Abstract
The Mediator complex is an essential, multisubunit transcriptional coactivator that is highly conserved in eukaryotes. Mediator interacts with gene-specific transcription factors, the RNA polymerase II transcriptional machinery, as well as several other factors involved in transcription, and acts as an integral hub to regulate various aspects of transcription. Recent studies of the plant Mediator complex have established that it functions in diverse aspects of plant development and fitness. Jasmonate (JA) is an oxylipin-derived plant hormone that regulates plant immunity and development. The basic helix–loop–helix transcription factor MYC2, which is a master regulator of JA signaling, orchestrates genome-wide transcriptional reprogramming of plant cells to coordinate defense- and growth-related processes. Here, we review the function of the plant Mediator complex in regulating JA signaling. We focus on the multifunctional Mediator subunit MED25, which emerges as an integrative hub for the transcriptional regulation of jasmonate signaling.
Keywords: Jasmonate signaling, MED25, Mediator, MYC2, transcriptional regulation
We present an update on the function of the plant Mediator complex in the regulation of jasmonate signaling, and focus on the role of the MED25 subunit in jasmonate-signaled transcriptional regulation.
Introduction
Dynamic changes in eukaryotic gene expression in response to intracellular or environmental stimuli are largely regulated by DNA-binding transcription factors (TFs). Mediator is a multiprotein complex that functions as a signal integrator transmitting information from the DNA-binding TFs to the RNA polymerase II (Pol II) transcriptional machinery (Malik and Roeder, 2010). Kornberg and colleagues first identified Mediator in yeast in 1990 (Kelleher et al., 1990). The addition of a crude protein fraction from yeast to a reconstituted Pol II transcriptional system relieved competition between activators for a common target that was present in a limited amount. The activity in this crude protein fraction was named Mediator. Subsequently, similar activities were reported in mammals, both in vitro (Kretzschmar et al., 1993; Merino et al., 1993) and in vivo (Keaveney et al., 1993). In 1994, Kornberg and colleagues purified the yeast Mediator as a large complex containing 20 subunits (Kim et al., 1994). The first mammalian Mediator-like complex was isolated from human HeLa cells by Roeder and colleagues (Fondell et al., 1996). Several laboratories used conventional chromatography to purify Mediator-like complexes from mouse, rat, and other species (reviewed in Conaway et al., 2005). The current evidence indicates that Mediator is an evolutionarily conserved multisubunit protein complex comprising 25 subunits in yeast and up to 30 subunits in human (Fig. 1; Soutourina, 2018).
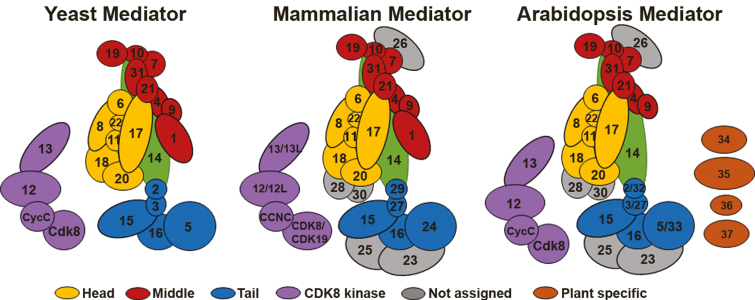
Fig. 1.
Subunit composition of the Mediator complex. The illustrations represent the modular organization of the Mediator complex in yeast (A), mammals (B), and Arabidopsis (C) based on a recently revised structure derived from electron microscopy data in yeast and human complexes. Mediator comprises four distinct modules: Head (yellow), Middle (red), Tail (blue), and CDK8 (purple). Mediator subunit 14 (MED14), which links the three main modules (Head, Middle, and Tail), is indicated in green. The exact module localization of five metazoan and plant conserved subunits (MED23, MED25, MED26, MED28, and MED30) remains to be assigned (subunits indicated in gray). Four plant-specific subunits are indicated in orange.
