Abstract
The conservation and development of chicken have received considerable attention, but the admixture history of chicken breeds, especially Chinese indigenous breeds, has been poorly demonstrated. In this study, we aimed to evaluate the genetic diversity and population structure of eight chicken breeds (including conserved chicken breeds) from different geographic origin and to identify admixture within these breeds using a 600-K single-nucleotide polymorphism panel for genotyping. Using the genotype of 580,961 single-nucleotide polymorphism markers scored in 1,200 animals, we evaluated the genetic diversity (heterozygosity and proportion of polymorphic markers), linkage disequilibrium decay, population structure (principal component analysis and neighbor-joining tree), genetic differentiation (FST and genetic distance), and migration events (TreeMix and f-statistics) of the eight domesticated chicken breeds. The results of population analytical methods revealed patterns of hybridization that occurred after divergence in Tibetan chicken. We argue that chicken migration and admixture, followed by trade, have been important forces in shaping the genomic variation in modern Chinese chicken. Moreover, isolation by distance might play a critical role in shaping the genomic variation within Eurasia continent chicken breeds. Moreover, genetic information provided in this study is valuable resources for production applications (genomic prediction, and breeding strategy) and scientific research (genetic basis detection, studying evolution, or domestication).
Keywords: admixture, Chinese indigenous chicken, genetic diversity, genome-wide single-nucleotide polymorphism, population structure
Introduction
Domestication of chicken starting with red junglefowl (Gallus gallus) began in China ∼10,000 years ago (Xiang et al. 2014). Because of biogeographic differences and selection within wild ancestral populations, China has the most prolific chicken genetic resource in the world (Chen et al. 2004, 2008; FAO 2016; Chiang et al. 2017). One hundred and eight indigenous chicken breeds (Chen et al. 2004), recorded in China with distinct phenotypes, such as behavior, reproduction, and feather color, play a crucial role in Chinese poultry industry and are an important breeding resource to meet the future market demands (Zhang et al. 2002; Chen et al. 2008; Chiang et al. 2017). The last few decades have seen a dramatic increase in the pace of genetic gain via organized breeding methods. However, an increase in production performance is always accompanied by a reduction in genetic diversity (Boettcher et al. 2010), and this can have dramatic consequences leading to an irreversible loss of untapped genotypic and phenotypic variations, including disease-resistance adaptability. Therefore, the conservation of locally adapted indigenous chicken breeds has become an important milestone in endangered animal protection and sustainable breeding.
Genetic diversity, an effective monitor for conservation purposes, within Chinese indigenous chicken populations has been evaluated using microsatellites (Abiye Shenkut et al. 2015; Azimu et al. 2018), random-amplified polymorphic DNA (Lynch and Milligan 1994), amplified fragment length polymorphisms (Christian 2004), and genome-wide single-nucleotide polymorphisms (SNPs) (Chen et al. 2018; Zhang et al. 2018). Large-scale genotyping technologies have enabled the analysis of admixture of various domestic animals, including dog (Vonholdt et al. 2010), sheep (Lawson Handley et al. 2007; Peter et al. 2007), cattle (Decker et al. 2014), and pig (Groenen et al. 2012).
As live animal import became organized after “reform and opening-up” (Carter et al. 1999), admixture has inevitably occurred in Chinese native chicken breeds. For example, the white leghorn chicken and Rhode Island Red chicken, two main foreign layers, were imported to produce high-production intercross breeds in the last few decades. Simultaneously, Chinese Government has made considerable efforts for conserving several indigenous chicken breeds (Chen et al. 2008; Zhang et al. 2018). However, the current status of Chinese local chicken conservation, effects of these exotic commercial breeds on the local populations, and the effect of this admixture in the protection of endangered local breeds remain unclear. In this study, we aimed to evaluate the genetic diversity and population structure of eight chicken breeds (including conserved chicken breeds) from different geographic origin and to identify admixture within these breeds using a 600-K SNP panel for genotyping. We used five Chinese native chicken breeds (three chicken breeds in the national conservation program) and three imported breeds (European and American highly modified domestic chicken breeds) in the present study. The genetic information could be useful for further genomic prediction, genetic basis detection (economically important traits), and breeding strategy establishment in China.
Materials and Methods
Ethics Statement
Sample collection was performed by strictly following the protocols approved by the Animal Welfare Committee of China Agricultural University (Approval Number: XK622).
Sample Selection
We used 1,200 chickens from eight breeds, namely, five distinct Chinese indigenous breeds (Beijing You [BY], Hongshan [HS], Shouguang [SG], Taihe Silkie [SK], and Tibetan [TB] Chickens), two European breeds (White Leghorn [WL] and Houdan [HD] chickens), and one North American breed (Rhode Island Red [RIR] chicken).
These Chinese native chicken breeds were chosen from five different provinces (table 1). Different geographical and environmental factors resulted in the distinct phenotype of these breeds (fig. 1A). Beijing You chicken, mainly produced in Beijing, has a unique appearance with yellow feathers, crest, and beard; polydactyl; and feathers on both shanks (Zhang et al. 2016). It is known for its high-quality meat and is an egg-type breed. Hongshan chicken is a classic dual-purpose breed that originated in Hubei Province. This breed has two distinctly different tail types (Wang et al. 2018). Shouguang chicken is a breed that originated in Shandong Province and has dual-purpose. They are uniquely marked by black feathers, face, eyes, and beak. Taihe Silkie chicken is an ancient breed, mainly produced in Jiangxi Province, and widely known by its black skin, meat, and bone. Tibetan chicken originates from the Qinghai–Tibet Plateau and has good adaptation to hypoxic conditions (Zhang et al. 2007; Jiang et al. 2018). In addition, BY, SK, and TB are listed in the national conservation program (which contains 28 Chinese chicken breeds).
