Abstract
Chondrosarcoma is a malignant tumor that arises from cartilaginous tissue and is radioresistant and chemoresistant to conventional treatments. The preferred treatment consists of surgical resection, which might cause severe disabilities for the patient; in addition, this procedure might be impossible for inoperable locations, such as the skull base. Carbon ion irradiation (hadron therapy) has been proposed as an alternative treatment, primarily due to its greater biological effectiveness and improved ballistic properties compared with conventional radiotherapy with X-rays. The goal of this study was to characterize the genetic mutations of a grade III chondrosarcoma cell line (CH2879) and examine the cellular responses to conventional radiotherapy (X-rays) and hadron therapy (proton and carbon ions) in the presence of the PARP inhibitor Olaparib. To better understand PARP inhibition, we first analyzed the formation of poly-ADP ribose chains by western blot; we observed an increase in its signal after irradiation, which disappeared on addition of the PARP inhibitor. PARPi enhanced ratio of approximately 1.3, 1.8, and 1.5 following irradiation of cells with X-rays, protons, and C-ions, respectively, as detected by clonogenic assay. The decrease in cell survival was confirmed by proliferation assay. The radiosensitivity of CH2879 cells was associated with mutations in homologous recombination repair genes, such as RAD50, SMARCA2 and NBN. This study demonstrates the capacity of the PARP inhibitor Olaparib to radiosensitize mutated chondrosarcoma cells to conventional photon irradiation, proton and carbon ion irradiation.
Keywords: PARP inhibitor, Olaparib, Chondrosarcoma, Carbon ions irradiation, Mutation status
Graphical abstract

1. Introduction
Emerging radiotherapy protocols with protons or heavier particles, such as carbon ions (C-ions), in advanced medical facilities have revolutionized our views on local tumor control and the impact on healthy tissues [1], [2], [3], [4]. Particle therapy (hadron therapy) with protons has the advantage of a minimal exit dose after energy deposition in the target volume and thus greater sparing of critical structures in the vicinity of the tumor [5], [6], [7], [8]. Moreover, hadrontherapy with C-ions is an attractive radiation modality that combines the physical advantages of protons with a higher radiobiological effectiveness and less dependence of radiation sensitivity on the cell cycle and oxygen [5], [8], [9]. Because the high linear energy transfer (LET) of C-ion irradiation is densely ionizing, the resulting DNA damage in a cell occurs more often, making it more difficult for the cell to repair it, increasing the efficiency of killing such cells that are typically resistant to conventional radiotherapy [2], [8], [10]. Unlike photons, high LET charged particles form a Bragg peak, a sharp rise in energy deposition at the end of their range, with a steep decline in dose downstream [5]. Ion beam therapy with protons or C-ions is beneficial for the treatment of radiation-resistant tumors and was recently established as the first-line treatment for chondrosarcoma [11], [12], [13], [14]. According to an expert committee of the European Science Foundation (the Nuclear Physics European Collaboration Committee), the highest priority for hadrontherapy is given to those who are affected by chordomas/chondrosarcomas of the skull base, soft tissue and bone sarcomas, and large uveal and mucosal melanomas and most pediatric patients who are eligible for radiotherapy [15].
Sarcomas are a diverse group of tumors that arise from connective tissue that is mesenchymal in origin [16]. These tumors, which can affect almost any part of the body, have high rates of mortality (poor prognosis), metastasis, and recurrence. Histologically, they are divided in 2 groups: soft tissue sarcomas and bone sarcoma (chondrosarcoma, osteosarcoma, chordoma, and Ewing sarcoma). In the latter, chondrosarcoma develops from cartilage and produces a significant amount of extracellular matrix [17]. If the primary chondrosarcoma is uncommon, secondary chondrosarcoma can emerge from benign cartilage defects, such as osteochondroma and enchondroma. Surgery remains the primary treatment for chondrosarcoma, when it is anatomically possible, but it often causes major disabling sequelae [18]. Although local control is generally high with hadron therapy, for most malignancies, radiotherapy must be combined with systemic therapies to control metastasis and increase survival [19]. However, few radiobiology studies have specifically examined the potential synergy between drugs and ion irradiation. Emerging drugs, such as DNA repair protein inhibitors, are promising, especially when coupled with the specific mutation status of the cancer cells that are to be treated.
The development PARP inhibitors accelerated when the concept of synthetic lethality appeared in homologous recombination-deficient cells [20], [21], [22], wherein the targeted proteins, poly (ADP-ribose) polymerases (PARPs), detect damaged DNA and then activate signaling pathways that promote the appropriate cellular responses [23]. Cancer cells with impaired capacity to repair double-strand DNA breaks by homologous recombination (with defects in BRCA1 and BRCA2) are highly sensitive to inhibitors of PARP that block the repair of single-strand DNA breaks [24], [25]. PARPs are involved in base excision repair (BER), mediating the recruitment and activation of BER proteins and consequently facilitating the single-strand break (SSB) repair [26]. PARP1, 2, and 3 are equally involved in other cellular mechanisms, such as chromatin remodeling or DNA double-strand break repair [23]. Homologous recombination repair (HRR) and non-homologous end joining (NHEJ) are the main types of repair mechanisms for double-strand breaks (DSBs). HRR is an accurate but slow process that incorporates a homologous DNA sequence and occurs exclusively during the S and G2 phases of the cell cycle. In contrast, NHEJ is rapid and imprecise, wherein broken DNA ends are joined directly. Whereas HRR normally predominates in the repair of DSBs, its deficiency (driven by defects in BRCA1, BRCA2, or other pathway components) often stimulates NHEJ. Consequently, some of the genetic alterations that are introduced by NHEJ promote carcinogenesis by cancer-driver genes.
Several PARP inhibitors are being evaluated in clinical trials [27], [28], [29]. The loss of DNA repair capacity in the presence of these inhibitors has led to their evaluation as single agents and as enhancers of cytotoxic agents that provoke DNA damage, such as alkylating agents and radiation therapy [30]. Olaparib was the first PARP inhibitor to gain US FDA approval and has been examined in phase I/II/III studies as monotherapy in BRCA1/2-mutated tumors in combination with cytotoxic chemotherapy for breast cancer, ovarian cancer, and BRCA-like tumors (tumors with dysfunctional homologous recombination that is not caused by BRCA1/2 mutations—ie, BRCAness) and with radiotherapy for several BRCA-mutated and tumors with BRCAness [21], [22], [31]. Olaparib (trade name Lynparza, formerly known as AZD2281) was initially developed by Kudos Pharmaceuticals and was later acquired by AstraZeneca (Macclesfield, UK). This compound is a pan-PARP inhibitor, because it has inhibitory activity against PARP-1, PARP-2, and PARP-3. PARP-1 accounts for nearly 90% of total PARP activity and is an important protein in DNA base excision repair and the repair of DNA single-strand breaks (SSBs). PARP-1 binds to SSBs and catalyzes the polymerization of ADP-ribose (PARylation), resulting in the production of polymers of ADP-ribose (PAR). We have reported that the combination of depletion of PARP-1 and C-ion exposure sensitizes HeLa cell and induces apoptosis to a greater extent than C-ion exposure alone [32].
