Atypical hemolytic uremic syndrome (aHUS) is a rare thrombotic microangiopathy (TMA) characterized by the triad of microangiopathic hemolytic anemia, thrombocytopenia, and acute kidney injury (AKI).1,S1–S5 The disease features overactivity of the alternative pathway of the complement system, most often arising from loss-of-function mutations in regulators (Factor H, Factor I, and membrane cofactor protein).S4–S7 Less frequently, a gain-of-function mutation in 1 of 2 activators (C3, Factor B) is identified.S8–S10 Approximately 60% of patients with aHUS carry a rare variant in one of the complement proteins. However, <50% of these rare variants have a known functional consequence or one that is readily predicted from the DNA sequence alteration (such as nonsense, splice-site, or frame-shift mutations).1,S4 Most of the clinically identified variants have not been functionally characterized. Instead, computational prediction algorithms are utilized to predict the potential impact of these variants on the mature protein, taking into consideration the following: (i) evolutionary conservation of an amino acid or nucleotide; (ii) location and context within the protein sequence; (iii) biochemical consequence of the amino acid substitution, and, in some cases, (iv) topology of a previously solved structural domain or subdomain. Based on these criteria, most variants are reported as variants of uncertain clinical significance (VUSs). The presence of such rare variants is particularly vexing for clinical management.
Genetic variants in Factor I (FI) have been reported in 5%–15% of patients with aHUS.2,S4 The likely clinical outcome of patients with an FI mutation is dismal, and the risk of recurrence after kidney transplantation is high.3,S11–S15 FI is a plasma glycoprotein regulator of the alternative pathway of the complement system (Figure 1).4,S16–S18 It is a 2-chain 88-kDa serine protease synthesized predominantly by the liver. FI regulates complement activation by inactivating C3b and C4b through proteolytic cleavage (i.e., cofactor activity) in the presence of one of its “cofactor proteins”—Factor H (FH), membrane cofactor protein (MCP; CD46), C4b-binding protein (C4BP), or complement receptor 1 (CR1; CD35).
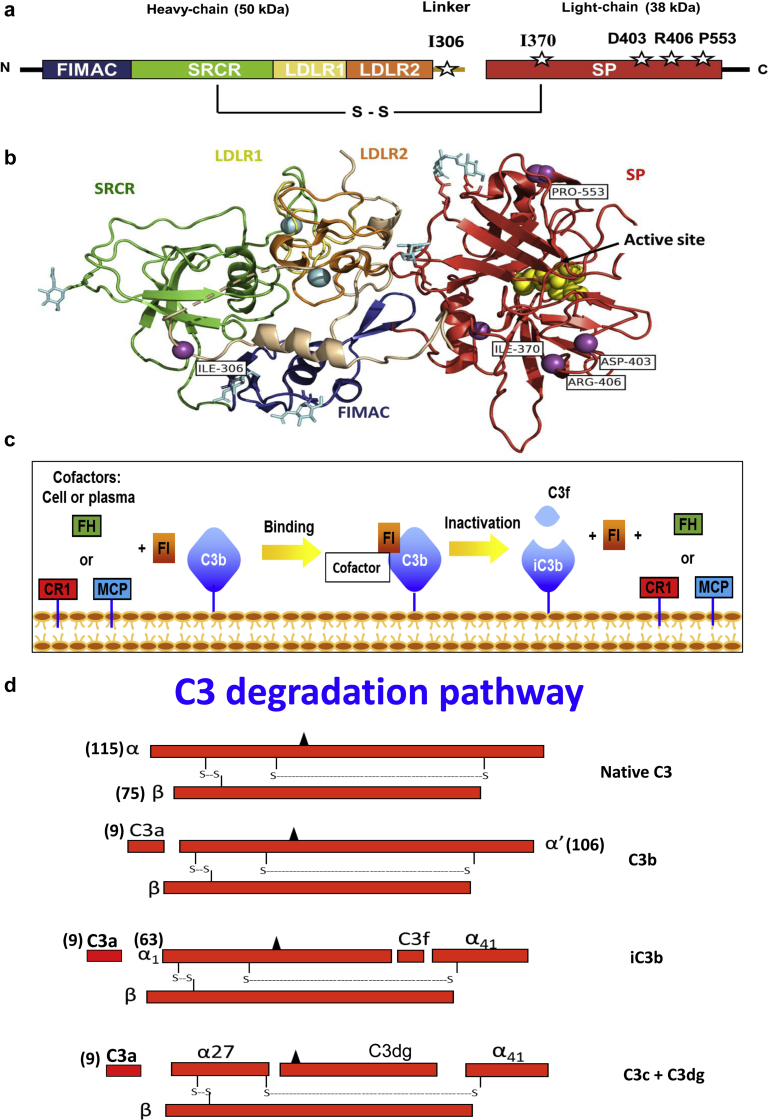
Figure 1.