Table 1.
Provenance and Genetic Diversity Measurements of All Breeds Included in the Present Study
| Breed | Abbr. | No. Samples | Continent | Geographic Origin | Central Site Latitude/Longitudef | Sampling Location | Conservation Generation | He | PN (%) |
|---|---|---|---|---|---|---|---|---|---|
| Beijing You | BY | 77 | Asia | Beijing, China | 39°55′06.2″N, 116°23′49.2″E | Beijing | >20g | 0.2834 | 81.44 |
| Hongshan | HS | 96a | Asia | Hubei Province, China | 30°35′04.9″N, 114°17′49.5″E | Hubei Province | ∼11 | 0.3171 | 92.56 |
| Shouguang | SG | 109 | Asia | Shandong Province, China | 37°00′33.7″N, 118°49′24.8″E | Beijing | >18g | 0.2839 | 68.22 |
| Taihe Silkies | SK | 90 | Asia | Jiangxi Province, China | 26°47′29.4″N, 114°54′22.7″E | Beijing | >20g | 0.3012 | 73.04 |
| Tibetan | TB | 41b | Asia | Tibet, China | 29°38′53.9″N, 91°10′31.5″E | Beijing and Tibet | >13g | 0.3083 | 94.42 |
| Rhode Island Red | RIR | 469c | America | United States | 43°33′08.8″N, 10°18′30.5″E | Beijing | — | 0.2693 | 75.66 |
| Houdan | HD | 86d | Europe | France | 48°47′29.2″N, 1°36′21.1″E | Anhui Province | — | 0.2963 | 70.97 |
| White Leghorn | WL | 232e | Europe | Tuscany, Italy | 41°49′51.2″N, 71°24′53.4″W | Beijing | —g | 0.2904 | 68.26 |
Note.—He, expected heterozygosity; PN, proportion of polymorphic SNPs.
Data of 48 samples have been published (Wang et al. 2018).
Data of 15 samples (TB-CAU) were collected from the Experimental Chicken Farm at the China Agricultural University, 10 samples (TB-NM) were collected from Nimu, Tibet, 13 samples (TB-ND) were collected from Naidong, Tibet, and 3 samples (TB-LZ) were collected from Lingzhi, Tibet.
Data of 78 samples have been published (Nie et al. 2016).
Data of all 86 samples have been published (Zhang et al. 2016).
Data of 40 samples (WL-CAU) were collected from the Experimental Chicken Farm at China Agricultural University (CAU) and 192 samples (WL-YQ) were collected from a commercial company in Yanqing, Beijing.
The central site of origin as the geographic information for further analysis.
Samples (BY, SG, SK, partial TB, and WL) were raised in a conservation farm before maintaining in the Experimental Chicken Farm (CAU).
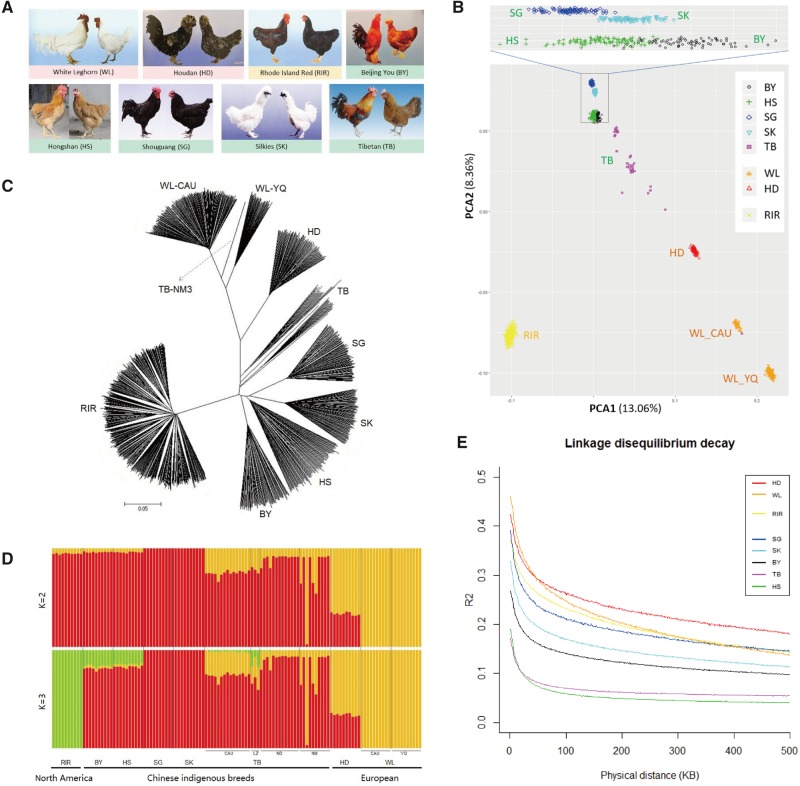
Fig. 1.
—Population genetic structure of the eight chicken breeds. (A) Eight different chicken breeds with distinct phenotypes (Chen et al. 2004). (B) The PCA plot of chicken populations. PCA1 and PCA2 explained 13.06% and 8.36% of the observed variance, respectively. (C) Neighbor-joining tree constructed using MEGA. (D) The admixture plot for breeds analyzed based on different number of assumed ancestors (K). (E) Linkage disequilibrium (LD) decay for the eight breeds. LD decay determined by r2 against distance between polymorphic sites.
White Leghorn chicken, which originated in Italy, is a commonly used layer that is characterized by high growth and egg production rates (Kerje et al. 2003); it has white feathers. Houdan chicken is an old French breed. It is a dual-purpose breed and is unusually marked by mottled feathers, different comb shapes, and polydactyl. Rhode Island Red chicken, which originated in America, is known for its prolific egg-laying ability.