PARP is also emerging an important factor in metastasis, perhaps explaining why the PARP inhibitor Olaparib has shown promising results against various metastatic cancer cells in clinical trials. Our previous report demonstrated that PARP-1 inhibition synergizes with the C-ion-induced reduction in metastatic potential in HeLa cells [32]. Based on these findings, a better understanding of the molecular abnormalities in BRCA-like tumors can guide the development of novel therapeutic strategies and drug combinations [31], [33], [34]. According to previous analyses, we showed that PARP inhibitors can induce a radiosensitization in several different chondrosarcoma cell lines [35].
To go further, in this study, we evaluated the radiosensitizing effects of Olaparib on a Grade 3 chondrosarcoma cell line (CH2879 [36]), in association with conventional low-LET radiotherapy and high-LET particle therapy, such as carbon ion beam. In relation with a radiotherapy context, we focused our interest on cell growth and cell divisions (tumour control), following irradiations with or without Parp inhibitors using clonogenic assays and CellTrace strategies, to get an accurate vision of the behavior of each cell following treatment. Our results reveal the ability of Olaparib to radiosensitize this chondrosarcoma cell line with X-rays, protons, and C-ions exposure.
2. Materials and methods
2.1. Cell culture
The CH2879 chondrosarcoma cell line [36] was initiated from a primary grade III chondrosarcoma of the chest wall from a 35-year-old female and cultured in RPMI 1640 (Merck KGaA, Darmstadt, Germany), supplemented with 10% fetal calf serum (Merck KGaA, Darmstadt, Germany), 2 mM glutamine (Merck KGaA, Darmstadt, Germany), and 1% antibiotics (penicillin-streptomycin, Merck KGaA, Darmstadt, Germany), at 37 °C in a humidified atmosphere with 5% CO2.
2.2. Irradiation
For the X-rays irradiation, doses of between 0.5 and 2 Gy were used. The photon beam was delivered with a tube tension of 225 kV and an intensity of 10 mA, corresponding to a dosage of 2 Gy/min, using a Pxi XradSmart 225cX irradiator.
Proton irradiation experiments were performed using the clinical 62 MeV beams at the INFN-LNS (INFN-LNS, Catania, Italy) proton therapy facility. The beam reached the sample with a circular spot, 30 mm in diameter, and a dose homogeneity of over 2%. Reference dosimetry was conducted using a Markus-type, free-air ionization chamber that was positioned in the middle of a clinical spread out Bragg peak (SOBP) per the IAEA TRS 398 Code of Practice. The overall precision in the absolute dose was estimated to be 3%. To irradiate the samples with protons at a fixed LET of 11 keV/µm, a calibrated and certified 9-mm-thick plastic slab was inserted between the final beam collimator and sample holder. The average dose-LET distribution at that position was estimated by Geant4 Monte Carlo simulation of the beamline. An automatized and remotely controlled device that was connected to the dosimetry and beam control system of the facility allowed accurate irradiation of the samples.
For C-ions irradiations, experiments were performed at GANIL (Caen, France), using the IRABAT beam line, according to [37] with a native 12C C-ions beam of 95 MeV/A. A 16.9-mm-thick PMMA was inserted between the exit of the beam and sample holder; the LET was 73 keV/µm (2 Gy = 1.71 × 107 particles/cm2).
2.3. PARP inhibitor treatment
Olaparib (AZD-2281; AstraZeneca, Cambridge, RU) was purchased from Tebu-bio (Ref 21,910–2154–25 mg, Le Perray en Yvelines, France) and used in vitro at 2 μM in the culture medium, according to a dose response experiment (Sup Fig. 1). Olaparib powder was first dissolved in DMSO at 10 mM, diluted to its final concentration with culture medium, and finally added to the cells 2 h before irradiation and left in the cell culture medium for 24 h. Negative control samples were treated with the DMSO concentration that was used for the test samples (0.02%).
2.4. Western blot
Following the irradiation and treatment, the cells were washed with PBS, and the cell pellet was lysed in RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA USA), supplemented with 1X protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA USA), by 10 strokes in insulin syringes (29 G, Myjector, VWR, Briare, France) at 4 °C. The lysate was then centrifuged (28,000 g, 1 h), and 20 µl of the supernatant was mixed with Laemmli 4X (Bio-Rad, Marnes-la-Coquette, France) and denatured 5 min at 95 °C. The samples were then separated by SDS-PAGE on a TGX 4–15% gradient polyacrylamide gel (Bio-Rad, Marnes-la-Coquette, France). Proteins were transferred to a nitrocellulose membrane and blocked with 5% skim milk powder in TBS-T (150 mM NaCl, 10 mM Tris-HCl pH 8, 0.05% Tween-20) for 45 min at 20 °C. The blots were incubated with mouse monoclonal anti-PADPR (anti-poly-ADP-ribose, clone 10H, 1:1000, ref GTX14459, GeneTex, Irvine, CA, USA) or mouse monoclonal anti-b-actin (loading control, 1/1000, Merck KGaA, Darmstadt, Germany) in TBST with 1% skim milk powder overnight at 4 °C and then washed 3 times for 10 min with TBS-T. Next, the blots were incubated with secondary HRP-conjugated goat anti-mouse (1:10,000, GE Healthcare) in TBS-T with 1% skim milk powder for 45 min at 20 °C and washed with TBS-T. The blots were treated with ECL chemiluminescence reagent (Merck KGaA, Darmstadt, Germany) for 1 min before exposure to Hyperfilms (VWR, Fontenay-sous-Bois, France) for 1 to 2 min. The films were developed and scanned as JPEGs on a GS 700 Bio-Rad scanner.
2.5. Clonogenic assay
CH2879 cells were irradiated in T25 cm2 flasks at confluence. A sham control was included to evaluate the plating efficiency. At 18–24 h after irradiation, the cells were harvested and re-plated at the appropriate dilutions in the multiwell plates. After an incubation of 8 days, the colonies were stained with crystal violet solution (0.3% w/v crystal violet in 20% v/v ethanol). Only colonies that comprised over 50 cells were counted by eye under a stereomicroscope. The results were expressed as the percentage of the sham-irradiated control.
2.6. Proliferation assay
Cell proliferation was evaluated with the CellTrace® Far Red Cell Proliferation Kit (ThermoFisher Scientific, Waltham, MA USA). On the day before irradiation, the cells were stained with CellTrace dye, per the manufacturer's instructions. Stained cells were seeded in flasks at a density that allowed 5 days of growth. The cells were irradiated 24 h after staining, and a control (non-irradiated and non-divided) was fixed in PFA solution (4% in PBS, Thermo Fisher Scientific, Waltham, MA USA). After irradiation, the flasks were placed in the incubator. Five days after irradiation, all samples (irradiated, treated with PARPi, and non-irradiated) were fixed with PFA solution (4% in PBS, Thermo Fisher Scientific, Waltham, MA USA). Non-divided samples, controls, and treated samples were then analyzed by flow cytometry. Experiments with X-rays and C-ions were analyzed on an ICORE platform (Caen, France) with a Gallios flow cytometer (Beckman Coulter, Villepinte, France). Calibration beads (Flowcheck Pro and Flowset Pro Fluorospheres, Beckman Coulter) were used to verify the optical alignment, fluidics system, and sensitivity. The data were acquired using Gallios software (Beckman Coulter). The excitation light was provided by a 25-mW red diode (wavelength 638 nm), and the fluorescence was measured in the FL6 channel with a 660-nm bandpass filter. Experiments with proton irradiation were analyzed on a Cytoflex flow cytometer (Beckman Coulter, Brea, CA, USA); data were acquired using a 638-nm excitation wavelength in the 660/10 BP fluorescence channel. Data were analyzed with FCS Express 6+ (DeNovo®), and proliferation index (p.i.) was calculated by the software using the non-divided control as reference. Control and irradiated samples were compared with the corresponding PARPi treated samples to estimate the radiosensitization effect; a parameter, EF (Enhancement Factor,%) was calculated accordingly, using each sample couples.
