Most of the great diversity of oxylipins in plants is produced by a group of specialized cytochrome P450 enzymes, among which is ALLENE OXIDE SYNTHASE (AOS). Many AOSs generate precursors of the defense hormone jasmonate. As a consequence, aos mutants fail to defend themselves against herbivores and do not display restriction of vegetative growth when wounded. These links between growth and defense that are controlled by AOS-derived oxylipins are ancient. Here, we focus on oxylipin-regulated coordination of growth/defense, how this optimizes defense, and how a plant’s need for light can override jasmonate activity. AOS-derived oxylipins are candidate regulators throughout land plant evolution.
Oxylipins form a vast and ancient superfamily of oxygenated fatty acid derivatives of enzymatic and non-enzymatic origin, and are found in plants, animals, fungi, protists, and microbes. Many, if not most, enzymatically synthesized oxylipins in plants originate from fatty acid hydroperoxides that are formed by the action of lipoxygenase. These hydroperoxides are then subject to metabolism by a group of closely related enzymes, among which is ALLENE OXIDE SYNTHASE (AOS; Brash, 2009; Lee et al., 2008). AOS produces short-lived intermediates that rearrange in various ways, for example cyclizing to form compounds such as oxo-phytodienoic acid (OPDA) and dinor-OPDA, or reorganizing to produce fatty acid ketols and many other oxylipin variants. In brief, AOS produces most of the non-volatile oxylipins in plants, whereas closely related HYDROPEROXIDE LYASES (HPLs) produce the bulk of fragmented, volatile fatty acid-derivatives (Ameye et al., 2018; Box 1). Together, the atypical cytochrome P450 enzymes AOS, HPL, and a related enzyme DIVINYL ETHER SYNTHASE create a highly diverse pool of oxylipins. Jasmonates and their precursors make up the most widespread part of this pool whereas many other oxylipin components tend to be more genus-specific. Due to their relatively large masses, AOS-derived oxylipins can display great structural diversity; new examples of these compounds are frequently discovered. For example, Grechkin et al. (2018) discovered that young roots of wheat and rice produce a remarkable group of AOS-derived branched-chain oxylipins that they term ‘graminoxins’. Together with an earlier report of cyclic, pathogen-induced AOS products known as ‘death acids’ in maize (Christensen et al., 2015), these studies reveal the enormous potential to discover novel oxylipins.
Box 1. AOS and HPL, non-volatile and volatile oxylipin generators, respectively.
Fatty acid hydroperoxides are metabolized by closely related cytochrome P450 oxidases, namely ALLENE OXIDE SYNTHASE (AOS), HYDROPEROXIDE LYASE (HPL), and DIVINYL ETHER SYNTHASE (DES, not shown). Examples of AOS-derived plant oxylipins are OPDA and dinor-OPDA, and jasmonates such as JA-Ile. AOS produces the bulk of non-volatile oxylipins in plants, whereas HPL produces many volatile oxylipins such as green leaf volatiles (GLVs, a family of short-chain aldehydes, alcohols, and their derivatives; Ameye et al., 2018) that are not discussed in the text. It appears that most land plants make OPDA and/or dinor-OPDA as well as volatile aldehydes such as hexenals. Most, if not all angiosperms, make JA and biologically active JA-derivatives such as JA-Ile. Together, AOS and HPL play dominant roles in enzymatic fatty acid oxidation. The products they produce help to make plants distinct from other organisms.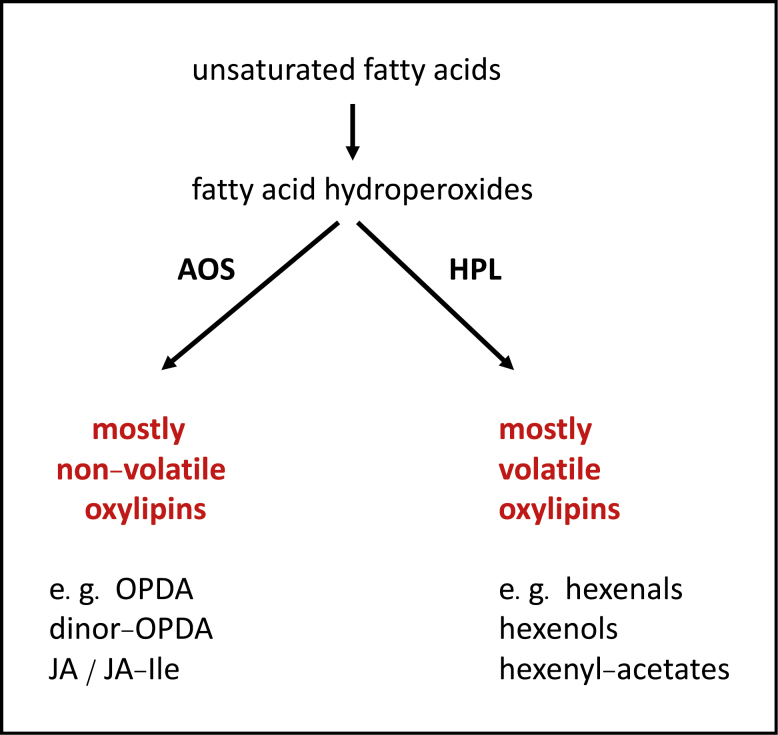