Herein, the samples are identified by breed abbreviation. Breed and sampling information are summarized in table 1. Partial data used in this research were obtained from previous studies (Nie et al. 2016; Zhang et al. 2016; Wang et al. 2018). Two milliliters of blood samples were collected using an injection via the wing vein into centrifuge tubes containing DNA anticoagulating agent and stored at −20 °C for further analysis.
Genotyping and Preparation
The genomic DNA was extracted using the standard phenol/chloroform method from blood samples (Green and Sambrook 2017) and genotyped using the 600 K Affymetrix Axiom Chicken Genotyping Array (Affymetrix, Inc. Santa Clara, CA) (Kranis et al. 2013). Axiom Analysis Suite v4.0.1 (AxAS) software (Applied Biosystems 2017) was then used for quality control and genotype calling (chicken genome version: Gallus_gallus v5.0). Specifically, only samples with a dish quality control (DQC) of >0.82 and call rate of >98% were used for subsequent analysis.
We filtered SNPs with unknown genomic positions or redundant genomic coordinates. Using PLINK (v1.90) (Purcell et al. 2007), we removed SNPs with the following criteria: missing rate of >0.01 and minor allele frequency of <0.01.
Calculation of Genetic Diversity
After sample and SNP quality control, sample pools of the eight breeds (namely, BY, HS, SG, SK, TB, WL, HD, and RIR) were used to calculate genetic diversity. Two diversity indicators, namely, expected heterozygosity (He) and proportion of polymorphic markers (PN), were calculated using PLINK with the default settings.
Population Structure
The principal component analysis (PCA) (Price et al. 2006) as implemented in PLINK (Purcell et al. 2007) was used to detect the population structure (with parameters: –pca). A neighbor-joining (NJ) tree was built using the MEGA (v6.0) pipeline with standard settings and 1,000 bootstrap replicates (Tamura et al. 2007).
We further estimated the ancestry (from K = 2 to K = 21) of each individual using the genome-wide SNP data set and the model-based assignment software program ADMIXTURE (v1.3) to quantify admixture among the eight chicken breeds (Alexander et al. 2009). The optimal K should be determined using the complete data set. However, with an increasing difference in sample size between populations, both estimators deteriorate quickly (Wang 2017). In present analysis, cross-validation was utilized to determine the optimal K value using all TB chicken samples and 10 randomly selected samples from the other populations (supplementary table S1, Supplementary Material online). This analysis was replicated ten times over (Martiniano et al. 2017). Here, we obtained the lowest mean CV error for K = 8 (supplementary table S2, Supplementary Material online). Plots have been performed using package “ggplot2” in R.
Linkage Disequilibrium Decay
The square of the correlation coefficient (r2), based on the genotype frequency, between alleles at two separate SNP loci was used for linkage disequilibrium (LD) estimates (Vanliere and Rosenberg 2008). Within each population, the SNPs (MAF < 0.01, Hardy–Weinberg equilibrium <10E-6) were used to calculate r2 using Plink with the following equation:
where, fA1_B1, fA2_B2, fA1_B2, fA2_B1, fA1, fA2, fB1, and fB2 are the frequency of haplotypes (A1B1, A2B2, A1B2, and A2B1) and alleles (A1, A2, B1, and B2) in the population, respectively (Khanyile et al. 2015; Seo et al. 2018).
Estimation of Genetic Differentiation
Artificial selection has resulted in a wide range of phenotypes among domestic chicken breeds. An unbiased genetic differentiation estimate, FST (Weir and Cockerham 1984), was calculated using a self-developed code in R (http://www.R-project.org/; Accessed 23 June, 2019) with the filtered SNP data set to estimate genetic differentiation among populations. Pairwise geographic distances were calculated from origin information obtained using Google map. We used Pearson correlation to test the association between genetic distance, which is calculated as (FST/1−FST), and geographic distance.
TreeMix Analysis
To build population trees in the presence of admixture, we modified the TreeMix model (Pickrell and Pritchard 2012). This software models the relationship between the tested populations with their ancestral population using genome-wide allele frequency data and a Gaussian approximation of genetic drift. We unrooted the graph because we did not include wild population in our study (Chen et al. 2004, 2008; Chiang et al. 2017). TreeMix was used to create an ML tree of the eight breeds. We used the -m2 option to add migration events on the built phylogeny and the -se option to calculate the SE of migration proportions. Migration edges were added until 99.8% of the variance in ancestry between populations was explained by the model (Decker et al. 2014).
The f3 and f4 statistics (Reich et al. 2009; Patterson et al. 2012), performed using TreeMix (THREEPOP and FOURPOP programs), support admixture in the sampled populations. In the f3 test (A; B, C), calculated with all possible triplets from the eight breeds, a significantly negative value of the f3 statistic implies that population A is a result of admixture of B and C. In the f4 test (A, B; C, D), which reveals the tree topology of four populations, a significant nonzero value indicates gene flow in the tree.
where a′, b′, c′, and d′ were the allele frequency in populations A, B, C, and D at an SNP, respectively (Patterson et al. 2012).
Results
Genetic Diversity among the Eight Breeds
A total of 1,193 individuals from the eight chicken populations with 542,872 SNPs were included in the final data set after applying the quality control filters. Hongshan chicken breed exhibited the highest genetic diversity, He (0.3171), and PN (92.56%) (table 1). The highest PN was observed in TB (94.42%), whereas SG chicken exhibited the lowest PN (68.22%). The lowest He was observed in RIR chicken (0.2693).