Structure and function of Factor I (FI). Cartoon schematic of FI. White stars represent the location of the variants (a). Crystal structure of FI (generated using Pymol). Symbols indicate the following: purple circles = variant sites; blue circles = calcium atoms; yellow circles = serine protease (SP) site; light-blue hexagons = N-glycosylation sites. Thick arrow-shaped structures in the various domains are beta-pleated sheets. The dotted beige line in the linker represents a structure that is not visible in the crystal structure, and its location is therefore hypothetical (b). C3b binding and cofactor activity. FI, in the presence of a cofactor protein (factor H [FH], membrane cofactor protein [MCP; CD46], or complement receptor 1 [CR1; CD35]), cleaves the αˈ chain of C3b to obtain α41 and α43 (liberating C3f, a 2-kDa fragment), forming iC3b. Next, CR1 achieves further cleavage of iC3b to C3c and C3dg (c,d). Arg, arginine; Asp, aspartic acid; C, carboxyl-terminus; FIMAC, FI membrane attack complex; Ile, isoleucine; LDLR1, low-density lipoprotein receptor class 1; LDLR2, low-density lipoprotein receptor class 2; N, amino-terminus; Pro, proline; SRCR, scavenger receptor cysteine-rich.
Our study hypothesized that the clinical presentation and pathology of aHUS can be better understood and treated by examining critical clinical information, and by profiling the functional repertoire along with structurally modeling genetic variants. This approach integrates information derived from several types of analyses in order to create a more informed model. The strategy described in this report thus provides a model for how to characterize these rare variants clinically based upon functional evidence, which is one of the interpretive categories of the American College of Medical Genetics (ACMG)5 guidelines for variant interpretations related to pathogenicity. This strategy may be useful for guiding more-appropriate medical therapeutic decisions and generating further insights relative to the pathogenic mechanisms underlying aHUS.
Results
This report describes 5 patients in whom aHUS was considered as a possible diagnosis. Genetic testing revealed a variant in FI (Table 1). These patients were referred to us for assistance in clinical decision-making relative to therapeutic options for renal transplant and/or targeted therapy using a humanized monoclonal antibody that blocks C5 activity (eculizumab).6,S19–S21 In order to obtain a more comprehensive assessment of each patient’s risk, we wanted to evaluate their clinical history and genetic data in relation to a functional assessment of each variant. Our hypothesis was that the additional results would provide critical information for preparing a more rational assessment for individualized therapeutic decisions for each patient. Therefore, we produced recombinant proteins, assessed regulatory function (cofactor activity), and performed structural modeling of each variant protein (see Supplementary Methods, STROBE Statement, and Figure 27).
Table 1.
Summary of clinical data of patients with Factor I variants
| Patient | Variant | Age/sex | Age of onset | Outcomes | Transplant | Treatments | aHUS-risk SNPs | Additional serum complement levels |
|---|---|---|---|---|---|---|---|---|
| 1 | R406H | 22 yr/M | 3 yr | ESRD | Two failed transplants | Eculizumab x 1 year after second transplant | None reported | C3:72 mg/dl C4: 15 mg/dl |
| 2 | I306V | 17 yr/F | 17 yr | CR | None | Plasmapheresis, eculizumab, Cellcept, and Prednisone | FH: V62I, H402Y FI: T300A FB: R32Q, R32W |
C3: 45 mg/dl C4: 6 mg/dl |
| 3 | P553S | 20 yr/F | 19 yr | ESRD | Awaiting transplant | Supportive treatment | FH: V62I, H402Y FI: T300A C3: R102G, P314L |
C3: 103 mg/dl C4: 24.9 mg/dl |
| 4 | D403N | 57 yr/M | 57 yr | CR | None | Eculizumab x 6 years | FH: H402Y FI: T300A C3: R102G, P314L FB: G252S |
C3: 95 mg/dl C4: 34 mg/dl CH50: 50 U/ml |
| 5 | I370N | 24 yr/M | 19 yr | CKD | None | Supportive treatment | FH: V62I, H402Y, E936D FI: T300A C3: R102G, P314L |
Not available |
aHUS, atypical hemolytic uremic syndrome; CKD, chronic kidney disease; CR, complete remission; ESRD, end-stage renal disease; F, female; FB, Factor B; FH, Factor H; FI, Factor I; M, male; SNP, single-nucleotide polymorphisms.