AOS is a focus of attention in large part due to its key role in the synthesis of jasmonic acid (JA) and biologically active jasmonoyl-isoleucine (JA-Ile). In angiosperms, JA-Ile is the best-studied bioactive jasmonate, and the rich transcriptional response of Arabidopsis to treatment with methyl jasmonate (Hickman et al., 2017) appears to depend on its conversion to this molecule. Importantly, not all land plants produce jasmonic acid and its derivatives, but where jasmonates are absent their AOS-derived precursors such as OPDA are present. As an example, the aerial parts (microphylls) of the lycopod Selaginella martensii produce various isomers of OPDA and dinor-OPDA (Ogorodnikova et al., 2015) while no JA or JA-Ile are detected in this plant. The even more ancient liverwort Marchantia polymorpha produces biologically active AOS-derived dinor-OPDA isomers that regulate defense and growth (Monte et al., 2018) through a signaling pathway remarkably similar to the jasmonate pathway in angiosperms (Howe et al., 2018). Here, we focus on the principal roles of jasmonates and OPDA/dinor-OPDA in coordinating defense, development, and growth, although the breath of action of jasmonates in plants is larger than this. For example, carnivorous sundews (Drosera species) first capture their prey on trichome-like tentacles and then the leaf closes around the prey to digest it. Interestingly, feeding Drosera capensis with fruit flies caused localized increases in JA and JA-Ile. Exogenous JA elicits the production of digestive enzymes including cysteine and aspartic proteases that liberate nutrients from captured invertebrates (Krausko et al., 2017). This jasmonate-dependent induction of proteases is seen as a parallel to herbivore-induced jasmonate-dependent defense induction in other plants (reviewed in Pavlovič and Mithöfer, 2019). Additional roles for OPDA and jasmonate outside the defense, development, and growth trio are likely to be discovered in land plants.
Turnover of jasmonates in vivo
The mechanisms underlying the turnover of certain AOS-derived oxylipins such as JA and JA-Ile have been studied in depth. A major metabolite produced from both of these molecules is 12-hydroxy-JA (12-OH-JA; reviewed in Wasternack and Feussner, 2018). Production of 12-OH-JA via the oxidation of JA-Ile and the subsequent removal of isoleucine by amidohydrolases is well characterized (Wasternack and Feussner, 2018). However, an unexpected result is observed when JA-Ile oxidation is impeded through mutation. Rather than leading to a stronger wound response, blocking JA-Ile hydoxylation produces JA-Ile deficiency symptoms (Poudel et al., 2016). Further work will be required to understand why this occurs. Recently, four Arabidopsis JASMONATE-INDUCED OXYGENASES (JOXs)/JASMONIC ACID OXIDASES (JAOs) have been found to oxygenate the JA-Ile precursor JA to produce 12-OH-JA (Caarls et al., 2017; Smirnova et al., 2017). Deficiency in JOX2/JAO2 increases resistance to Botrytis cinerea (Smirnova et al., 2017) and a quadruple JOX/JAO mutant is highly resistant to this fungus as well as to the insect herbivore Mamestra brassicae (Caarls et al., 2017). Notably, the undamaged quadruple-mutant is far smaller than the wild type (Caarls et al., 2017). In summary, JA oxidation to produce 12-OH-JA can modulate defense and growth.