2.7. Characterization of cell line by sequencing
The sequences of 69 genes that have been implicated in DNA repair and, specifically, homologous recombination were analyzed for mutations (Supplementary Data 1). Regions of interest were captured with the SureSelect XT protocol (Agilent, Santa Clara, CA, USA) and sequenced on an Ilumina NextSeq (Illumina, San Diego, CA USA), using the 2 × 75 pb paired-end technique. Bioinformatics analysis was performed using CASAVA Suite v1.8 for demultiplexing, followed by BWA 0.7.12 for alignment and the GATK v3.3 pipeline to produce BAM files, according to Broad Institute recommendations. The variant-calling step was performed using HaplotypeCaller (HC) [38], LoFreq v2.1.1 [39], and Outlyzer v1.0 [40] for small variants and OncoCNV v6.4 [40] for large-scale rearrangements.
2.8. Statistics
The statistical analysis was performed using the statistical module in Origin (V 6.0) by t-test (2 populations) with an independent type and a 0.05 significance level. Data were considered to be significantly different when p < 0.05 (*).
3. Results
3.1. Olaparib delays proliferation in CH2879 cells
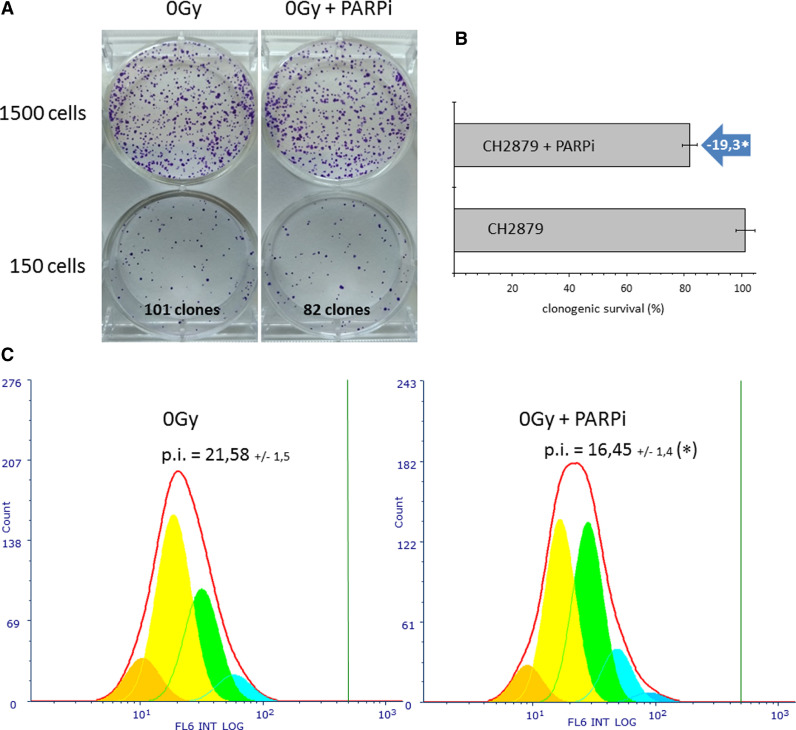
The effect of Olaparib alone was tested first. CH2879 chondrosarcoma cells were incubated with 2 µM Olaparib, and clonogenic survival and proliferation index were measured per Fig. 1. Olaparib reduced the survival of CH2879 cells by 19.3% (Fig. 2A and B). This experiment was repeated several times to obtain accurate values of the effect of Olaparib on CH2879 cells. The proliferation index (p.i.) also declined under the same treatment conditions (Fig. 2C). In control unirradiated cells, the proliferation index was 21.58 (+/−1.5), compared with 16.45 (+/−1.4) with Olaparib, corresponding to a significant reduction of 23.7%. This variation in proliferation index was attributed primarily to several shifts in the distribution of cells between generations (Table 1). With Olaparib treatment, the youngest generations (generation 6 in orange and generation 5 in yellow) decreased, and the oldest generations (generation 4 in green, generation 3 in light blue, and generation 2 in blue) rose, reflecting lower cell proliferation with Olaparib compared with nontreated cells.
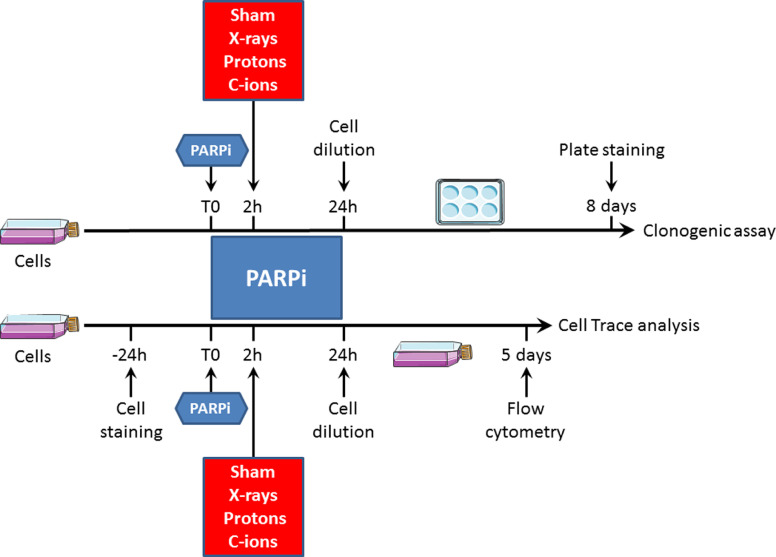
Fig. 1.
Experimental strategy combining PARP inhibition and cell irradiation.
In the clonogenic assay (top part), cells at 80% confluence were incubated with PARPi 2 h before irradiation, and after 24 h, cells were seeded in 6-well plates. The plates were left at 37 °C for 8 days, and the clones were stained and counted. In the CellTrace analysis (bottom part), cells at 50% confluence were first stained with CellTrace reagent 24 h before being incubated with PARPi. Two hours after PARPi incubation, the cells were irradiated, and after 24 h, they were diluted to allow cell growth without contact inhibition. The flasks were left at 37 °C for 5 days, and the cells were analyzed by flow cytometry.
Fig. 2.
Olaparib reduces chondrosarcoma cell survival and proliferation.
A. CH2879 cells were treated for 24 h with 2 µM Olaparib, and living cells were counted; then, 150 or 1500 cells/well were re-seeded in drug-free medium in a 6-well plate. Colony formation was assessed after 8 days by crystal violet staining, and colonies with more than 50 cells were counted macroscopically. The number of colonies is expressed as a percentage of untreated control; representative images are shown. B. Clonogenic survival of CH2879 cells. Data are shown as mean +/- SD of 3 independent experiments performed in triplicate; *, P < 0.05. C. CH2879 cells were first stained with CellTrace and then treated for 24 h with 2 µM Olaparib; living cells were counted and subsequently diluted in drug-free medium. Proliferation assay was performed 5 days later by flow cytometry, and proliferation index (p.i.) was calculated using the non-divided control as reference (vertical green bar). Proliferation index is shown as mean +/- SD of 3 independent experiments performed in triplicate. Each generation of cells is shown in a different color, as presented in Table 1.