Population Structure Analysis
Population structure of the eight domesticated chicken breeds was inferred using the PCA, NJ tree analysis, and Bayesian ancestry models. The PCA showed that the first two principal components account for 13.06% (PC1) and 8.36% (PC2) of the total variability (fig. 1B). With the exception of TB chicken, the Chinese, European, and North American breeds separated into distinct clusters reflecting their geographic origin. Further, WL chicken from the two subpopulations (table 1) was clearly grouped into the respective clusters. Additionally, despite being grouped together, TB chicken from the four subpopulations (table 1) clustered more loosely with each other indicating more genetic variation in this breed and probably reflecting admixture. Moreover, other four Chinese indigenous chicken breeds (BY, HS, SG, and SK) were clustered more closely. The results of the NJ tree (fig. 1C) were consistent with the PCA results (fig. 1B). All birds from the same population clustered together, except one TB chicken (fig. 1C).
Subsequently, in the ADMIXTURE analysis, two Chinese indigenous breeds (SG, SK) and WL formed the two distinct populations obtained at K = 2. At K = 3, individuals clustered strongly into the three groups of origin (North America, China, and Europe), which is consistent with the PCA results (fig. 1D). Moreover, the five Chinese indigenous breeds were always grouped into one main population until K = 6. Furthermore, SG and SK formed different ancestors at K = 7–8. For K = 8, both BY, HS and TB appear as admixed groups (supplementary fig. S1, Supplementary Material online). However, the admixture proportion in BY and HS is consistent but TB is diverse.
LD Decay
As expected, LD estimated by the decay of genotypic association between markers (r2) was lower in the five Chinese indigenous breeds than in the other three breeds. In particular, the LD value of BY, HS, SG, SK, and TB was 56, 11, 42, 37, and 17 kb, respectively, whereas that of European breeds were 131 (HD) and 84 (WL). The highest LD value was observed in RIR (169 kb).
Meanwhile, two breeds, namely, WL and TB, decayed faster than the other chicken breeds (fig. 1E: orange and purple lines).
Population Differentiation Analysis
Artificial selection has resulted in a wide range of phenotypes in domesticated chickens. To investigate population differentiation among different breeds, FST was calculated using the filtered SNPs. The FST values, shown in table 2, varied from 0.03 to 0.19. The FST values are expected to be significantly higher between breeds from different continents than between breeds within a continent. All the FST values between breeds in China were <0.11 (from 0.03 to 0.10). In contrast, the FST values between the two European breeds (WL and HD) and other breeds were >0.12 (from 0.12 to 0.19). Interestingly, the FST values between RIR and the European breeds were in the same range as those between the two European breeds (0.14 and 0.19). Moreover, the FST values between RIR and the Chinese breeds ranged from 0.08 to 0.11, indicating that gene flow may have occurred between RIR and the Chinese breeds.
Table 2.
Matrix Showing Pairwise Differentiation (FST) and Genetic Distance (FST/1−FST) Estimates among the Eight Breeds
| BY | HS | SG | SK | TB | WL | HD | RIR | |
|---|---|---|---|---|---|---|---|---|
| BY | 0.05 | 0.10 | 0.09 | 0.07 | 0.17 | 0.16 | 0.09 | |
| HS | 0.06 | 0.03 | 0.06 | 0.03 | 0.16 | 0.14 | 0.08 | |
| SG | 0.11 | 0.03 | 0.07 | 0.07 | 0.13 | 0.15 | 0.11 | |
| SK | 0.10 | 0.06 | 0.07 | 0.08 | 0.19 | 0.17 | 0.10 | |
| TB | 0.07 | 0.03 | 0.07 | 0.09 | 0.12 | 0.13 | 0.08 | |
| WL | 0.20 | 0.19 | 0.15 | 0.23 | 0.13 | 0.13 | 0.19 | |
| HD | 0.19 | 0.16 | 0.17 | 0.21 | 0.14 | 0.15 | 0.14 | |
| RIR | 0.10 | 0.09 | 0.12 | 0.11 | 0.08 | 0.23 | 0.16 |
Note.—Upper triangle: FST, lower triangle: FST/1−FST.
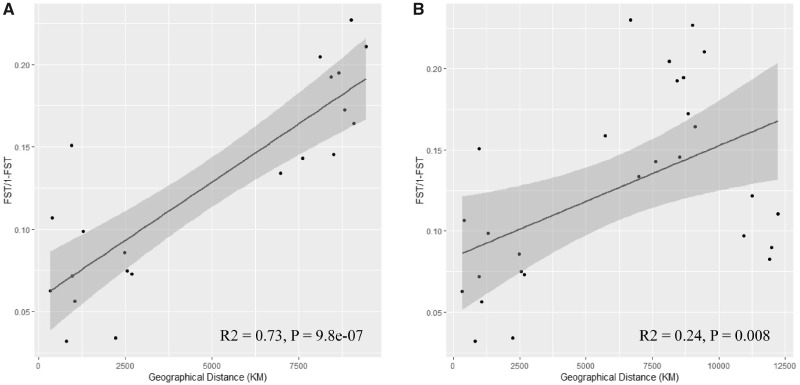
Pairwise genetic distance between breeds was highly correlated with geographic distance in Eurasian breeds, suggesting a strong signal of isolation by distance (r2 = 0.73, P = 9.8e-07) (fig. 2A). However, this correlation strongly decreased in the North American breed (RIR), which is consistent with the FST results shown above (r2 = 0.24, P = 0.008) (fig. 2B).
Fig. 2.
—Scatterplots show that pairwise genetic distance is associated with geographic distance. (A) Eurasian breeds and (B) eight domesticated breeds.