Jasmonate signaling
Arabidopsis is currently the most advanced model for the study of jasmonate signaling. The canonical signal pathway in this plant involves binding of JA-Ile (and other ligands such as the bacterial toxin coronatine: Zhang et al., 2017) to the jasmonate receptor CORONATINE INSENSITIVE 1 (COI1). How COI1 is regulated at the transcriptional and post-transcriptional levels is under intense investigation (Williams et al., 2019). Bound JA-Ile strongly increases the affinity of COI1 for members of the JASMONATE-ZIM-DOMAIN (JAZ) co-receptor proteins. This then leads to JAZ ubiquitination and to proteolytic JAZ destruction. In the absence of JA-Ile, JAZ proteins are relatively stable and they sequester a variety of transcriptional regulators including transcription factors (Howe et al., 2018). When JAZ proteins are degraded in the presence of JA-Ile, these transcription factors are liberated to activate RNA polymerase II (Pol II)-dependent transcription. Among these transcription factors is MYC2 (Box 2A), an essential component of jasmonate signaling (Howe et al., 2018). An et al. (2017) recently showed that COI1 is enriched on promoters of MYC2 targets. Mediator subunit MED25 brings COI1 to these target genes by essentially forming a molecular bridge between COI1 and MYC2. But MYC2 is inactive, in part because it is bound to JAZ proteins that themselves are bound to transcriptional co-repressor complexes. While JA-Ile-mediated destruction of JAZ liberates MYC2 from co-repressor proteins, MED25 helps to recruit a massive transcriptional complex of at least 50 proteins that includes the MED complex and RNA polymerase II (Pol II) as well as chromatin modifiers. In a new review, Zhai and Li (2019) describe in detail what we know about these fascinating events.
Box 2. Environmental modulation of jasmonate pathway inducibility.
(A) Summary of molecular control over jasmonate biosynthesis as illustrated by the key oxylipin biosynthesis gene ALLENE OXIDE SYNTHASE (AOS). In shaded plants, phytochrome B (phyB) is inactive and JAZ proteins are stabilized. Complexes of JAZ, NINJA (NOVEL INTERACTOR of JAZ), and TPL (TOPLESS) proteins repress transcription. In the case of the AOS gene, JAV1, JAZ8, and WRKY51 (W51) complexes also restrain transcription. In sunlight phyB is active, JAZ proteins are readily degraded, and jasmonate synthesis is highly damage-inducible. JA-Ile production causes destruction of JAZ, which liberates NINJA and TPL. In addition, JAV1 complexes are destroyed. The full Mediator complex is recruited by Med25 together with RNA Pol II to initiate MYC2-mediated transcription. The figure represents JAV1/JAZ8/W51 complexes as being separate from the complex of proteins around MYC2. The exact configuration of the complexes and the stoichiometry of several components within them are unknown. (B) Different levels of defense in shade and sun. In the shade, the photoreceptor phyB is inactive. Plants display high apical dominance and elongation growth. Jasmonate biosynthesis genes such as AOS are not strongly inducible, levels of JA-Ile are low, and JAZ proteins restrain the activity of the JA pathway. Under these conditions, jasmonate-inducible defenses are not strongly activated when plants are attacked by herbivores. In contrast, plants exposed to the sun show reduced apical dominance and reduced elongation growth due to phyB activation. Activity of the jasmonate pathway is highly inducible, causing JA-Ile production and subsequent JAZ protein destruction. These unshaded plants have a high defense capacity. Intermediate situations must be predominant in nature. Figure based on Zhai and Li (2019), Wang et al. (2019), Williams et al. (2019), and Ballaré and Austin (2019).