Table 1.
Proportion of cells (% of total) in each generation in control (0 Gy) and treated (0 Gy + PARPi) samples by flow cytometry proliferation assay (CellTrace).
| Generation (color) | 0 Gy (% cells) | 0 Gy + PARPi (% cells) |
|---|---|---|
| # 1 (violet) | 0.0 | 0.0 |
| # 2 (blue) | 0.0 | 1.9 |
| # 3 (cyan) | 7.0 | 11.4 |
| # 4 (green) | 30.6 | 39.1 |
| # 5 (yellow) | 50.8 | 39.7 |
| # 6 (orange) | 11.6 | 7.8 |
3.2. Accumulation of PAR chains in response to irradiation
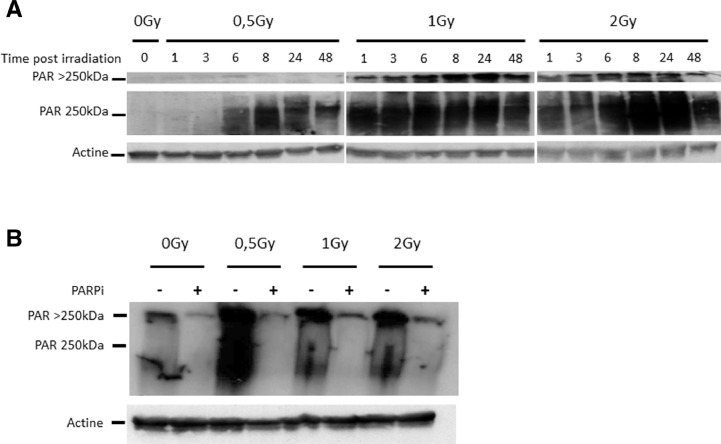
PARP-1 activity can be measured by monitoring the PARylation status of cellular proteins by western blot using anti-PAR. Generally, PARP-1 is activated immediately after breaks in DNA and begins PARylation of its target proteins. We analyzed the kinetics of PAR chain formation in CH2879 cells after treatment with various doses of X-rays (Fig. 3A). After 6 h of exposure to 0.5 Gy X-rays or after 1 h with 1 Gy or 2 Gy X-rays, appreciable PARylation of cellular proteins was observed. A specific and characteristic signal was observed as a smear, with a maximum of intensity at approximately 250 kDa. In the western blot experiments, the wells of the polyacrylamide gels (4% to 16% acrylamide gradient gels) were also transferred to the membrane for analysis. Following 1 Gy and 2 Gy irradiation with X-rays, a signal was observed in the wells, demonstrating significant polymerization of the PAR chains that prevented the protein from entering the top of the 4% acrylamide gel. In addition, maximum PARylation was reached after 24 h at each dose. PARylation was inhibited in CH2879 cells by Olaparib, as shown in Fig. 3B. PAR chains were analyzed 24 h after irradiation with 0.5 Gy, 1 Gy, and 2 Gy X-rays. The signal that corresponded to the PAR chains (in the wells of the acrylamide gel and the smear at approximately 250 kDa) nearly disappeared with the PARPi, reflecting the efficacy of Olaparib after irradiation of CH2879 cells with X-rays.
Fig. 3.
Quantification of PAR chains according to irradiation dose, time point, and PARPi.
A. CH2879 cells were irradiated with X-rays (0, 0.5 Gy, 1 Gy, and 2 Gy), and PAR chains were analyzed by western blot following irradiation at various time points (0, 1, 3, 6, 8, 24, and 48 h); actin served as the loading control. B. CH2879 cells were irradiated with X-rays (0, 0.5 Gy, 1 Gy, and 2 Gy), and PAR chains were analyzed by western blot following irradiation with or without PARPi at 24 h after irradiation; actine served as the loading control.
3.3. Olaparib reduces cell survival in response to irradiation
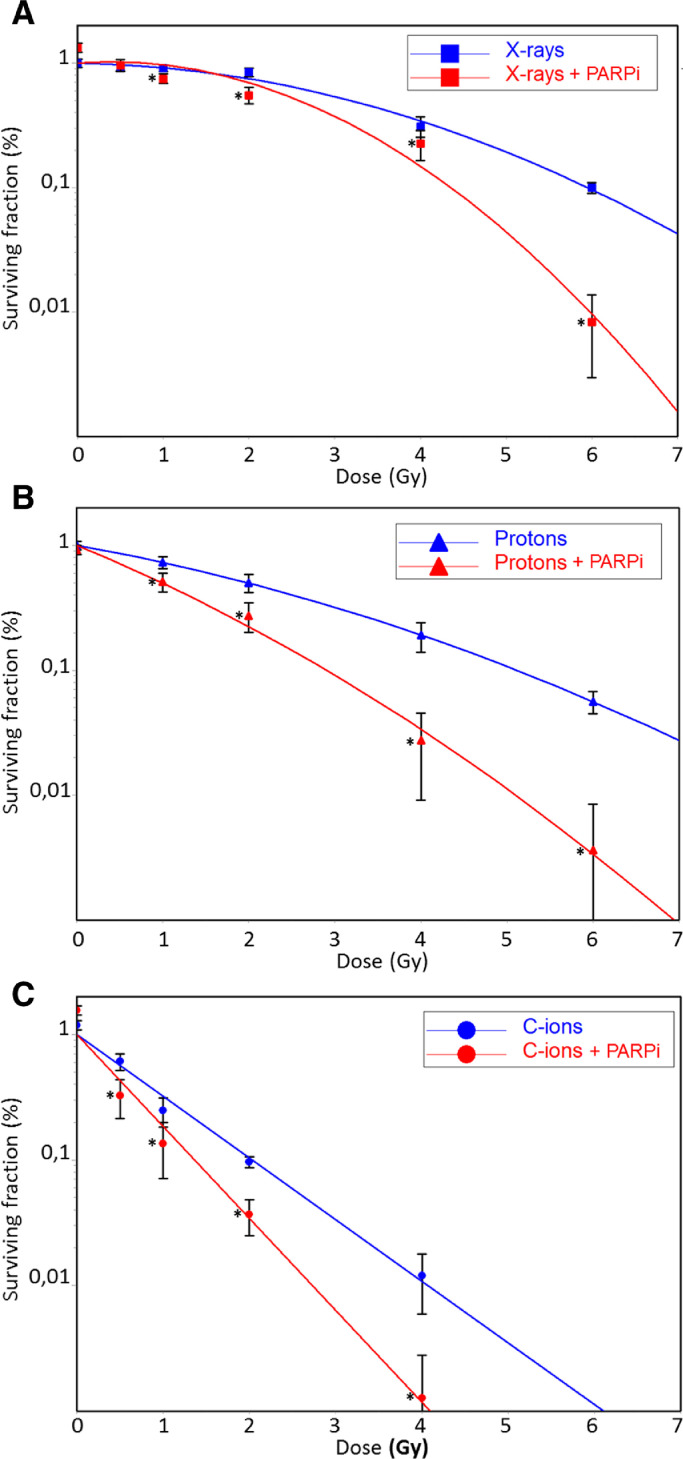
CH2879 cells were irradiated with various types of radiation with and without Olaparib according to the protocol (Fig. 1). Clonogenic survival of CH2879 cells after irradiation was expressed as a function of dose, and the results were plotted in CS Cal (Fig. 4). Surviving fractions of X-rays- and proton-irradiated samples were plotted using a linear quadratic model; a linear model was used for those after C-ion irradiations. All Olaparib treatments significantly decreased the surviving fractions of CH2879 cells at all irradiation doses (except the low X-rays dose) and all irradiation types. Enhancement ratio (ER) was calculated for each irradiation type using D10 and D37 values (Table 2). The dose that was necessary to obtain 10% survival (D10) fell by 1.37, 1.76, and 1.43 using Olaparib with X-rays and proton and C-ions irradiation, respectively. The surviving fraction at 2 Gy (SF2) were significantly different between irradiations qualities and between samples treated with and without Olaparib for the same doses. According to ER (D37), it was interesting to notice that Proton + PARPi was significantly more radiosensitizing than X-rays + PARPi; ER values differed with D37, yielding dose reductions of 1.93 and 1.27, respectively (p < 0.05).