Admixture in Asian Breeds
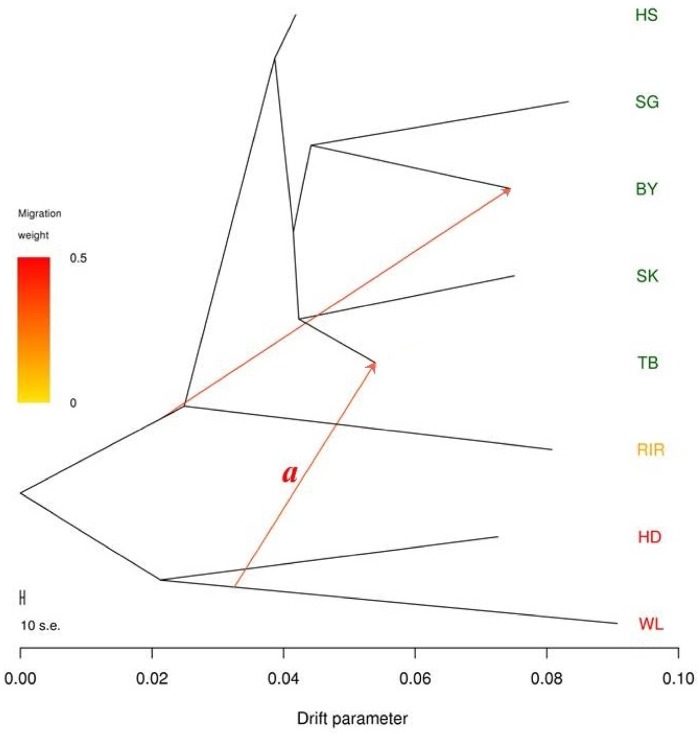
We used ancestry graphs implemented in TreeMix to analyze the admixture events and genetic relationships between the eight breeds of domesticated chickens. Sampled chickens were collected from different farms in China, which imported commercial birds from Europe (WL and HD) and North America (RIR). The migration edge a estimate (gene flow event) in the phylogenetic network (fig. 3 and supplementary fig. S2, Supplementary Material online) indicates that the Chinese indigenous breed TB was admixed as a result of introgression in European chicken (WL). Dominant white feather is a unique characteristic of WL. White-feathered chicken in TB population (supplementary fig. S3, Supplementary Material online) strongly supported this gene flow hypothesis.
Fig. 3.
—Phylogenetic network of the inferred relationships between the eight chicken breeds. Breeds were colored according to their geographic origin: green, Asia; orange, North America; red, Europe. Migration edge a signals introgression of White Leghorn chicken into Tibetan chicken.
We also used f-statistics to explore the evidence for European and North American chicken introgression into Chinese chicken. We observed that five significant tests out of 168 possible tests contained TB. The three most negative and significant f3 statistics for TB chicken (Chinese chicken breed) also support WL (European chicken breed) introgression into TB (supplementary table S3, Supplementary Material online). Moreover, the 20 most positive and negative significant f4 statistics from 210 statistics for the eight breeds clearly showed that gene flow occurred within Eurasian breeds (table 3). The first 10 negative and 20 positive significant f4 tests indicating gene flow appeared in the two European chicken breeds (WL and HD).
Table 3.
Twenty Most Positive and Negative Significant f4 Statistics for the Eight Breeds
| Breed A | Breed B | Breed C | Breed D | f4 Statistic | SE | Z-Score |
|---|---|---|---|---|---|---|
| White Leghorn | Shouguang | Hongshan | Houdan | −0.0608401 | 0.00106716 | −57.0111 |
| White Leghorn | Silkies | Hongshan | Houdan | −0.0578532 | 0.00102133 | −56.6447 |
| White Leghorn | Beijing You | Hongshan | Houdan | −0.0530342 | 0.00102241 | −51.8717 |
| White Leghorn | Rhode Island Red | Hongshan | Houdan | −0.0467017 | 0.00107123 | −43.5965 |
| White Leghorn | Shouguang | Rhode Island Red | Houdan | −0.0462463 | 0.00120241 | −38.4614 |
| White Leghorn | Hongshan | Rhode Island Red | Houdan | −0.0451155 | 0.00112782 | −40.0023 |
| White Leghorn | Beijing You | Rhode Island Red | Houdan | −0.0443478 | 0.00117477 | −37.7502 |
| White Leghorn | Silkies | Rhode Island Red | Houdan | −0.0438055 | 0.00116908 | −37.47 |
| White Leghorn | Tibetan | Hongshan | Houdan | −0.0387665 | 0.000801083 | −48.3925 |
| White Leghorn | Tibetan | Rhode Island Red | Houdan | −0.0292413 | 0.000882704 | −33.1269 |
| White Leghorn | Shouguang | Silkies | Tibetan | −0.0271946 | 0.000573204 | −47.4431 |
| White Leghorn | Silkies | Shouguang | Tibetan | −0.0268721 | 0.000620136 | −43.3326 |
| White Leghorn | Hongshan | Shouguang | Tibetan | −0.0266428 | 0.000578868 | −46.0256 |
| White Leghorn | Beijing You | Shouguang | Tibetan | −0.0257752 | 0.000605496 | −42.5687 |
| White Leghorn | Shouguang | Hongshan | Tibetan | −0.0246132 | 0.000494063 | −49.8178 |
| White Leghorn | Hongshan | Silkies | Tibetan | −0.0242663 | 0.000491401 | −49.3819 |
| White Leghorn | Beijing You | Silkies | Tibetan | −0.0233398 | 0.000525144 | −44.4445 |
| Houdan | Silkies | Shouguang | Tibetan | −0.022303 | 0.000540078 | −41.2959 |
| Houdan | Shouguang | Silkies | Tibetan | −0.022015 | 0.000510751 | −43.1031 |
| White Leghorn | Silkies | Hongshan | Tibetan | −0.0219142 | 0.000440134 | −49.7899 |
| White Leghorn | Shouguang | Houdan | Silkies | 0.0634215 | 0.00110297 | 57.5007 |
| White Leghorn | Silkies | Houdan | Shouguang | 0.062811 | 0.00112482 | 55.8412 |
| White Leghorn | Hongshan | Houdan | Shouguang | 0.0614296 | 0.00105974 | 57.9666 |
| White Leghorn | Hongshan | Houdan | Silkies | 0.0590531 | 0.000996507 | 59.2601 |
| White Leghorn | Beijing You | Houdan | Shouguang | 0.0574748 | 0.00111229 | 51.6724 |
| White Leghorn | Shouguang | Houdan | Beijing You | 0.0564516 | 0.00112923 | 49.9913 |
| White Leghorn | Beijing You | Houdan | Silkies | 0.0550394 | 0.00105076 | 52.3804 |
| White Leghorn | Silkies | Houdan | Beijing You | 0.0534057 | 0.00109536 | 48.7564 |
| White Leghorn | Hongshan | Houdan | Beijing You | 0.0526004 | 0.00103346 | 50.8974 |