Structural studies reveal that Mediator subunits form stable subcomplexes, and the structure of the whole complex can be divided into three main modules (Head, Middle, and Tail) plus a transiently associated cyclin-dependent kinase 8 (CDK8) module (Asturias et al., 1999; Dotson et al., 2000; Bourbon, 2008; Tsai et al., 2014; Wang et al., 2014). The modular organization of Mediator may reflect the different functions of Mediator components. Structural and biochemical studies in yeast and human indicate that the subunits comprising the Head and Middle modules are tightly associated with each other and constitute a stable core (Cevher et al., 2014; Plaschka et al., 2015). The Head module binds to Pol II and stabilizes the transcription initiation complex. The Middle module extends to the Pol II foot and may influence polymerase conformation (Cevher et al., 2014; Plaschka et al., 2015). Different from the Head and Middle modules, the individual subunits of the Tail module are relatively loosely associated, and are considered to be targeted by specific TFs (Blazek et al., 2005). Mediator subunit14 (MED14) contacts all three main modules (Head, Middle, and Tail) and has a critical role in Mediator organization (Cevher et al., 2014; Plaschka et al., 2015). The CDK8 module reversibly associates with the main modules and broadly tends to have a regulatory function (Knuesel et al., 2009; Tsai et al., 2013). The Mediator structure can shift dramatically on binding to other proteins or protein complexes (Allen and Taatjes, 2015). These structural rearrangements are essential for Pol II holoenzyme formation, and could explain the capacity of Mediator to integrate multiple regulatory signals (Tsai et al., 2017).
The most well-studied function of Mediator is its ability to regulate the formation of the transcriptional pre-initiation complex (PIC). Mediator is recruited to the target promoter and enhancer regions via direct interactions with specific TFs, while also directly interacting with Pol II and other PIC components. Thus, Mediator acts as a molecular bridge that communicates regulatory signals from DNA-binding TFs to the Pol II enzyme (Holstege et al., 1998; Asturias et al., 1999; Myers et al., 1999; Davis et al., 2002; Bernecky et al., 2011). After PIC formation, Mediator is required for phosphorylation of the carboxy-terminal domain of Pol II, which in turn disrupts the Mediator–Pol II interaction and results in the escape of Pol II from the promoter (Nair et al., 2005; Boeing et al., 2010; Wong et al., 2014). Structural and biochemical data also implicate Mediator in regulating the functions of Pol II subunit M (POLR2M) and the TFIIF TF during Pol II pausing and elongation (Cheng et al., 2012; Jishage et al., 2012; Wu et al., 2012). The metazoan-specific MED26 subunit of Mediator is reported to interact with the super-elongation complex to facilitate Pol II elongation. CDK8 is reported to positively regulate Pol II elongation during serum and hypoxia responses (Donner et al., 2010; Galbraith et al., 2013). Newly emerging functions are continuously ascribed to yeast and metazoan Mediator in regulating almost every step of Pol II-dependent gene transcription, including chromatin remodeling and modification, non-coding RNA activation, mRNA processing, chromatin loop formation, and super-enhancer-dependent transcriptional regulation (Malik and Roeder, 2010; Poss et al., 2013; Hnisz and Young, 2017; Jeronimo and Robert, 2017; Soutourina, 2018). Here, we review the function of Mediator complex in plants, especially its role in jasmonate (JA) signaling.