Fig. 4.
Clonogenic survival of chondrosarcoma cells after X-rays, proton, and C-ions irradiation +/- PARPi.
A. CH2879 cells were irradiated with X-rays (blue squares) or X-rays with PARPi (red squares). B. CH2879 cells were irradiated with protons (blue triangles) or protons with PARPi (red triangles). C. CH2879 cells were irradiated with C-ions (blue circles) or C-ions with PARPi (red circles). Cell survival (%) is shown as mean +/- SD of 3 independent experiments performed in triplicate for X-rays and C-ions. In the case of proton irradiation, cell survival (%) is shown as mean +/- SD of triplicate points corresponding to a single irradiation experiment. For each dose, clonogenic survival was considered to be significantly different (with and without PARPi) when p < 0.05 (*).
Table 2.
Calculated parameters of CH2879 cell survival after irradiation with X-rays, protons, and C-ions with and without PARPi (from Fig. 4).
| D10a | D37b | SF2c | ER (D10)d | ER (D37)e | |
|---|---|---|---|---|---|
| X-rays | 5.9 | 3.8 | 0.75 | / | / |
| X-rays + PARPi | 4.3 | 3 | 0.7 | 1.37 | 1.27 |
| Protons | 5.1 | 2.7 | 0.5 | / | / |
| Protons + PARPi | 2.9 | 1.4 | 0.2 | 1.76 | 1.93 |
| C-ions | 2 | 0.9 | 0.1 | / | / |
| C-ions + PARPi | 1.4 | 0.6 | 0.03 | 1.43 | 1.5 |
the D10 dose gives a surviving fraction of 0.1.
the D37 dose gives a surviving fraction of 0.37.
the SF2 fraction is observed at 2 Gy irradiation.
ER (D10) values are calculated as: D10 (with PARPi) / D10 (without PARPi) for each irradiation quality.
ER (D37) values are calculated as: D37 (with PARPi) / D37 (without PARPi) for each irradiation quality.
3.4. Olaparib decreases cell proliferation in response to irradiation
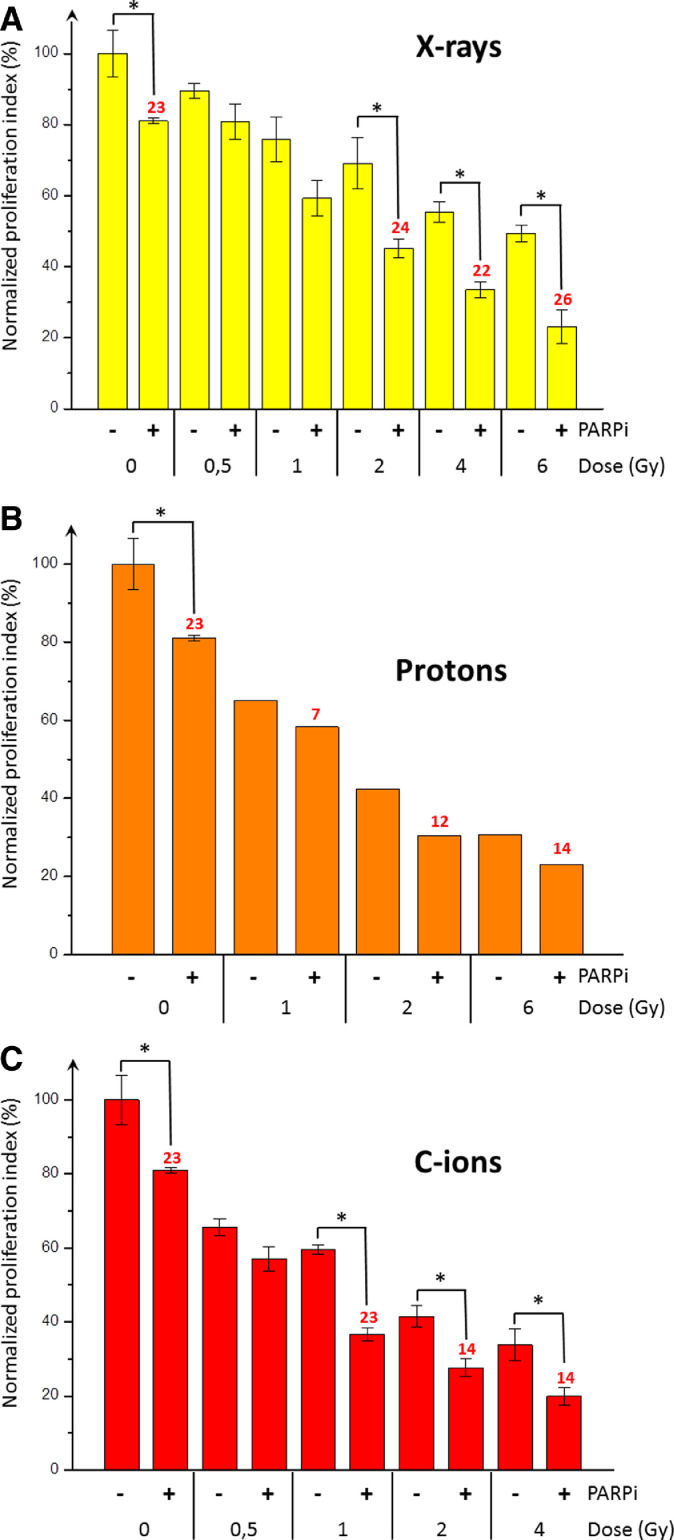
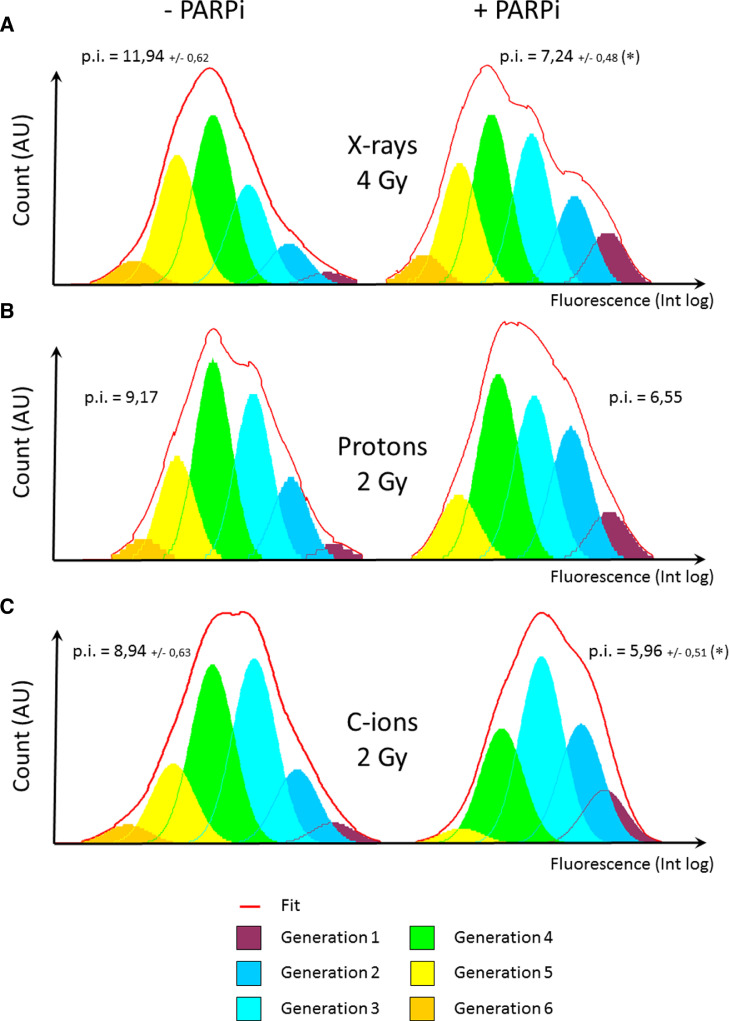
As in of the analysis of clonogenic survival, CH2879 cells were irradiated with and without Olaparib, per the protocol (Fig. 1). Normalized cellular index values (against a sham-irradiated control) of CH2879 cells after irradiation were expressed as a function of the dose with or without Olaparib (Fig. 5). Following X-rays irradiation at 0.5 and 1 Gy, Olaparib did not significantly decrease the cellular index. But, after 2 Gy, 4 Gy, and 6 Gy X-rays, the enhancement factors (EFs) were 24%, 22%, and 26% when combined with Olaparib. For proton irradiation, the EF was approximately 10% at both doses. The lowest cellular index (roughly 20%) was observed with the combination of 6 Gy protons + Olaparib, approximating the value with the same combination of X-rays. Following C-ions irradiation at 1 Gy, 2 Gy, and 4 Gy, the EFs were 23%, 14%, and 14%, respectively with Olaparib. Cell trace proliferation assay allows one to perform a detailed analysis of cells in each generation following treatment. To compare irradiation types and treatments, we selected a dose that reduced the proliferation index of control cells by approximately 50% (Fig. 6), corresponding to 4 Gy for X-rays (p.i. = 12.38 and 55% of control), 2 Gy for protons (p.i. = 9.17 and 42% of control), and 2 Gy for C-ions (p.i. = 8.50 and 41% of control). In the absence of Olaparib, the 6 generations appeared with all irradiation type, but the proportions of generation 5 cells (yellow) with X-rays and generation 3 cells (light blue) with C-ions were higher compared with protons. When Olaparib was combined irradiation, the proportion of cells in generations 1, 2, and 3 generally increased, and those in generation 6 disappeared with protons and C-ions. The proportion of generation 5 cells (yellow) was low with C-ions and Olaparib. In contrast, the proportion of generation 6 cells (orange) was stable with X-rays irradiation and Olaparib compared with X-rays alone.