For patients 1, 2, and 3, the reference range for C3 is 82–167 mg/dl; the reference range for C4 is 10–40 mg/dl. For patient 4, the reference range for C3 is 98–201 mg/dl; the reference range for C4 is 18–56 mg/dl; and the reference range for CH50 is 30–75 U/ml.
Figure 2.
Mapping of the Factor I (FI) variants on the surface of C3b in a complex with FI and Factor H (FH; complement control repeats 1–4 and 19–20]. Color coding is as follows: C3b, gray; FH, cyan; serine protease, red; FI membrane attack complex, blue; linker, beige; low-density lipoprotein receptor (LDLR) class 1, yellow; LDLR class 2, orange; scavenger receptor cysteine-rich, green. FI domains are color coded to match those in Figure 1. Refer to the text for further explanation of the structural evaluation for each variant. D, aspartic acid; H, histidine; I, isoleucine; N, asparagine; P, proline; R, arginine; S, serine; V, valine. Adapted with permission from Forneris F, Wu J, Xue X, et al. Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 2016;35:1133–1149.7 Copyright © 2016 The Authors. Published under the terms of the CC BY-NC-ND 4.0 license.
Case 1
This 22-year-old Caucasian male developed end-stage renal disease at the age of 3 years, thought to be secondary to Escherichia coli–triggered HUS. In 2006, he underwent a renal transplant at age 11 years. In 2007, he developed renal failure due to Banff 1B acute cellular rejection and returned to dialysis in 2009. He was evaluated for a second renal transplant in 2014. Given his history of HUS, genetic testing was performed, which showed a missense variant (p.R406H) in FI. The patient received eculizumab for 1 year after his second transplant in May 2015. In April 2017, an allograft biopsy for worsening renal function again revealed Banff 1B acute cellular rejection, in addition to antibody-mediated rejection. As of this writing, he is undergoing evaluation for a third renal transplant.
Genetic Analysis
The patient’s heterozygous variant is within the serine protease domain and occurs at an amino acid position that is not conserved. This variant has been reported in patients with age-related macular degeneration,S22 and at the time the variant was identified in this patient, the frequency in the general population was reported at 0.12%. Due to the availability of larger cohorts in gnomAD, this variant is now reported to occur at an overall frequency of 1.6% (minor allele frequency: Asians, 2.4%–4.3%; Europeans, 0.1%–2.4%; Africans and Latinos, <1%). Several in silico models (SIFT, PolyPhen2, Mutation Assessor, Condel) predict this variant to be benign; taken together, the clinical interpretation reported for this variant was “likely benign.”
Antigenic Analysis
The secretion of the recombinantly produced variant protein in the supernatant (determined by enzyme-linked immunosorbent assay) was comparable to wild type (WT, 2.3 ± 0.75 μg/ml; R406H, 2.1 ± 0.97 μg/ml). The serum FI antigenic level in this patient was normal: 31 μg/ml (reference range for the outside laboratory: 24–49 μg/ml).
Functional Analysis
This FI variant had defective proteolytic activity with FH and CR1 as cofactor proteins, as evidenced by a decreased rate of cleavage of the αʹ chain of C3b and thereby reduced generation of the α1 fragment compared to WT (Figure 3). P values for the difference in functional activity between the WT and variant were statistically significant. No defect was observed with MCP as the cofactor protein.
Figure 3.
Functional evaluation of Factor I (FI) variant R406H: cofactor activity. The fluid-phase C3b cofactor activity of the variant FI with its cofactor proteins (Factor H [FH], membrane cofactor protein [MCP], or complement receptor 1 [CR1]) was assessed by cleavage of purified C3b to iC3b and compared to wild type (WT). The percentage of alpha chain remaining and the generation of an α1 fragment indicates cleavage of C3b to iC3b. A kinetic analysis of cofactor activity was conducted at time 0, 10, 20, and 30 minutes. Cleavage rate was measured by densitometric analysis of the alpha chain remaining or the generation of α1 relative to the β chain. Representative Western blot for FH, MCP, and CR1 (a–c). Densitometric quantification of the Western blot for FH, MCP, and CR1 (d–f). Data represent 3 separate experiments with bars corresponding to the SEM. Upon comparison to WT FI, the cofactor activity of variant R406H is defective with both FH and CR1. For FH, the P value for the difference in the percentage of alpha chain remaining between WT and variant was 0.05, and for the difference in α1 generation, it was 0.0037. For CR1, the P value for the difference in the percentage of alpha chain remaining was 0.028, and for the difference in α1 generation, it was 0.0081. No defect was observed with MCP as the cofactor protein. H, histidine; R, arginine.