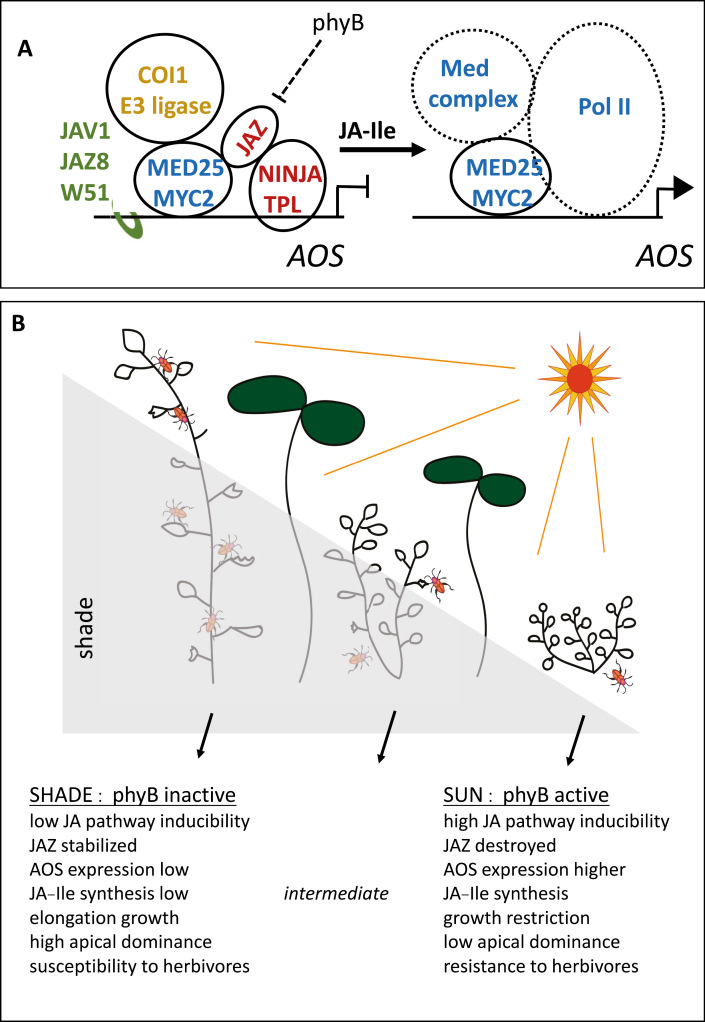
Many jasmonate biosynthesis genes are jasmonate-inducible and among these are many AOSs. The structure of the Arabidopsis AOS protein has been resolved (Lee et al., 2008), but how is expression of the AOS gene regulated in this plant? Yan et al. (2018) showed that the promoter of the single Arabidopsis AOS gene is bound by a complex of at least three proteins: JASMONATE-ASSOCIATED VQ MOTIF GENE 1 (JAV1), JAZ8, and the transcription factor WRKY51. Two of these proteins, JAV1 and WRKY51, bind to a W-box DNA sequence region in the AOS promoter. Upon wounding, JAV1 is phosphorylated through a Ca2+/calmodulin-dependent mechanism and this leads to destruction of JAV1, which then allows AOS expression. Consistent with a role of JAV1, JAZ8, and WRKY51 acting together to repress AOS transcription, plants with reduced JAV1 and WRKY51 gene activity are more resistant to herbivory than the wild type. Yan et al. (2018) point out that still more mechanisms must exist to regulate wound-induced AOS expression. They note that undamaged plants with lowered levels of JAV1 and WRKY51 still grow similarly to the wild-type, indicating that the jasmonate pathway is not highly up-regulated in these plants.
Jasmonate involvement in development and growth
Jasmonates regulate development in both reproductive and vegetative tissues in angiosperms. In tomato, jasmonate pathway activity is necessary for both female and male functions (Dobritzsch et al., 2015). With regards to male development, jasmonates function to inhibit ethylene synthesis and this prevents premature desiccation of stamens (Dobritzsch et al., 2015). In Arabidopsis, defects in JA synthesis or signaling impair filament elongation, pollen viability, and dehiscence. Interestingly, restoring COI1 expression specifically in the epidermises of the anther and stamen filament rescues these events in male-sterile coi1 mutants. This suggests that a JA-dependent cell-to-cell signal produced in the epidermis coordinates the major events associated with pollen development and release (Jewell and Browse, 2016). Importantly, jasmonates also act as developmental signals in wounded vegetative tissues. In Arabidopsis leaf explants, wounding causes induction of JA-dependent ETHYLENE-RESPONSE FACTOR 109 (ERF109) that leads to enhanced auxin synthesis, which drives the production of roots (Zhang et al., 2019). In roots, Zhou et al. (2019) have shown that nematode attack and even soil penetration activate the JA pathway to increase the expression of ERF109. This in turn activates the expression of ERF115 to promote root stem cell regeneration. These studies provide good examples of how AOS-derived signal molecules function along with ethylene in plant development.