Mediator complex in plants
The first plant Mediator complex was isolated from Arabidopsis in 2007 (Bäckström et al., 2007). Björklund and colleagues performed biochemical purification of the complex and identified 21 conserved and six plant-specific Mediator subunits. Although the CDK8 module was not identified in that study, Arabidopsis has homologs to MED12, MED13, CDK8, and CycC genes encoding subunits of the CDK8 module (Bäckström et al., 2007). Bourbon performed a bioinformatics analysis to identify an Arabidopsis homolog of metazoan-specific MED30, a subunit that was not purified from the Arabidopsis complex. That study also identified that the plant-specific MED32 and MED33 subunits were homologs of the yeast/metazoan MED2/29 and MED5/24 subunits, respectively (Bourbon, 2008). Recent evidence suggests that the metazoan-specific MED26 subunit may have a homolog in land plant species (Mathur et al., 2011). By contrast, the conserved yeast and metazoan MED1 subunit was not biochemically purified or identified by bioinformatics analysis in Arabidopsis. Therefore, the Arabidopsis Mediator complex currently comprises 29 conserved and four plant-specific subunits (Fig. 1).
Comparative genomics analyses indicate that homologs of the Arabidopsis Mediator subunits are expressed in diverse plants, such as Oryza sativa and Physcomitrella patens (Mathur et al., 2011). The subunit sequences displayed high similarity in different plants, suggesting that the function of the corresponding subunits is conserved in different plant species (Yang et al., 2016). Several Mediator subunits were studied genetically before biochemical purification of the Arabidopsis Mediator complex, including PHYTOCHROME and FLOWERING TIME1 (PFT1)/MED25 and STRUWWELPETER (SWP)/MED14 (Autran et al., 2002; Cerdán and Chory, 2003). A growing body of evidence suggests that plant Mediator subunits participate in multiple biological processes including flowering, defense, abiotic stress, non-coding RNA production, genomic stability, metabolism homeostasis, and growth and development (Kidd et al., 2011; Kim and Chen, 2011; An and Mou, 2013; Samanta and Thakur, 2015; Yang et al., 2016; Dolan and Chapple, 2017).
MYC-orchestrated transcriptional regulation is a central theme of JA signaling
Plant oxylipins such as JAs are a major class of immunity hormone that functions in diverse aspects of plant immunity and development (Creelman and Mullet, 1997; Turner et al., 2002; Howe and Jander, 2008; Browse, 2009; Sun et al., 2011; Kazan and Manners, 2013; Wasternack and Hause, 2013; Zhai et al., 2017b). Extensive studies in Arabidopsis have revealed a core JA signaling module consisting of the F-box protein CORONATINE INSENSITIVE 1 (COI1) that forms a functional SCF E3 ubiquitin ligase complex (Devoto et al., 2002; Xu et al., 2002), a group of Jasmonate ZIM-domain (JAZ) proteins that function as transcriptional repressors (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007), and a battery of TFs that differentially regulate diverse aspects of JA responses (Gimenez-Ibanez et al., 2015; Zhai et al., 2017b). Among these TFs, the basic helix–loop–helix TF MYC2 is the most important and well-studied and differentially regulates two branches of JA-responsive genes (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Kazan and Manners, 2013; Zhai et al., 2013). A breakthrough in the mechanistic understanding of JA signal transduction was the discovery that jasmonoyl-isoleucine (JA-Ile) is the receptor-active form of the hormone and that JA-Ile perception involves the formation of a SCFCOI1–JAZ co-receptor complex (Fonseca et al., 2009; Yan et al., 2009; Pauwels et al., 2010; Sheard et al., 2010). In the absence of the hormone, JAZ proteins interact with and prevent MYC2 from activating JA-responsive gene expression. In response to stimuli that trigger JA-Ile synthesis, JA-Ile promotes direct binding of JAZ repressors to the F-box protein COI1. SCFCOI1-dependent degradation of JAZ repressors via the ubiquitin–proteasome system leads to de-repression of MYC2-directed transcription of JA-responsive genes. In addition to MYC2, other members of the bHLH TFs including MYC3, MYC4, and MYC5 work redundantly to regulate the expression of JA-responsive genes (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011; Figueroa and Browse, 2015; Qi et al., 2015). These data indicate that MYC-orchestrated transcriptional reprogramming is a central theme of JA signaling (Kazan and Manners, 2013; Wasternack and Hause, 2013; Zhai et al., 2017b).