Structural Analysis
The side chain of R406 in FI forms a salt bridge with the carboxyl group of E123 of FH7,S23 (Figure 2). This interaction stabilizes the FI–FH binding interface. Residue E123 is a unique signature of FH and is lacking in MCP, possibly accounting for the fact that the R406H mutation impaired FH- but not MCP-mediated cofactor activity. The effect of the mutation on CR1 cofactor activity is more difficult to interpret because residue E123 is also absent in CR1, being replaced by a histidine. However, given that a histidine residue can be protonated under physiological values of pH and ionic strength conditions,S24 a histidine–arginine like-charged ion pair is expected to occur at the FI–CR1 binding interface.
Implications
Although this variant was synthesized normally, it had a reduced rate of C3b cleavage by FH and CR1 compared to WT FI. This variant is therefore “functionally impaired.” However, taken together with the frequency of the p.R406H variant, which is more common than would be expected based on the prevalence of aHUS, lack of consistency between in silico tools and now our data supporting pathogenicity, this variant is being reclassified as a VUS according to ACMG guidelines. Of note, although glomerular endothelial cellsS25 do not express CR1 in the resting state, there is evidence that hypoxia or inflammatory mediators may induce the expression of this CR1 on endothelial cells. This phenomenon could therefore be of significance during disease states such as aHUS. We also know that aHUS is a disease with incomplete penetrance and that the penetrance is age-related, being reported as being as high as 65% by age 70 years for individuals carrying a single genetic mutation.S26 Therefore, although this patient did not develop TMA, given his clinical course with end-stage renal disease secondary to HUS, 2 allograft rejections, and our newly derived data, we believe that the patient is at risk of developing a TMA and/or an accelerated antibody-mediated rejection. Although future studies will affect the decision on how long to continue treatment, we would recommend that he receive eculizumab at the time of his third transplant and remain on it indefinitely. Additionally, we would advise that his family members undergo genetic testing.
Case 2
This 17-year-old African American female presented with pain and swelling in her extremities along with shortness of breath and palpitations. Laboratory evaluation revealed mild anemia, elevated erythrocyte sedimentation rate, multiple positive autoantibodies (anti-Ro, anti-La, Smith), and antinuclear antibodies of 1:640. She was treated with high-dose steroids for possible systemic lupus erythematosus, and her symptoms improved. A year later, she presented with altered mental status, slurred speech, and shortness of breath due to septic shock secondary to a pneumonia. Over the next 3 days, she developed AKI, worsening anemia, thrombocytopenia, elevated lactate dehydrogenase, and a low haptoglobin level. A diagnosis of thrombotic thrombocytopenic purpura/aHUS was considered, and treatment with plasmapheresis and eculizumab was initiated. Test results for ADAMTS13 levels came back normal. Renal biopsy demonstrated a TMA. Genetic workup identified a missense variant (p.I306V) in FI. She had complete renal and hematologic recovery after 6 weeks. Eculizumab was discontinued and she was treated with mycophenolate mofetil.
Genetic Analysis
This heterozygous variant is in the serine protease–linker domain of FI. It occurs at an amino acid position that is not highly conserved and has a minor allele frequency of ∼0.5% within the African American population. In silico tools predict a benign effect on the translated protein. This particular variant has been identified in a patient in the setting of de novo TMA following kidney transplantation.8 On the basis of these collective findings, the variant is categorized as a VUS.
Antigenic Analysis
The secretion of the recombinantly produced variant protein in the supernatant (determined by enzyme-linked immunosorbent assay) was comparable to WT (WT, 2.2 ± 1.07 μg/ml; I306V, 2.7 ± 0.5 μg/ml). The serum FI antigenic level in our patient is not available. However, the serum FI level in the patient previously reported in the literature was normal: 61 μg/ml (reference range for the outside laboratory: 42–78 μg/ml).
Functional Analysis
The variant had a normal degree of activity with all 3 cofactors (FH, MCP, and CR1) (Figure 4a). P values for the difference in degradation of the αʹ chain of C3b and for the generation of the α41 and α43 fragments between WT and variant were not statistically significant.