Roles of AOS-derived oxylipins in growth responses have been studied in increasing detail since it was found that whilst wild-type plants that have been wounded repetitively have short petioles and small leaves, repetitively wounded aos mutants fail to show growth restriction (Yan et al., 2007). The oxylipin-dependent vegetative growth inhibition response is not unique to angiosperms. A similar phenomenon has recently been discovered in gametophytes of the liverwort Marchantia, where wound-induced growth inhibition is dependent on COI1–JAZ interaction promoted by dinor-OPDA (Monte et al., 2018). Oxylipin-dependent growth restriction therefore dates back to some of the earliest land plants. Returning to Arabidopsis, what molecular events cause wound-response growth restriction that is seen in wild-type plants? The results of a number of studies have been consistent with a growth/defense trade-off whereby plants redirect resources from growth to defense when there are simply not enough to do both (Huot et al., 2014). A recent paper by Guo et al. (2018) sheds more light on this.
Numerous regulatory proteins including many transcription factors can bind to JAZ proteins. This strongly interlinks the jasmonate pathway to signal pathways controlled by major regulators such as auxin, gibberellin, salicylate, and ethylene (Howe et al., 2018), making it particularly difficult to predict what would happen if multiple members of the 13-member Arabidopsis JAZ gene family were mutated. To investigate this, Guo et al. (2018) generated an Arabidopsis decuple jaz mutant (jazD). Unwounded jazD plants had short roots, short petioles, and reduced seed production. Concomitantly, they had higher levels of jasmonate/wound-response transcripts than the wild type and showed strong defense against a chewing herbivore. Not surprisingly, among the proteins up-regulated in jazD was AOS. Interestingly, leaves of jazD produced higher levels of herbivore defense-associated endoplasmic reticulum (ER) bodies and accumulated higher levels of several indole glucosinolates than the wild type. These results indicate that JAZ genes, central components of jasmonate signaling, coordinate jasmonate pathway activity so that plants can direct carbon into the production of defense metabolites and, at the same time, ensure that this does not completely eliminate growth and reproduction (Guo et al., 2018).
Jasmonate and environmental control of growth
How can we integrate what is known about jasmonate action into the larger context of plant responses to the environment? Ongoing work aimed at understanding natural variations that affect jasmonate pathway activity in Nicotiana attenuata (Ray et al., 2019) promise to greatly extend our appreciation of jasmonate activity in nature. In parallel, there is constant progress in understanding the molecular basis of defense against insects (reviewed in Erb and Reymond, 2019; Wang et al., 2019). Here, we consider one environmental input, namely light, on plant growth and defense. Box 2B illustrates the growth and defense status of plants in nature where light quality modulates the activity of the jasmonate pathway, specifically in a situation where far-red light is increased relative to red light as a result of the use of the latter by neighbouring plants. In direct sunlight, phytochrome B (phyB) signaling has an activating effect on the jasmonate pathway, and plants growing in sunlight have strongly inducible defenses against herbivores. However, in the shade of other plants (characterized by a low red to far-red light ratio), phyB is inactivated locally, and shaded parts of plants display a suite of developmental responses including elongation growth, organ reorientation, and strong apical dominance. This has consequences for herbivore success: plants in the shade have a reduced ability to defend themselves (Cerrudo et al., 2017). Ballaré and Austin (2019) see this somewhat differently than as a ‘growth–defense trade-off’. They argue that jasmonate affects growth in a directly different way to the effect of competition between plants for light that triggers shade avoidance (elongation) morphology. Whereas stunted growth due to jasmonate signaling may limit tissue availability to herbivores, elongation growth and apical dominance in shaded plants that seek light are seen as maladaptive in the face of attack. These authors name this as ‘configurational conflict’. The crux of the argument is that plants have fundamentally different growth plans that either favour light-gathering or reduce availability to herbivores.
