liquid chromatography with electrochemical detection for determination of neurotransmitters. Clin. Chem. 1984;30:1477–1488. [PubMed] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995;57:289–300. [Google Scholar]
- 45.Pons, P. & Latapy, M. Computing communities in large networks using random walks. In Proc. 20th International Conference on Computer and Information Sciences. 284–293 (Springer-Verlag, Istanbul, Turkey, 2005).
- 46.von Elm E, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann. Intern. Med. 2007;147:573–577. doi: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 47.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 49.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golino HF, Epskamp S. Exploratory graph analysis: a new approach for estimating the number of dimensions in psychological research. PLoS ONE. 2017;12:e0174035. doi: 10.1371/journal.pone.0174035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Do KT, et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabol.: Off. J. Metabol. Soc. 2018;14:128. doi: 10.1007/s11306-018-1420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinheiro J. C., Bates D. J. D., DebRoy S., Sakar D. The Nlme Package: Linear and Nonlinear Mixed Effects Models, R Version 3, vol. 6 (2012).
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
- 54.Conesa A1, Nueda MJ, Ferrer A, Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 55.van Borkulo C. D., Jonas H., Epskamp S., Millner A. Network Comparison Test: statistical comparison of two networks based on three invariance measures (R package Version 2.0.1) [Computer Software]. (2016).
- 56.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst. Biol. 2011;5:21. doi: 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hubbard TD, Murray IA, Perdew GH. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 2015;43:1522–1535. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitfield-Cargile CM, et al. The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes. 2016;7:246–261. doi: 10.1080/19490976.2016.1156827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tilg H. Diet and intestinal immunity. N. Engl. J. Med. 2012;366:181–183. doi: 10.1056/NEJMcibr1113158. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 63.Schiering C, et al. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242–245. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rothhammer V, et al. Microglial control of astrocytes in response to microbial metabolites. Nature. 2018;557:724–728. doi: 10.1038/s41586-018-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosas HD, et al. A systems-level “misunderstanding”: the plasma metabolome in Huntington’s disease. Ann. Clin. Transl. Neurol. 2015;2:756–768. doi: 10.1002/acn3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karu N, et al. Tryptophan metabolism, its relation to inflammation and stress markers and association with psychological and cognitive functioning: Tasmanian Chronic Kidney Disease pilot study. BMC Nephrol. 2016;17:171. doi: 10.1186/s12882-016-0387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evenepoel P, Meijers BK, Bammens BR, Verbeke K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. Suppl. 2009;114:S12–S19. doi: 10.1038/ki.2009.402. [DOI] [PubMed] [Google Scholar]
- 69.Coppen A, Brooksbank BW, Eccleston E, Peet M, White SG. Tryptophan metabolism in depressive illness. Psychol. Med. 1974;4:164–173. doi: 10.1017/S0033291700041994. [DOI] [PubMed] [Google Scholar]
- 70.Sallee M, et al. The aryl hydrocarbon receptor-activating effect of uremic toxins from tryptophan metabolism: a new concept to understand cardiovascular complications of chronic kidney disease. Toxins. 2014;6:934–949. doi: 10.3390/toxins6030934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaudhari K, et al. Clinical correlation of alteration of endogenous antioxidant-uric acid level in major depressive disorder. Indian J. Clin. Biochem.: IJCB. 2010;25:77–81. doi: 10.1007/s12291-010-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kesebir S, Tatlidil Yaylaci E, Suner O, Gultekin BK. Uric acid levels may be a biological marker for the differentiation of unipolar and bipolar disorder: the role of affective temperament. J. Affect. Disord. 2014;165:131–134. doi: 10.1016/j.jad.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 73.Wen S, et al. Serum uric acid levels and the clinical characteristics of depression. Clin. Biochem. 2012;45:49–53. doi: 10.1016/j.clinbiochem.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 74.Agren H, Niklasson F, Hallgren R. Brain purinergic activity linked with depressive symptomatology: hypoxanthine and xanthine in CSF of patients with major depressive disorders. Psychiatry Res. 1983;9:179–189. doi: 10.1016/0165-1781(83)90042-2. [DOI] [PubMed] [Google Scholar]
- 75.Ali-Sisto T, et al. Purine metabolism is dysregulated in patients with major depressive disorder. Psychoneuroendocrinology. 2016;70:25–32. doi: 10.1016/j.psyneuen.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 76.George J, Struthers AD. Role of urate, xanthine oxidase and the effects of allopurinol in vascular oxidative stress. Vasc. Health Risk Manag. 2009;5:265–272. doi: 10.2147/VHRM.S4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sautin YY, Johnson RJ. Uric acid: the oxidant–antioxidant paradox. Nucleosides, Nucleotides Nucleic Acids. 2008;27:608–619. doi: 10.1080/15257770802138558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orru M, et al. Psychostimulant pharmacological profile of paraxanthine, the main metabolite of caffeine in humans. Neuropharmacology. 2013;67:476–484. doi: 10.1016/j.neuropharm.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russell WR, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013;57:523–535. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- 80.Hubkova B, Velika B, Birkova A, Guzy J, Marekova M. Hydroxybenzoic acids and their derivatives as peroxynitrite scavengers. Free Radic. Biol. Med. 2014;75(Suppl 1):S33–S34. doi: 10.1016/j.freeradbiomed.2014.10.770. [DOI] [PubMed] [Google Scholar]
- 81.Weinshilboum RM, Raymond FA, Elveback LR, Weidman WH. Correlation of erythrocyte catechol-O-methyltransferase activity between siblings. Nature. 1974;252:490–491. doi: 10.1038/252490a0. [DOI] [PubMed] [Google Scholar]
- 82.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu. Rev. Pharmacol. Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 83.Miller AL. The methylation, neurotransmitter, and antioxidant connections between folate and depression. Altern. Med. Rev. 2008;13:216–226. [PubMed] [Google Scholar]
- 84.Shi M, et al. Methylation status of the serotonin transporter promoter CpG island is associated with major depressive disorder in Chinese Han population: a case-control study. J. Nerv. Ment. Dis. 2017;205:641–646. doi: 10.1097/NMD.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 85.Bilkei-Gorzo A, Racz I, Michel K, Zimmer A. Diminished anxiety- and depression-related behaviors in mice with selective deletion of the Tac1 gene. J. Neurosci.: Off. J. Soc. Neurosci. 2002;22:10046–10052. doi: 10.1523/JNEUROSCI.22-22-10046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruzzo EK, et al. Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron. 2013;80:429–441. doi: 10.1016/j.neuron.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cocchi E, et al. Genome-wide association study of antidepressant response: involvement of the inorganic cation transmembrane transporter activity pathway. BMC Psychiatry. 2016;16:106. doi: 10.1186/s12888-016-0813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 89.Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.