Figure 4.
Cofactor activity of I306V (a), P553S (b), and D403N (c), compared to wild type. Densitometric quantification of the Western blot for percentage of alpha chain remaining for wild type and each variant. Data represent 3 separate experiments with bars corresponding to the SEM. No significant differences were observed. Due to the limited cleavage of C3b with D403N using membrane cofactor protein (MCP) as the cofactor, we conducted the experiment under low ionic strength conditions (25 mM NaCl) and obtained the same results (see Supplementary Figure S1). CR1, complement receptor 1; D, aspartic acid; FH, Factor H; I, isoleucine; N, asparagine; P, proline; S, serine; V, valine.
Structural Analysis
In the crystal structure of C3b-FH-FI, I306 is located >30 Å and 57 Å away from the FI–C3b and FI–FH binding interfaces, respectively. Hence, I306 is not involved in protein–protein interactions (Figure 2). Furthermore, Ile to Val is a conservative mutation, usually well tolerated by the protein fold. On these structural bases, the I306V mutation is expected to have no detrimental effects on cofactor activity.
Implications
This variant is of “no functional consequence” based on normal expression and cofactor activity. Given our current findings, in silico tools that support a benign effect of this variant on FI, the frequency among those with African ancestry, this variant would be reclassified as likely benign per ACMG guidelines. The patient remains stable on mycophenolate mofetil and off anti-complement therapy. We believe that the TMA was most likely secondary to lupus or infection and was not related to the variant in FI.
Case 3
This 20-year-old Caucasian female developed preeclampsia during her second pregnancy and underwent a cesarean section. The postpartum course was complicated by the development of anemia (Coombs negative hemolytic anemia), thrombocytopenia, and AKI. Over the next few days, her hematologic parameters improved, but her kidney function did not recover and she progressed to end-stage renal disease. A differential diagnosis of HELLP (hemolysis, elevated liver enzymes, low platelets) versus aHUS was considered. A year later, she was referred to us for evaluation for a kidney transplant. In order to elucidate a more definitive cause of the kidney failure (that is, to determine if the patient had aHUS and pregnancy was a likely second hit) and to further define risk of recurrence after transplant, genetic testing was performed. A missense variant (p.P553S) in FI was identified.
Genetic Analysis
This heterozygous variant is in the serine protease domain. It has been described in age-related macular degeneration and is more common in controls than patients (minor allele frequency of 0.2 vs. 0.06). The variant has also been reported in a 30-year-old patient with end-stage renal disease from aHUS9; however, that patient carried variants in FH (p.R1210C) and in MCP (p.Y29X; see Implications section). The variant p.P553S is predicted to be “likely benign” by the computational programs.
Antigenic Analysis
The secretion of the recombinant variant protein in the supernatant (determined by enzyme-linked immunosorbent assay) was comparable to WT (WT, 3.1 ± 0.5 μg/ml; P553S, 2.1 ± 0.28 μg/ml). The serum FI antigenic level in this patient was normal at 34 μg/ml (reference range: 24–49 μg/ml).
Functional Analysis
This variant had normal activity with FH, MCP, and CR1 serving as the cofactor protein (Figure 4b). There was no statistically significant difference in the cleavage of the αʹ chain or in the production of the α1, α41, and α43 fragments, compared with WT.
Structural Analysis
P553 is located >20 Å from the catalytic triad and is part of the short loop exposed to the solvent. P553 is part of neither the substrate-enzyme nor the cofactor-enzyme binding interfaces (Figure 2).
Implications
This variant is of “no functional consequence” based on normal expression and proteolytic activity. Considering the findings of these studies, ACMG now classifies this variant as benign. Although FI mutations have been identified in patients with preeclampsia,S27 and evidence from previous experiments has shown that dysregulation of complement activation is involved in the development of preeclampsia, this does not seem to be true for this patient. In the previously reported case of aHUS with this variant, the patient also carried the pathogenic variants, p.R1210C in FH and p.Y29X in MCP.9 The combination of R1210C and Y29X was likely synergistic in manifestation of disease in the previously reported patient. In our patient, however, it is unlikely that P553S is causative of the renal disease.
Case 4
This 57-year-old Caucasian male presented with generalized fatigue and weakness ongoing for 10 days. The patient was found to be in septic shock secondary to a hepatic abscess. Three days later, he experienced a rapid critical decline characterized by AKI requiring dialysis, hemolytic anemia, and profound thrombocytopenia. A diagnosis of thrombotic thrombocytopenic purpura/aHUS was made, and he was treated with plasmapheresis and eculizumab, with complete hematologic and renal recovery over the next 6 weeks. Test results for ADAMTS13 level came back normal. Genetic testing revealed a rare variant (p.D403N) in FI. He remains on eculizumab (for the past 6 years).