Fig. 5.
Impaired proliferation of chondrosarcoma cells after X-rays, proton and C-ions irradiation +/- PARPi.
A. CH2879 cells were irradiated with X-rays +/- PARPi (yellow). B. CH2879 cells were irradiated with protons +/- PARPi (orange). C. CH2879 cells were irradiated with C-ions +/- PARPi (red). Proliferation indexes (%) are shown as mean +/- SD of 3 independent experiments performed in triplicate for X-rays and C-ions. In the case of proton irradiation, proliferation indexes (%) corresponded to a single irradiation experiment. For each dose, proliferation indexes were considered to be significantly different (with and without PARPi) when p < 0.05 (*). An enhancement factor (EF%) was added to the PARPi-treated samples (red numbers) when significant differences were observed. In the case of proton irradiation, no statistical analysis was performed, and EF was proposed to be exploratory.
Fig. 6.
Changes in cell generations after X-rays, proton, and C-ions irradiation +/- PARPi.
A. Repartition of cells between 6 generations following 4 Gy X-rays irradiation. B. Repartition of cells between 6 generations following 2 Gy proton irradiation. C. Repartition of cells between 6 generations following 2 Gy C-ions irradiation. Proliferation index is shown as mean +/- SD of 3 independent experiments performed in triplicate for X-rays and C-ions irradiations. For each dose, proliferation indexes were considered to be significantly different (with and without PARPi) when p < 0.05 (*).
3.5. Genetic characterization of CH2879 chondrosarcoma cells
To determine the mechanism of the radiosensitivity of CH2879 cells, a panel of 69 genes that have been implicated in DNA repair were sequenced and analyzed for mutations (Supplementary Data 1). Of the 69 genes that were analyzed, 12 were mutated by at least 2 callers (Table 3): by substitution or deletion, creating nonsense, missense, or frameshift mutation or possible splice (ARID1A; ARID1B, BRIP1, FANCA, FANCG, FANCI, GATA3, HDAC3, NBN, PARP1, PTEN, and RAD50) and loss of sequence (SMARCA2). Of the 12 genes with a single mutation, 3 had a frequency of up to 10%. According to Table 3, PTEN was mutated (c.697C > T; nonsense) in 100% of alleles, RAD50 was mutated (c.2165del; frameshift) at 44%, NBN was mutated (c.2249T > A; nonsense) at 31% and SMARCA2 locus was lost in 100% of alleles. The frequencies of all other mutations were below 10%, having a moderate effect on the entire cell population.
Table 3.
Analysis of specific mutations in CH2879 cells.
| Gene | transcript | Description1 | Expected consequence2 | Mutated allele frequency | Interpretation | detectable by |
|---|---|---|---|---|---|---|
| NBN | NM_002485.4 | c.2249T>A | p.Leu750* | 31% | probably inactive protein with premature termination codon (Nonsens mediated decay may be not active because the variant is located at the very end of the coding sequence) | HC; Lofreq; OutLyser |
| PTEN | NM_000314.6 | c.697C>T | p.Arg233* | 100% | inactive protein due to the introduction of premature termination codon | HC; Lofreq; OutLyser |
| RAD50 | NM_005732.3 | c.2165del | p.Lys722Argfs*14 | 54% | inactive protein due to the introduction of premature termination codon | HC; Lofreq; OutLyser |
| SMARCA2 | N.A. | Loss of locus | 100% | Loss of the entire tumor suppressor gene (probable Loss of heterozygoty) | OncoCNV |
Mutation nomenclature per HGVS recommendations; nucleotide position +1 corresponds to the A of the ATG translation initiation codon.
Expected consequence on protein level. 3Algorithms used for prediction: SIFT, PolyPhen, MutationTaster.