Tomato is another plant model system that has been extensively studied to identify the mechanisms underlying JA-triggered plant immunity (Ryan, 2000; Schilmiller and Howe, 2005; Howe and Jander, 2008; Sun et al., 2011; Rosli and Martin, 2015). The major molecular components that constitute the core JA signaling pathway in tomato (i.e. COI1, JAZs, and MYC2) are largely conserved with those in Arabidopsis (Boter et al., 2004; Li et al., 2004; Sun et al., 2011). However, the action modes of specific molecular components differ in the two species. For example, in both tomato and Arabidopsis, the active hormone JA-Ile promotes COI1-dependent degradation of JAZ repressors and thereby de-represses the master TF, MYC2. In Arabidopsis, MYC2 positively regulates wounding-responsive genes and negatively regulates pathogen-responsive genes (Lorenzo et al., 2004; Dombrecht et al., 2007). By contrast, in tomato, MYC2 positively regulates both wounding- and pathogen-responsive genes. Tomato MYC2 directly regulates the transcription of its downstream intermediate TFs, which in turn regulate the expression of late wounding- or pathogen-responsive genes. MYC2 and its downstream intermediate TFs form a hierarchical transcriptional cascade that initiates and amplifies JA-mediated transcriptional output in tomato (Du et al., 2017). The functional divergence of the two MYC2 TFs might be due to recruitment of different cofactors while regulating the transcription of pathogen-responsive genes (Du et al., 2017). Nevertheless, MYC-mediated transcriptional regulation is a central theme of JA signaling in both Arabidopsis and tomato.
MED25 is an integrator of JA-mediated transcriptional activation
The COI1-dependent degradation of JAZ repressors leads to the ‘de-repression’ of MYC TFs (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009; Sheard et al., 2010). In turn, free MYC TFs form a transcriptional activation complex with the MED25 subunit of the plant Mediator to activate the expression of JA-responsive genes (Çevik et al., 2012; Chen et al., 2012; Zhang et al., 2015; An et al., 2017; Zhai et al., 2017a). First, MED25 functionally interacts with both MYC2 and Pol II, thereby promoting PIC assembly during MYC2-dependent transcription of JA-responsive genes (Chen et al., 2012). Second, MED25 physically recruits COI1 to MYC2 target promoters in the resting state, and facilitates COI1-dependent degradation of JAZ repressors in the presence of JA-Ile (An et al., 2017). Third, MED25 physically and functionally interacts with HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1 (HAC1), which selectively regulates hormone-induced acetylation of lysine 9 (K9) in histone H3 (H3K9ac) at MYC2 target promoters (An et al., 2017). These combined results indicate that MED25 acts as an integrative hub to coordinate the actions of multiple regulators during hormone-triggered MYC2 activation.
MED25 promotes PIC assembly on MYC2 target promoters
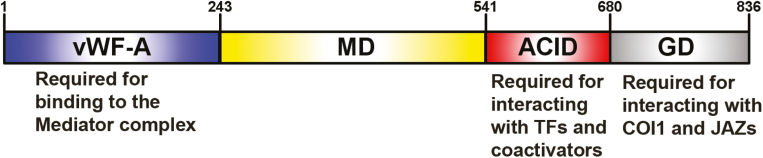
The MED25 subunit has not been identified in yeast, and the location of MED25 within the Mediator complex is unknown (Soutourina, 2018). However, human MED25 interacts extensively with different Tail subunits and gene-specific TFs, suggesting that MED25 is most likely associated with the Tail module of the Mediator complex (Fig. 1; Tsai et al., 2014; Soutourina, 2018). Consistently, Arabidopsis MED25 also interacts with the Tail module MED16 subunit and several TFs (Elfving et al., 2011; Ou et al., 2011; Çevik et al., 2012; Chen et al., 2012; Yang et al., 2014; Ito et al., 2016). Arabidopsis MED25 contains a von Willebrand factor type A (vWF-A) domain (MED251–242), a non-conserved middle domain (MD, MED25243–540), an activator-interacting domain (ACID, MED25541–680), and a glutamine-rich domain (GD, MED25681–836) (Fig. 2). The amino-terminal vWF-A domain, which is conserved in mammalian and plant MED25, is required for MED25 binding with the Mediator complex (Mittler et al., 2003; Yang et al., 2014). The MED25 ACID is essential for its interaction with different TFs and coactivators, such as MYC TFs (Çevik et al., 2012; Chen et al., 2012; Zhang et al., 2015), APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) TFs (Ou et al., 2011), and histone acetyltransferase HAC1 (An et al., 2017). The C-terminal GD is necessary for MED25 interaction with the JA receptor COI1 and JAZ repressors (An et al., 2017; Zhai et al., 2017a).