Genetic Analysis
This heterozygous variant is in the serine protease domain and is present in population databases at very low allele frequencies (0.008% in NHLBI Exome Variant server and 0.00003% in the ExAC browser). In silico models predict a benign effect on protein function. This variant was also identified in 1 of 202 patients with aHUS in a French cohort and was absent from 100 healthy controls.9 Based on the limited evidence available, this variant is classified as a VUS.
Antigenic Analysis
The secretion of the variant in the supernatant (determined by enzyme-linked immunosorbent assay) was comparable to WT (WT, 4.8 ± 0.55 μg/ml; D403N, 3.8 ± 0.97 μg/ml). Factor I antigenic level in our patient was normal at 28.3 μg/ml (normal range: 16–40 μg/ml).
Functional Analysis
The variant had normal activity with the 3 cofactors analyzed (Figure 4c and Supplementary Figure S1). Compared to WT, there was no statistically significant difference in cleavage of the αʹ chain of C3b or in the production of the α1, α41, or α43 fragments.
Structural Analysis
D403 is located just 3 amino acids downstream of R406 (Case 1; Figure 2). Unlike R406, the side chain of D403 does not make direct contact with FH. The mutation D403N replaces a hydroxyl with an amino group, which retains the H-bonding capabilities. This substitution is, therefore, expected to have minimal impact on the structural properties of the loop and thus on cofactor activity.
Implications
This variant is of “no functional consequence” based on normal expression and proteolytic activity with all 3 cofactor proteins and a normal FI antigenic level. Although this variant would be expected to be classified as benign, this variant remains a VUS due to its rarity in the general population. The patient has been stable on eculizumab for 6 years. Although categorization of a variant as VUS is not recommended for use in clinical decision-making, given these new functional data, a trial of eculizumab may be attempted along with close follow-up and monitoring.
Case 5
This 24-year-old Caucasian male underwent a heart transplant at the age of 9 years for arrhythmogenic ventricular dysplasia. He developed AKI at the age of 19 years. Laboratory values (platelet count 40,000/μl, creatinine 1.5 mg/dl [baseline 0.9 mg/dl] and presence of schistocytes on peripheral smear) were concerning for aHUS, but a precipitating event was not apparent. During the course of his hospitalization, his creatinine level did not improve and he remained thrombocytopenic. Genetic testing revealed the presence of a novel variant (p.I370N) in FI.
Genetic Analysis
This heterozygous novel missense variant is in the serine protease domain and occurs at an amino acid position that is highly conserved. In silico algorithms predict this change to be deleterious to protein function. The p.I370N variant is not reported in population variant databases. On the basis of this evidence, this variant was characterized as “likely pathogenic.”
Antigenic Analysis
This variant was synthesized but was not secreted, as evident from the Western blot analysis of the cell lysates and the supernatants (Figure 5). The serum FI antigenic level in this patient was not available.
Figure 5.
Biosynthetic evaluation of I370N. Western blots of supernatants and cell lysates from transfected wild type and I370N are shown. Supernatants: the reduced blot for wild type demonstrates a band at 50 kDa that is the heavy chain, and a band at 38 kDa that is the light chain. The nonreduced blot shows a single band of full length Factor I (FI) at ∼88 kDa. No bands are seen for I370N, indicating that the protein either is not synthesized or not secreted. Purified FI differs slightly in molecular weight from the recombinant wild type (particularly under non-reducing conditions), most likely secondary to a difference in the glycosylation (a). Lysates: the reduced and nonreduced blots show a predominant single band at 88 kDa. This represents Pro-FI. Mutant protein is observed in the lysate, indicating that the protein is synthesized but is not secreted, as it is absent in the supernatants. The polyclonal antibody to FI predominantly binds to the heavy chain at the expected molecular weight (50 kDA) but variably detects the light chain under reducing conditions (b). I, isoleucine; N, asparagine.
Structural Analysis
Ile370 is buried into a hydrophobic pocket (Figure 2) surrounded by I368, Y369, L376, V454, V540, F577, and I569. Introduction of a polar amino acid such as an asparagine is expected to disrupt the hydrophobicity of this pocket, thereby leading to protein instability and misfolding.