4. Discussion
In this study, we analyzed the effects of PARP inhibition in association with various irradiation modalities on CH2879 chondrosarcoma cells using the PARP inhibitor Olaparib. X-rays irradiation of CH2879 cells induced the significant accumulation of PAR chains by western blot (Fig. 3.A)—reflecting the typical response to X-rays and the normal activity of PARPs. PAR chains facilitate the recruitment of SSB repair scaffolding proteins, such as X-rays repair cross-complementing protein 1 (XRCC1), DNA ligase III, and DNA polymerase beta. The most notable effects of PARP inhibition is the blockade of PAR chain synthesis (Fig. 3.B) and the accumulation of SSBs, leading to DNA double-strand breaks (DSBs) through collapse of the replication fork. The synthetic lethal interactions between PARP inhibition and BRCA1 or BRCA2 mutations suggest a unique strategy for treating patients with BRCA-mutant tumors. BRCA-mutant tumor cells are more sensitive to PARPi than BRCA-normal wild-type cells [32]. Combined with irradiation, PARPis amplify unrepaired DNA damage, SSBs, and DSBs. In a favorable genetic context that prevents HRR, synthetic lethality occurs, accompanied by large-scale genomic rearrangements, often leading to cell death. Based on the genetic characterization of CH2879 cells, several genes that are involved in HRR were mutated. The first main mutation was observed in PTEN, which was disrupted in the entire cell population (Table 1). PTEN is essential in the maintenance of chromosomal stability [41], and its deficiency sensitizes cancer cells with PARPis and downregulates RAD51 [42]. But, PARP inhibition as monotherapy has no sensitizing effect on PTEN-deficient cells—only the combination of PARP and PI3K inhibitors prevented tumor growth in a mouse model of PTEN-deficient endometrioid endometrial cancer [43]. In the absence of irradiation, 2 µM Olaparib induced a roughly 20% decrease in clonogenic survival in CH2879 cells (Fig. 2A,B). By cell proliferation assay, the underlying mechanism was suggested to be a delay in cell growth (Fig. 2C). The proportion of cells in generations 2, 3, and 4 following Olaparib treatment increased, and that in generations 5 and 6 declined compared with untreated cells (Table 1). Two other genes were mutated—RAD50 and NBN—at a frequency of 44.2% and 30.66%, respectively. RAD50 and NBN (nibrin) are components of the MRN complex (with MRE11), which is central in DNA repair machinery, double-strand break signaling, and the chromatin template. These mutations, which caused a frameshift and nonsense mutation, generated nonfunctional proteins. SMARCA2, which presented a loss of the entire tumor suppressor gene, is involved in chromatin remodeling at DNA damage sites and/or replication forks during double-strand break (DSB) response, allowing access to several complexes to damage sites. Because several genes in HRR were mutated in CH2879 cells, synthetic lethality could be linked to PARPis and irradiation in the so-called BRCAness concept (in the absence of a BRCA mutation). Such cells, with defective HRR, should be more sensitive to PARPis [33]. By clonogenic survival assay, we observed greater sensitivity of CH2879 cells to a PARPi with and without irradiation (Fig. 2, Fig. 4). In addition, we noted the effectiveness of ion beam irradiation. The radiosensitization effects of Olaparib were conserved with proton and C-ion irradiation. Even with C-ions irradiation of CH2879 cells, which significantly improved cell death alone, the addition of Olaparib amplified the effects of the particle. As observed by CellTrace assay (Fig. 5), this outcome was related to a delay in proliferation. Our earlier report showed that depletion of PARP-1 results in a delay in S-phase in HeLa cells [44]. The combination of Olaparib and C-ions had a significant impact on cell cycle time course in this cell type, affecting a significant and reproducible shift in generations throughout the entire population (Fig. 6). The same result was obtained with proton irradiation but to a lesser extent. These preclinical data demonstrated the potential of Olaparib against cells with mutation in HRR genes. This is the first study to include chondrosarcoma in this category of cells, establishing the possibility of treating such radioresistant cancer cells.
Acknowledgments
Author contributions
FC and UG conceived the study and its design. MCé, JBA, UG, and FC performed the experiments. FC performed the statistical analysis. EM and LC performed the gene sequencing analyses. FPC, GP, and GAPC performed the INFN-LNS irradiation. MG and MCa performed the flow cytometry experiments. FC drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by “Agence Nationale de la Recherche,” in the framework of “Investments for the Future” under references France HADRON (ANR-11-INBS-0007) and Equipex Rec-Hadron (ANR-10-EQPX-1401); “Fondation ARC pour la Recherche sur le Cancer” (FC, PJA 20,151,203,177); a short-term EMBO fellowship (UG, ASTF 282 – 2016); and the ENSAR2 program for experiments and travel in Italy.
Conflicts of interest
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2019.100246.
Appendix. Supplementary materials
References
- 1.Pommier P., Hu Y., Baron M.-H., Chapet O., Balosso J. [Particle therapy: carbon ions] Bull. Cancer. 2010;97:819–829. doi: 10.1684/bdc.2010.1151. [DOI] [PubMed] [Google Scholar]
- 2.Thariat J., Bolle S., Demizu Y., Marcy P.-Y., Hu Y., Santini J., Bourhis J., Pommier P. New techniques in radiation therapy for head and neck cancer: IMRT, CyberKnife, protons, and carbon ions. improved effectiveness and safety? Impact on survival? Antic. Drugs. 2011;22:596–606. doi: 10.1097/CAD.0b013e328340fd2b. [DOI] [PubMed] [Google Scholar]
- 3.Kamada T., Tsujii H., Blakely E.A., Debus J., De Neve W., Durante M., Jäkel O., Mayer R., Orecchia R., Pötter R., Vatnitsky S., Chu W.T. Carbon ion radiotherapy in Japan: an assessment of 20 years of clinical experience. Lancet Oncol. 2015;16:e93–e100. doi: 10.1016/S1470-2045(14)70412-7. [DOI] [PubMed] [Google Scholar]
- 4.Glowa C., Karger C.P., Brons S., Zhao D., Mason R.P., Huber P.E., Debus J., Peschke P. Carbon ion radiotherapy decreases the impact of tumor heterogeneity on radiation response in experimental prostate tumors. Cancer Lett. 2016;378:97–103. doi: 10.1016/j.canlet.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durante M., Debus J. Heavy charged Particles: does improved precision and higher biological effectiveness translate to better outcome in patients? Semin Radiat Oncol. 2018;28:160–167. [Google Scholar]
- 6.Durante M. New challenges in high-energy particle radiobiology. Br. J. Radiol. 2014;87 doi: 10.1259/bjr.20130626. 20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraft G. Tumor therapy with heavy charged particles. Progress Part. Nuclear Phys. 2001;46:I. [Google Scholar]
- 8.Suzuki M., Kase Y., Yamaguchi H., Kanai T., Ando K. Relative biological effectiveness for cell-killing effect on various human cell lines irradiated with heavy-ion medical accelerator in Chiba (HIMAC) carbon-ion beams. Int. J. Radiat. Oncol. Biol. Phys. 2000;48:241–250. doi: 10.1016/s0360-3016(00)00568-x. [DOI] [PubMed] [Google Scholar]
- 9.Paganetti H. Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys. Med. Biol. 2014;59:R419–R472. doi: 10.1088/0031-9155/59/22/R419. [DOI] [PubMed] [Google Scholar]