If plants that are well adapted to their characteristic environment are considered, it would be expected that the majority of them would be in the ‘intermediate’ category illustrated in Box 2B. Their statures will allow them to survive in their co-adapted community. Having just sufficient access to light, their jasmonate-dependent defenses will be relatively well functioning. Moving these plants into an adjacent environment that is more sunny will perhaps allow them to defend themselves fully. However, moving them into a shaded area within the same environment will trigger elongation growth and suppress jasmonate signaling. How these events affect plant fitness is not simple to predict. Firstly, herbivores that may thrive on shade-grown plants might, due to plant elongation growth, be more visible to predators than they would be on compact plants. Secondly, other as yet unknown environmental factors may exert powerful control over JA pathway activity and this will probably depend on the plant species. A challenge will be to identify further factors that affect the activity of the jasmonate pathway and to relate this to plant growth modes. Finally, the consequences of jasmonate pathway activity for plant fitness are far from understood. For example, if tall-growing invasive plants come into a new habitat and they shade well-adapted indigenous plants, they will probably reduce jasmonate pathway inducibility in those plants. In this context, the human manipulation of environments, for example through plant introductions, would lead indirectly to manipulation of jasmonate pathway activity. This may have consequences for entire ecosystems.
Conclusions
Recent progress continues to highlight the great diversity of AOS-derived oxylipins in plants. These oxylipins include jasmonates, which play remarkable roles in defense, development, growth and in other processes. Jasmonates link defense and growth, and much effort over the last decade has concentrated on this link. In parallel, the molecular events underlying jasmonate signaling are becoming increasingly clear, although many of the roles of jasmonates in nature may still be unknown to us. There is much more to discover about the roles of AOS-derived oxylipins and the enzymes that produce them in all plant groups. For example, new interactions between oxylipins and other hormone signal pathways are likely to be revealed, and it will be interesting to investigate how environmental inputs impinge on oxylipin activities at different evolutionary levels. In particular, oxylipin research in lineages such as lycophytes, ferns, and gymnosperms clearly requires more attention and is likely to yield valuable insights into plant evolution. AOS-derived oxylipins help make all plants what they are.
Acknowledgements
We thank O. Michaud and Y-Q. Gao (University of Lausanne) for critical reading of the original manuscript.
References
- Ameye M, Allmann S, Verwaeren J, Smagghe G, Haesaert G, Schuurink RC, Audenaert K. 2018. Green leaf volatile production by plants: a meta-analysis. New Phytologist 220, 666–683. [DOI] [PubMed] [Google Scholar]
- An C, Li L, Zhai Q, You Y, Deng L, Wu F, Chen R, Jiang H, Wang H, Chen Q, Li C. 2017. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proceedings of the National Academy of Sciences, USA 114, E8930–E8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Austin AT. 2019. Recalculating growth and defense strategies under competition: key roles for photoreceptors and jasmonates. Journal of Experimental Botany 70, 3425–3435. [DOI] [PubMed] [Google Scholar]
- Brash AR. 2009. Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes. Phytochemistry 70, 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Elberse J, Awwanah M, Ludwig NR, de Vries M, Zeilmaker T, Van Wees SC, Schuurink RC, Van den Ackerveken G. 2017. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proceedings of the National Academy of Sciences, USA 114, 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Caliri-Ortiz ME, Keller MM, Degano ME, Demkura PV, Ballaré CL. 2017. Exploring growth-defence trade-offs in Arabidopsis: phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shade-avoidance responses. Plant, Cell & Environment 40, 635–644. [DOI] [PubMed] [Google Scholar]