Implications
Based on these functional studies showing defective secretion and prior supporting information, this novel variant is reclassified as pathogenic per ACMG. The patient is at high risk of recurrent aHUS. Of note here is that the patient did not develop aHUS at the time of the heart transplant, which would be considered a precipitating event, but rather developed the disease at a later age without a clear trigger. This patient should be treated with eculizumab. Unfortunately, this patient has been lost to follow up.
Discussion
Recent technological advances in genetic testing have enabled laboratories to expand the data obtained from DNA analyses. However, correct functional interpretation of such analyses continues to be a challenge.S28 As a result, the clinical significance of many genetic variants remains unresolved and raises a number of problematic questions. Should they inform clinical management? What follow-up studies should be performed? Should the data be disclosed to patients and how should the patients be counseled? In an attempt to help resolve such issues, the ACMG has issued guidelines for clinicians.5 The ACMG’s variant classification guidelines state that “A variant of uncertain significance should not be used in clinical decision making. Efforts to resolve the classification of the variant as pathogenic or benign should be undertaken through further testing.”
One way to further address this issue is to test family members to determine whether variants are shared by other affected and unaffected relatives. In our cases, the family histories were negative for a similar syndrome. Also, even if suggestive, this approach can be time consuming and labor intensive, and it relies on family availability, cooperation, and consent. Additionally, insurance companies refuse to pay for this testing. Another approach is to model the genetic variant using mutagenesis, protein expression, and structure–function analyses to more definitively identify the clinical implications of each genetic variant.
Consequently, we performed analyses of the 5 variants in FI identified in aHUS (2 were VUS; 2 were likely benign; and 1 was likely pathogenic). Using these “real-world” cases, we obtained functional and structural data on each variant. The results were utilized to reclassify the variants per the ACMG guidelines (Table 2).8 They changed the clinical interpretation for 4 of the 5 variants.
Table 2.
Summary of genetic and functional analyses of Factor I variants in aHUS
| Variant | dbSNP ID | g. syntax | Domain | Serum levels | Recombinant secretion | CA (FH) | CA (MCP) | CA (CR1) | Previous ACMG interpretation | Modified ACMG interpretation |
|---|---|---|---|---|---|---|---|---|---|---|
| R406H | rs74817407 | chr4: g.110667590C>T | SP | NL | NL | DF | NL | DF | LB | VUS |
| I306V | rs113273712 | chr4: g.110673648T>C | LNK | NAa | NL | NL | NL | NL | VUS | LB |
| P553S | rs113460688 | chr4: g.110662144G>A | SP | NL | NL | NL | NL | NL | LB | Benign |
| D403N | rs139881195 | chr4: g.110667600C>T | SP | NL | NL | NL | NL | NL | VUS | VUS |
| I370N | novel | chr4: g.110670413A>T | SP | NA | Protein synthesized; not secreted | ND | ND | ND | LP | Pathogenic |
ACMG, American College of Medical Genetics; aHUS, atypical hemolytic uremic syndrome; CA, cofactor activity; CR1, complement receptor 1; dbSNP, database of single-nucleotide polymorphisms; DF, defective; FH, Factor H; LB, likely benign; LNK, linker; LP, likely pathogenic; MCP, membrane cofactor protein; NA, not available; ND, not done; NL, normal; SP, serine protease; VUS, variant of uncertain significance.
Normal in the one other case reported in Le Quintrec et al.8
Putative limitations to our analyses are as follows: (i) Incomplete penetrance of a variant can confound the standard ACMG guideline interpretation. For example, despite our functional data demonstrating no functional defect, the interpretation for p.D403N remained a VUS due to the rarity of the variant. Investigating the effect of this variant on the cell surface might be additionally instructive. (ii) Assays could also incorporate the cofactor for C4b cleavage, i.e., C4b-binding protein. However, rather than the classical pathway, it is the alternative pathway that mediates aHUS. (iii) The relative levels of the fully functional form of FI versus the level of the rare variant are unknown. (iv) Longer-term follow-up is needed to assess the outcome of the clinical course for each patient. (v) Given that these functional studies were not conducted in a College of American Pathologists (CAP)–accredited and Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory, routine use of these methods to facilitate clinical decision-making will require substantial investment in developing high-throughput assays.
Nevertheless, our results identify a useful model (Figure 6) illustrating how a sequential and systematic analysis can provide more accurate investigation of the clinicopathologic significance of genetic variants in aHUS. Additional studies of complement parameters, including, for example, activation fragments, could add important information to inform therapeutic decisions. Such a model provides a means to further define an individualized treatment plan as well as the risk of recurrence after a kidney transplant.