Fig. 2.
Schematic representation of the MED25 protein structure. ACID, activator-interacting domain; GD, glutamine-rich domain; MD, non-conserved middle domain; vWF-A, von Willebrand factor type A domain.
MED25 was originally identified in Arabidopsis (previously designated as PFT1) as a factor of the phytochrome B (phyB) signaling pathway, which promotes flowering in response to shade (Cerdán and Chory, 2003). Biochemical purification of the Arabidopsis Mediator revealed that PFT1 was homologous to the MED25 subunit of the metazoan Mediator complex (Bäckström et al., 2007). Early evidence that plant MED25 participates in JA signaling came from a study in Kazan’s lab. They found that an Arabidopsis pft1/med25 mutant exhibited reduced expression of JA-responsive genes and attenuated resistance in response to necrotrophic pathogen infection, suggesting that MED25 positively regulates JA-mediated plant defense (Kidd et al., 2009). In addition, they found that a wheat homolog of MED25 could complement the defense and the developmental defects of the Arabidopsis pft1/med25 mutant, suggesting that the function of MED25 in regulating JA signaling is conserved in higher plants (Kidd et al., 2009). Further evidence for the role of MED25 in JA signaling came from our genetic analysis of the bestatin resistant 6 (ber6) mutant, which exhibits attenuated sensitivity to JA-mediated root growth inhibition (Zheng et al., 2006) and reduced expression of the two branches of JA-responsive genes (Chen et al., 2012). Map-based cloning combined with complementation analysis revealed that the defective JA response phenotype of ber6 resulted from a mutation in MED25 (Chen et al., 2012). Very recently, tomato MED25 has been implicated in the regulation of JA-mediated wounding and pathogen responses (Liu et al., 2019), further confirming the conserved function of MED25 in higher plants.
The fundamental role of MED25 in JA signaling is to promote PIC assembly on MYC2 target promoters. In both Arabidopsis and tomato, MED25 physically interacts with MYC2 (Çevik et al., 2012; Chen et al., 2012; Liu et al., 2019). The MED25 ACID and the MED25-interacting domain (MID) of MYC2 are necessary for MED25–MYC2 interactions (Chen et al., 2012; Liu et al., 2019). The MID is located on the transcriptional activation domain of MYC2 (Chen et al., 2012; Liu et al., 2019). MYC2 recruits MED25 to its target promoters in a JA-dependent manner, and in turn, MED25 recruits Pol II to the promoter of MYC2 targets (Chen et al., 2012). Thus, during hormone-triggered transcription of JA-responsive genes, MED25 directly bridges communication between the gene-specific MYC2 TF and the Pol II general transcription machinery for PIC assembly (Chen et al., 2012).