- 10.Walenta S., Mueller-Klieser W. Differential superiority of heavy charged-particle irradiation to X-rays: studies on biological effectiveness and side effect mechanisms in multicellular tumor and normal tissue models. Front Oncol. 2016;6:30. doi: 10.3389/fonc.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Amorim Bernstein K., DeLaney T. Chordomas and chondrosarcomas-The role of radiation therapy. J. Surg. Oncol. 2016;114:564–569. doi: 10.1002/jso.24368. [DOI] [PubMed] [Google Scholar]
- 12.Hug E.B., Loredo L.N., Slater J.D., DeVries A., Grove R.I., Schaefer R.A., Rosenberg A.E., Slater J.M. Proton radiation therapy for chordomas and chondrosarcomas of the skull base. J. Neurosurg. 1999;91:432–439. doi: 10.3171/jns.1999.91.3.0432. [DOI] [PubMed] [Google Scholar]
- 13.Schulz-Ertner D., Nikoghosyan A., Hof H., Didinger B., Combs S.E., Jäkel O., Karger C.P., Edler L., Debus J. Carbon ion radiotherapy of skull base chondrosarcomas. Int. J. Rad. Oncol. Biol. Phys. 2007;67:171–177. doi: 10.1016/j.ijrobp.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Nikoghosyan A.V., Rauch G., Münter M.W., Jensen A.D., Combs S.E., Kieser M., Debus J. Randomised trial of proton vs. carbon ion radiation therapy in patients with low and intermediate grade chondrosarcoma of the skull base, clinical phase III study. BMC Cancer. 2010;10:606. doi: 10.1186/1471-2407-10-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NUPECC Hadron therapy : how nuclear research is improving human health for Medicine. Nuclear Phys. 2014 http://www.nupecc.org/pub/npmed2014_brochure.pdf [Google Scholar]
- 16.Ferrari A., Dirksen U., Bielack S. Sarcomas of soft tissue and bone. Prog Tumor Res. 2016;43:128–141. doi: 10.1159/000447083. [DOI] [PubMed] [Google Scholar]
- 17.Rozeman L.B., Hogendoorn P.C.W., Bovée J.V.M.G. Diagnosis and prognosis of chondrosarcoma of bone. Expert Rev. Mol. Diagn. 2002;2:461–472. doi: 10.1586/14737159.2.5.461. [DOI] [PubMed] [Google Scholar]
- 18.Gelderblom H., Hogendoorn P.C.W., Dijkstra S.D., van Rijswijk C.S., Krol A.D., Taminiau A.H.M., Bovée J.V.M.G. The clinical approach towards chondrosarcoma. Oncologist. 2008;13:320–329. doi: 10.1634/theoncologist.2007-0237. [DOI] [PubMed] [Google Scholar]
- 19.Jebsen N.L., Trovik C.S., Bauer H.C.F., Rydholm A., Monge O.R., Hall K.S., Alvegård T., Bruland O.S. Radiotherapy to improve local control regardless of surgical margin and malignancy grade in extremity and trunk wall soft tissue sarcoma: a scandinavian sarcoma group study. Int. J. Radiat. Oncol. Biol. Phys. 2008;71:1196–1203. doi: 10.1016/j.ijrobp.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 20.O'Neil N.J., Bailey M.L., Hieter P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 21.Lord C.J., Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashworth A., Lord C.J. Synthetic lethal therapies for cancer: what's next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018;15:564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 23.Ray Chaudhuri A., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 25.Sandhu S.K., Yap T.A., de Bono J.S. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur. J. Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Annunziata C.M., O'Shaughnessy J. PARP as a novel therapeutic target in cancer. Clin. Cancer Res. 2010;16:4517–4526. doi: 10.1158/1078-0432.CCR-10-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konecny G.E., Kristeleit R.S. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: current practice and future directions. Br. J. Cancer. 2016;115:1157–1173. doi: 10.1038/bjc.2016.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesueur P., Chevalier F., El-Habr E.A., Junier M.-P., Chneiweiss H., Castera L., Müller E., Stefan D., Saintigny Y. Radiosensitization effect of Talazoparib, a parp Inhibitor, on glioblastoma stem cells exposed to low and high linear energy transfer radiation. Sci. Rep. 2018;8:3664. doi: 10.1038/s41598-018-22022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Césaire M., Thariat J., Candéias S.M., Stefan D., Saintigny Y., Chevalier F. Combining PARP inhibition, radiation, and immunotherapy: a possible strategy to improve the treatment of cancer? Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19123793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.-M., Ledermann J.A., Kohn E.C. PARP inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann. Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.-M, Ledermann J.A., Kohn E.C. PARP inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann. Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghorai A., Sarma A., Chowdhury P., Ghosh U. PARP-1 depletion in combination with carbon ion exposure significantly reduces MMPs activity and overall increases TIMPs expression in cultured HeLa cells. Radiat Oncol. 2016;22:126. doi: 10.1186/s13014-016-0703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farmer H., McCabe N., Lord C.J., Tutt A.N.J., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M.B., Jackson S.P., Smith G.C.M., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 34.Lord C.J., Ashworth A. BRCAness revisited. Natl. Rev. Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 35.Lesueur P., Chevalier F., Austry J.-B., Waissi W., Burckel H., Noël G., Habrand J.-L., Saintigny Y., Joly F. Poly-(ADP-ribose)-polymerase inhibitors as radiosensitizers: a systematic review of pre-clinical and clinical human studies. Oncotarget. 2017;5 doi: 10.18632/oncotarget.19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gil-Benso R., Lopez-Gines C., López-Guerrero J.A., Carda C., Callaghan R.C., Navarro S., Ferrer J., Pellín A., Llombart-Bosch A. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: comparative histologic and genetic studies with its tumor of origin. Lab. Invest. 2003;83:877–887. doi: 10.1097/01.lab.0000073131.34648.ea. [DOI] [PubMed] [Google Scholar]
- 37.Durantel F., Balanzat E., Cassimi A., Chevalier F., Ngono-Ravache Y., Madi T., Poully J.-C., Ramillon J.-M., Rothard H., Ropars F., Schwob L., Testard I., Saintigny Y. Dosimetry for radiobiology experiments at GANIL, nuclear instruments and methods in physics research section A: accelerators, spectrometers. Detect. Assoc. Equ. 2016;816:70–77. [Google Scholar]
- 38.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis Toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller E., Goardon N., Brault B., Rousselin A., Paimparay G., Legros A., Fouillet R., Bruet O., Tranchant A., Domin F., San C., Quesnelle C., Frebourg T., Ricou A., Krieger S., Vaur D., Castera L. OutLyzer: software for extracting low-allele-frequency tumor mutations from sequencing background noise in clinical practice. Oncotarget. 2016;7:79485–79493. doi: 10.18632/oncotarget.13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin Y., Shen W.H. PTEN: a new guardian of the genome. Oncogene. 2008;27:5443–5453. doi: 10.1038/onc.2008.241. [DOI] [PubMed] [Google Scholar]
- 42.Mendes-Pereira A.M., Martin S.A., Brough R., McCarthy A., Taylor J.R., Kim J.-S., Waldman T., Lord C.J., Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bian X., Gao J., Luo F., Rui C., Zheng T., Wang D., Wang Y., Roberts T.M., Liu P., Zhao J.J., Cheng H. PTEN deficiency sensitizes endometrioid endometrial cancer to compound PARP-PI3K inhibition but not PARP inhibition as monotherapy. Oncogene. 2018;37:341–351. doi: 10.1038/onc.2017.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghorai A., Bhattacharyya N.P., Sarma A., Ghosh U. Radiosensitivity and induction of apoptosis by high LET carbon ion beam and low LET gamma Radiation: a comparative study. Scientifica (Cairo) 2014;2014 doi: 10.1155/2014/438030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.