| White Leghorn | Rhode Island Red | Houdan | Shouguang | 0.048422 | 0.00115074 | 42.0788 |
| White Leghorn | Rhode Island Red | Houdan | Silkies | 0.0465916 | 0.00109586 | 42.5162 |
| White Leghorn | Rhode Island Red | Houdan | Beijing You | 0.0455002 | 0.00112437 | 40.4672 |
| White Leghorn | Tibetan | Houdan | Silkies | 0.0411185 | 0.000820846 | 50.0929 |
| White Leghorn | Tibetan | Houdan | Shouguang | 0.0407961 | 0.00084949 | 48.0242 |
| White Leghorn | Shouguang | Houdan | Tibetan | 0.036227 | 0.000869125 | 41.6821 |
| White Leghorn | Silkies | Houdan | Tibetan | 0.0359389 | 0.000868802 | 41.3661 |
| White Leghorn | Tibetan | Houdan | Beijing You | 0.0352455 | 0.000847653 | 41.5801 |
| White Leghorn | Hongshan | Houdan | Tibetan | 0.0347868 | 0.00081795 | 42.5292 |
| White Leghorn | Beijing You | Houdan | Tibetan | 0.0316997 | 0.000849728 | 37.3056 |
| White Leghorn | Rhode Island Red | Houdan | Tibetan | 0.0268478 | 0.000843182 | 31.8411 |
Discussion
Our results indicate that most Chinese indigenous chicken breeds have higher genetic diversity (He and PN) than that of European or North American highly selected chicken breeds, which is in agreement with the findings of previous studies (Lujiang et al. 2006; Chen et al. 2018) and suggests that Chinese breeds have been less intensively selected since their domestication. However, the Chinese indigenous breeds also presented unequal polymorphisms among themselves. Three Chinese indigenous breeds, namely, SG, SK, and BY, exhibited relatively lower PN and higher r2 than those of HS and TB, which might be related to the low-priority conservation status of the HS breed (Zhang et al. 2018). The HS population (sampling time: 2014) raised in the Hubei Academy of Agricultural Sciences has been under conservation (∼11 generations) for a shorter period than either SG (>18 generations), SK (>20 generations), or BY (>20 generations) has been. Noticeably, the TB breed, which showed the highest PN, is still threatened in some areas of its origin, that is, the Qinghai–Tibet Plateau.
In the study of Zhang et al. (2018), relatively low He (0.2073 to 0.2281) and PN (75.83 to 82.41%) were observed in three Chinese chicken breed (BY, Langshan, and Baier chickens). Meanwhile, Chen et al. (2018) found different He (0.26–0.34) in seven Chinese native chicken breeds (Chen et al. 2018). In the present study, five indigenous chickens appeared to maintain relatively high levels of genetic diversity as evidenced with both He (0.2834–0.3171) and PN (68.22%–94.42%).
Chen et al. (2018) has reported similar He (0.29 vs. 0.3012) in SK but different He (0.22 vs. 0.2904) in WL chickens compared with that of our study. Moreover, in the study of Zhang et al. (2018), BY chicken of three different generations showed lower He (0.2091 to 0.2173) than that of our result (He = 0.2834) (Zhang et al. 2018).
Several factors might have contributed to the different results among our study, and those of Zhang et al. (2018) and Chen et al. (2018). First, WL used in our study was chosen from two different populations (table 1), and more sources of samples will provide higher heterozygosity. Second, different generation or conservation schemes in native chicken breeds result in distinct genetic diversity (Zhang et al. 2018). Final, biodiversity always shows different results in different populations; for instance, Baier chickens have shown different heterozygosities in the study of Zhang (0.2159) and Chen’s (0.33). Taken together, it is not surprising that the results of our study are different from those of previous studies.
We used unequal number of samples in some population structure analyses (PCA and NJ tree) for several reasons. First, we performed the preanalysis (relatively balance) using ∼10 samples from each population and all Tibetan chickens (supplementary table S1, Supplementary Material online). The results showed that chicken samples from different populations could be distinguished, except Tibetan chicken sample in the PCA plot (supplementary fig. S4, Supplementary Material online). Second, we conducted the NJ-tree (supplementary fig. S5, Supplementary Material online) analyses. The results revealed the Tibetan chicken population structure more clearly than the PCA analysis did. Taken together, we can use unequal number of samples per population to obtain the same results. However, the use of larger data in the analyses could provide relatively comprehensive results. Therefore, we used all ChIP-seq data in the analysis.