Figure 6.
Diagnostic algorithm for atypical hemolytic uremic syndrome (aHUS). ab, antibody; ADAMTS13, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13; C3Nef, C3 nephritic factor; FB, Factor B; FH, Factor H; FI, Factor I; MCP, membrane cofactor protein; STEC, Shiga toxin–producing Escherichia coli; TTP, thrombotic thrombocytopenic purpura.
In conclusion, given the current challenges to the clinician of reconciling genetic data with clinical management of aHUS, we believe our strategy of recombinant protein production followed by detailed functional and structural assessments is informative. The ability to more comprehensively define the functional repertoire of the variant protein provides critical guidance relative to the appropriate therapeutic regimen and thereby will ultimately pave the way for precision medicine in complement-mediated renal disease.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The authors thank Drs. Daniel C. Brennan (Johns Hopkins University) and Brent W. Miller (Indiana University) for their guidance; Drs. Anitha Vijayan and Tingting Li (both of Washington University School of Medicine) for their helpful suggestions during article preparation; and Drs. Corrine Benchimol (Mount Sinai Hospital, New York), Margaret S. Ray (Santa Barbara Cottage Hospital), and Ilene Weitz (Keck School of Medicine at the University of Southern California) for their assistance in providing further details on the clinical history for Cases 2, 4, and 5, respectively.
This study was supported by the Barnes-Jewish Hospital Foundation Fund (AJ) and U54 HL112303-03 (Sadler, Evan, Genetic Predisposition to the Thrombomicroangiopathies; JPA, Director of Genetics and Genomics Core).
This work was presented in the International Complement Workshop at Santa Fe (New Mexico) in September 2018, and the abstract was published in Molecular Immunology, 2018;102:166–167.
Author Contributions
AJ and JPA conceived and designed the research, analyzed data, and interpreted results of the experiments. AJ, ZH, LMC, PB, and ZR performed experiments. NP conducted the structural analysis. YJS performed the statistical analysis. LLG and JWH conducted ACMG re-interpretations. AJ prepared figures and drafted the manuscript. AJ, NP, JPA, MKL, LLG, and JWH edited and revised the manuscript.
Footnotes
Supplementary Methods.
Figure S1. Functional evaluation of D403N with MCP using 25 mM salt buffer.
Supplementary References.
STROBE Statement.
Supplementary material is linked to the online version of the paper at www.kireports.org.
Supplementary Material
Functional evaluation of D403N with MCP using 25 mM salt buffer.
References
- 1.Goodship T.H., Cook H.T., Fakhouri F. Atypical hemolytic uremic syndrome and C3 glomerulopathy: conclusions from a “Kidney Disease: Improving Global Outcomes“ (KDIGO) Controversies Conference. Kidney Int. 2017;91:539–551. doi: 10.1016/j.kint.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Kavanagh D., Richards A., Noris M. Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol. 2007;45:95–105. doi: 10.1016/j.molimm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Le Quintrec M., Zuber J., Moulin B. Complement genes strongly predict recurrence and graft outcome in adult renal transplant recipients with atypical hemolytic and uremic syndrome. Am J Transplantation. 2013;13:663–675. doi: 10.1111/ajt.12077. [DOI] [PubMed] [Google Scholar]
- 4.Roversi P., Johnson S., Caesar J.J. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc Natl Acad Sci U S A. 2011;108:12839–12844. doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nurnberger J., Philipp T., Witzke O. Eculizumab for atypical hemolytic-uremic syndrome. N Engl J Med. 2009;360:542–544. doi: 10.1056/NEJMc0808527. [DOI] [PubMed] [Google Scholar]
- 7.Forneris F., Wu J., Xue X. Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 2016;35:1133–1149. doi: 10.15252/embj.201593673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Quintrec M., Lionet A., Kamar N. Complement mutation-associated de novo thrombotic microangiopathy following kidney transplantation. Am J Transpl. 2008;8:1694–1701. doi: 10.1111/j.1600-6143.2008.02297.x. [DOI] [PubMed] [Google Scholar]
- 9.Bienaime F., Dragon-Durey M.A., Regnier C.H. Mutations in components of complement influence the outcome of Factor I-associated atypical hemolytic uremic syndrome. Kidney Int. 2010;77:339–349. doi: 10.1038/ki.2009.472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional evaluation of D403N with MCP using 25 mM salt buffer.