- Christensen SA, Huffaker A, Kaplan F, et al. 2015. Maize death acids, 9-lipoxygenase–derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proceedings of the National Academy of Sciences, USA 112, 11407–11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritzsch S, Weyhe M, Schubert R, Dindas J, Hause G, Kopka J, Hause B. 2015. Dissection of jasmonate functions in tomato stamen development by transcriptome and metabolome analyses. BMC Biology 13, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Reymond P. 2019. Molecular interactions between plants and insect herbivores. Annual Review of Plant Biology 70, 527–557. [DOI] [PubMed] [Google Scholar]
- Grechkin AN, Ogorodnikova AV, Egorova AM, Mukhitova FK, Ilyina TM, Khairutdinov BI. 2018. Allene oxide synthase pathway in cereal roots: detection of novel oxylipin graminoxins. ChemistryOpen 7, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Yoshida Y, Major IT, Wang K, Sugimoto K, Kapali G, Havko NE, Benning C, Howe GA. 2018. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115, E10768–E10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Van Verk MC, Van Dijken AJH, et al. 2017. Architecture and dynamics of the jasmonic acid gene regulatory network. The Plant Cell 29, 2086–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ. 2018. Modularity in jasmonate signaling for multistress resilience. Annual Review of Plant Biology 69, 387–415. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. 2014. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JB, Browse J. 2016. Epidermal jasmonate perception is sufficient for all aspects of jasmonate-mediated male fertility in Arabidopsis. The Plant Journal 85, 634–647. [DOI] [PubMed] [Google Scholar]
- Krausko M, Perutka Z, Šebela M, Šamajová O, Šamaj J, Novák O, Pavlovič A. 2017. The role of electrical and jasmonate signalling in the recognition of captured prey in the carnivorous sundew plant Drosera capensis. New Phytologist 213, 1818–1835. [DOI] [PubMed] [Google Scholar]
- Lee DS, Nioche P, Hamberg M, Raman CS. 2008. Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455, 363–368. [DOI] [PubMed] [Google Scholar]
- Monte I, Ishida S, Zamarreño AM, et al. 2018. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nature Chemical Biology 14, 480–488. [DOI] [PubMed] [Google Scholar]
- Ogorodnikova AV, Mukhitova FK, Grechkin AN. 2015. Oxylipins in the spikemoss Selaginella martensii: detection of divinyl ethers, 12-oxophytodienoic acid and related cyclopentenones. Phytochemistry 118, 42–50. [DOI] [PubMed] [Google Scholar]
- Pavlovič A, Mithöfer A. 2019Jasmonate signaling in carnivorous plants: copycat of plant defence mechanisms. Journal of Experimental Botany 70, 3379–3390. [DOI] [PubMed] [Google Scholar]
- Poudel AN, Zhang T, Kwasniewski M, Nakabayashi R, Saito K, Koo AJ. 2016. Mutations in jasmonoyl-L-isoleucine-12-hydroxylases suppress multiple JA-dependent wound responses in Arabidopsis thaliana. Biochimica et Biophysica Acta 1861, 1396–1408. [DOI] [PubMed] [Google Scholar]
- Ray R, Li D, Halitschke R, Baldwin IT. 2019. Using natural variation to achieve a whole‐plant functional understanding of the responses mediated by jasmonate signaling. The Plant Journal. doi: 10.1111/tpj.14331. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Marquis V, Poirier L, Aubert Y, Zumsteg J, Ménard R, Miesch L, Heitz T. 2017. Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea Infection. Molecular Plant 10, 1159–1173. [DOI] [PubMed] [Google Scholar]
- Wang J, Wu D, Wang Y, Xie D. 2019. Jasmonate action in plant defense against insects. Journal of Experimental Botany 70, 3391–3400. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Feussner I. 2018. The oxylipin pathways: biochemistry and function. Annual Review of Plant Biology 69, 363–386. [DOI] [PubMed] [Google Scholar]
- Williams C, Fernández-Calvo P, Colinas M, Pauwels L, Goossens A. 2019. Jasmonate and auxin perception: how plants keep F-boxes in check. Journal of Experimental Botany 70, 3401–3414. [DOI] [PubMed] [Google Scholar]
- Yan C, Fan M, Yang M, Zhao J, Zhang W, Su Y, Xiao L, Deng H, Xie D. 2018. Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Molecular Cell 70, 136–149.e7. [DOI] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. 2007. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li C. 2019. The plant Mediator complex and its role in jasmonate signaling. Journal of Experimental Botany 70, 3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhao F, Chen L, et al. 2019. Jasmonate-mediated wound signalling promotes plant regeneration. Nature Plants 5, 491–497. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. 2017. Jasmonate signaling and manipulation by pathogens and insects. Journal of Experimental Botany 68, 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B. 2019. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942–956.e14. [DOI] [PubMed] [Google Scholar]