In the present study, the PCA and NJ tree analysis revealed clear genetic divisions separating the Asian, European, and North American chickens. Moreover, the WL-CAU and WL-YQ populations always clustered closely showing that different populations of the same breed have unequal genetic background, but present similar results. Recently, Gholami et al. (2014) reported similar results in different WL and RIR populations using 600-K ChIP SNP data, indicating that genome-wide SNP data can effectively detect population genetic bias (Gholami et al. 2014). In addition, our results indicate that HS and BY are more genetically related to each other than with any other breed, as they always clustered together in both the NJ tree and PCA, and exhibited similar ancestry composition patterns. Furthermore, these two breeds share common phenotypes such as yellow feathers and body size. Taken together, this indicates that the two breeds are likely derived from a common ancestral population.
Linkage disequilibrium, the nonrandom association between pairs of alleles, is influenced by various factors. In the case of domestication, selection is expected to increase LD across the whole genome (Juang and Chiou 2004). The LD decay at a pairwise distance can be used to determine the evolutionary history of populations (Khanyile et al. 2015). Compared with foreign commercial breeds, the lower LD value in Chinese local chicken breeds shows lower selective intensity in China. Moreover, two breeds (WL and TB) decayed faster than the other chicken breeds (fig. 1E: orange and purple lines) were consistent with the findings of a previous study. The LD decays quicker in crossbred or multibreed populations (more sources) than in purebred populations (Fu et al. 2015).
Furthermore, some interesting findings were obtained using the pairwise genetic distance analysis between breeds. The high degree of geographic structure observed here in Eurasian domesticated chicken (r2 = 0.73) strongly suggests that isolation by distance is a powerful force structuring genome-wide variation in intracontinent chicken breeds. Our findings agree with those of a previous study on yellow warbler (migratory bird, r2 = 0.73) (Bay et al. 2018), but differ substantially from those observed in duck populations from China (Zhou et al. 2018), in which very low correlation (r2 = 0.03) between genetic and geographical distances exist. However, we also observed a rapidly reduced genetic–geographical correlation between breeds (r2 = 0.24), with the addition of RIR (North American breed). This result is consistent with the above NJ tree and FST analysis results.
Low-production performance is a well-known common disadvantage in Chinese indigenous chicken breeds. It is therefore foreseeable that Chinese farms are introducing nonnative variants to increase egg production (Bagust 1994). The migration pattern (TreeMix migration edge a in fig. 3), gene flow (f-statistics—supplementary table S3, Supplementary Material online and table 3), and admixture analysis (fig. 1D) obtained in our study strongly support an admixture history in TB breed. Furthermore, sample TB-NM-3 (collected in Nimu, Tibet) was clustered with WL, clearly showing genetic admixture of a commercial breed into TB Chinese indigenous breed. In addition, an impossible white feather phenotype (supplementary fig. S3, Supplementary Material online) observed in TB chicken from our field results seems to support this gene flow hypothesis. Taken together, TreeMix, f-statistics analyses and field results support our conclusion of commercial chicken admixture into Chinese local chicken breeds.
Although we collected experimental data as much as possible, limits on breed’s sources still affect comprehensive explanations in an overall situation. More data from wild populations and other breeds (different continent) are particularly important in future domestication or selection research.
Conclusions
In summary, we collected 1,200 samples of eight chicken breeds from three continents. We estimated the genetic diversity, population structure, and admixture events using a genome-wide SNP analysis. Our results suggest that some Chinese chicken breeds (namely, TB chicken) could be facing a high risk of admixture from European and North American breeds. In addition, the genetic–geographical correlation results showed that isolation by distance plays a critical role in structuring the genomic variation within these Eurasian chicken breeds. Moreover, genetic information provided in this study is valuable resources for production applications (genomic prediction, and breeding strategy) and scientific research (genetic basis detection, studying evolution, or domestication).
Ethics Approval and Consent to Participate
All procedures and protocols involving animals were conducted in accordance with the Guidelines for the Care and Use of Experimental Animals established by the Ministry of Agriculture of China (Beijing, China). All animal work was approved by the Animal Welfare Committee of China Agricultural University (Beijing, China) (Permit Number: XK622).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We gratefully thank two anonymous reviewers for insightful comments during the preparation of this article. We thank all members of the poultry team of the National Engineering Laboratory for Animal Breeding for insightful comments on the analysis and article. We thank Editage for offering professional English language editing to this study. This work was supported by funding from the National Nature Science Foundation of China (31811530296), National Scientific Supporting Projects of China (2015BAD03B03), National Nature Science Foundation of China (31672409), and Beijing Innovation Team of the Modern Agro-industry Technology Research System (BAIC04-2018).
Author Contributions
L.Q. and Z.N. conceived and designed the experiments. L.Q. and C.N. designed and performed bioinformatics analyses. C.N. analyzed the data and wrote the article. P.A. raised the article. Y.J. and H.B. provided partial of samples and genotypes. All authors read and approved the final article.
Data deposition: This project has been deposited at NCBI database under the accession GSE127968. R scripts has been deposited at Protocols.io database under the accession doi: dx.doi.org/10.17504/protocols.io.3xqgpmw.(Accessed 23 June, 2019).
Literature Cited
- Abiye Shenkut A, Sofia M, Johansson AM.. 2015. Genetic diversity of five local Swedish chicken breeds detected by microsatellite markers. PLoS One 10:e0120580.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimu W, et al. 2018. Genetic diversity and population structure analysis of eight local chicken breeds of Southern Xinjiang. Br Poult Sci. 59(6):629–635. [DOI] [PubMed] [Google Scholar]
- Bagust TJ. 1994. Improving health for poultry production in Asia: a developmental perspective. Avian Pathol. 23(3):395–404. [DOI] [PubMed] [Google Scholar]
- Bay RA, et al. 2018. Genomic signals of selection predict climate-driven population declines in a migratory bird. Science 359(6371):83–86. [DOI] [PubMed] [Google Scholar]
- Boettcher PJ, et al. 2010. Objectives, criteria and methods for using molecular genetic data in priority setting for conservation of animal genetic resources. Anim Genet. 41 (Suppl 1):64–77. [DOI] [PubMed] [Google Scholar]
- Carter CA, Li XH, Rozelle SD, Sumner DA.. 1999. Economic reform and the changing pattern of China’s agricultural trade. Working Papers. California Digital Library. [Google Scholar]
- Chen G, et al. 2008. Assessment of population structure and genetic diversity of 15 Chinese indigenous chicken breeds using microsatellite markers. Asian Australas J Anim Sci. 21(3):331–339. [Google Scholar]
- Chen GH, Wang KH, Wang JY, Ding C, Yang N.. 2004. Poultry genetic resources in China. Shanghai (China: ): Shanghai Scientific and Technological Press. [Google Scholar]
- Chen L, et al. 2018. Population genetic analyses of seven Chinese indigenous chicken breeds in a context of global breeds. Anim Genet. 50(1):82–86. [DOI] [PubMed] [Google Scholar]
- Chiang T-Y, et al. 2017. The genetic diversity of chicken breeds from Jiangxi, assessed with BCDO2 and the complete mitochondrial DNA D-loop region. PLoS One 12:e0173192.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian ST. 2004. The evolution of molecular markers—just a matter of fashion? Nat Rev Genet. 5:63–69. [DOI] [PubMed] [Google Scholar]
- Decker JE, et al. 2014. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 10(3):e1004254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2016. Status and trends of animal genetic resources. Commission on genetic resources for food and agriculture. 9th session: CGRFA/WG-AnGR-9/16/Inf.3
- Fu W, Dekkers JC, Lee WR, Abasht B.. 2015. Linkage disequilibrium in crossbred and pure line chickens. Genet Sel Evol. 47:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholami M, et al. 2014. Population genomic analyses based on 1 million SNPs in commercial egg layers. PLoS One 9(4):e94509.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J.. 2017. Isolation of high-molecular-weight DNA using organic solvents. Cold Spring Harb Protoc. (4):pdb.prot093450. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491(7424):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Xu HY, Shen ZN, Zhao CJ, Wu C.. 2018. Genome-wide association analysis reveals novel loci for hypoxia adaptability in Tibetan chicken. Anim Genet. 49(4):337–339. [DOI] [PubMed] [Google Scholar]
- Juang JG, Chiou HK.. 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20:103–111. [DOI] [PubMed] [Google Scholar]
- Kerje S, et al. 2003. The twofold difference in adult size between the red junglefowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim Genet. 34(4):264–274. [DOI] [PubMed] [Google Scholar]
- Khanyile KS, Dzomba EF, Muchadeyi FC.. 2015. Population genetic structure, linkage disequilibrium and effective population size of conserved and extensively raised village chicken populations of Southern Africa. Front Genet. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranis A, et al. 2013. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics 14:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, et al. 2007. Genetic structure of European sheep breeds. Heredity 99(6):620–631. [DOI] [PubMed] [Google Scholar]
- Lujiang Q, et al. 2006. Chinese indigenous chicken breeds using microsatellite markers. Sci China Ser C. 49(4):332–341. [DOI] [PubMed] [Google Scholar]
- Lynch M, Milligan BG.. 1994. Analysis of population genetic structure with RAPD markers. Mol Ecol. 3(2):91–99. [DOI] [PubMed] [Google Scholar]
- Martiniano R, et al. 2017. The population genomics of archaeological transition in west Iberia: investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 13(7):e1006852.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie C, et al. 2016. Genome-wide association study revealed genomic regions related to white/red earlobe color trait in the Rhode Island Red chickens. BMC Genet. 17:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, et al. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, et al. 2007. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim Genet. 38(1):37–44. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11):e1002967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, et al. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 461(7263):489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, et al. 2018. Estimation of linkage disequilibrium and analysis of genetic diversity in Korean chicken lines. PLoS One 13(2):e0192063.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S.. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24(8):1596–1599. [DOI] [PubMed] [Google Scholar]
- Vanliere JM, Rosenberg NA.. 2008. Mathematical properties of the measure of linkage disequilibrium. Theor Popul Biol. 74(1):130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonholdt BM, et al. 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464(7290):898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. 2017. The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol Resour. 17(5):981–990. [DOI] [PubMed] [Google Scholar]
- Wang Q, Pi J, Shen J, Pan A, Qu L.. 2018. Genome-wide association study confirms that the chromosome Z harbours a region responsible for rumplessness in Hongshan chickens. Anim Genet. 49(4):326–328. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Xiang H, et al. 2014. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci U S A. 111:17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chong L, Linsheng Z, Chunjiang HC.. 2007. Neuroglobin mutation associated with hypoxia adaptation in Tibet chicken. Prog Nat Sci Mater Int. 17(12):1419–1424. [Google Scholar]
- Zhang M, et al. 2018. Genomic diversity dynamics in conserved chicken populations are revealed by genome-wide SNPs. BMC Genomics 19:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Leung FC, Chan DK, Yang G, Wu C.. 2002. Genetic diversity of Chinese native chicken breeds based on protein polymorphism, randomly amplified polymorphic DNA, and microsatellite polymorphism. Poult Sci. 81(10):1463–1472. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2016. Parallel evolution of polydactyly traits in Chinese and European chickens. PLoS One 11(2):e0149010.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. 2018. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun. 9:2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.