MED25 facilitates COI1-dependent de-repression of JA signaling
In addition to interacting with the master TF, MYC2, MED25 also interacts with the hormone receptor protein COI1 and a subset of JAZ repressors. The MED25 GD is necessary for its interaction with both COI1 and JAZ proteins (An et al., 2017; Zhai et al., 2017a). In the absence of active hormone, MYC2 forms a ternary complex together with MED25 and JAZ proteins (An et al., 2017); JAZ repressors compete with MED25 for interaction with MYC TFs (Zhang et al., 2015). The MYC2–JAZ interaction is relatively strong and the MYC2–MED25 interaction is relatively weak. MED25 has a relatively strong interaction with COI1 and brings COI1 to MYC2 target promoters. In the presence of active hormone, JA-Ile acts as molecular glue to promote the formation of the COI1–JAZ co-receptor complex, which leads to proteasome-dependent degradation of JAZ repressors. During this stage, MED25 contributes to JA-Ile-mediated promotion of the COI1–JAZ interaction and subsequent JAZ degradation (An et al., 2017). Upon JAZ degradation, the MED25–COI1 interaction is weakened, whereas the MED25–MYC2 interaction is enhanced, which favors MYC2-mediated transcription of JA-responsive genes (An et al., 2017). These results indicate that MED25 has an important role in the dynamic changes of JA signaling from the repression stage to the de-repression stage (Fig. 3).
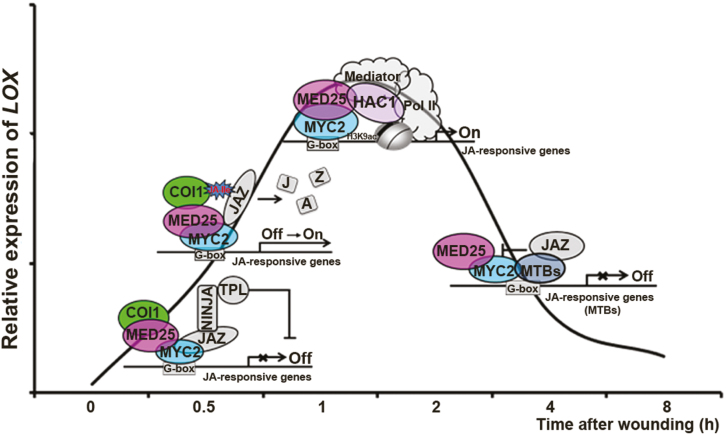
Fig. 3.
A model of MED25 regulation of different stages of JA signaling. In response to wounding or other stimulus, MED25 coordinates the actions of multiple components of JA signaling to regulate the expression of JA-responsive genes such as LOX. In the absence of stimulus, MED25 interacts with COI1 at the LOX promoter, whereas JAZ proteins interact with MYC2 and repress MYC2 activity. The expression level of LOX is low. In response to stimulus, MED25 promotes JA-triggered interaction of COI1 and JAZ proteins and subsequent degradation of JAZ repressors, which released the activity of MYC2. Then, MED25 interacts with MYC2 and recruits the coactivator HAC1 to activate LOX expression. The MYC2–MED25 functional complex activates the expression of MTBs, which form an autoregulatory negative feedback circuit to terminate JA signaling and thus reduce the LOX expression level. COI1, CORONATINE INSENSITIVE 1; JA, jasmonate; JA-Ile, jasmonoyl-isoleucine; JAZ, jasmonate-ZIM domain; LOX, LIPOXYGENASE; MED25, Mediator subunit 25; MTB, MYC2-TARGETED BHLH; NINJA, novel interactor of JAZ; TPL, TOPLESS.
The action mode of plant MED25 in regulating JA signaling exhibits a striking analogy to that of metazoan MED25, which participates in ligand-dependent interactions with several nuclear receptors (NRs) (Malik and Roeder, 2010; Fondell, 2013; Poss et al., 2013; Allen and Taatjes, 2015). Mammalian NRs are ligand-activated TFs that contain a conserved DNA-binding domain and a ligand-binding domain. After binding to hormone, NRs undergo significant conformational changes that create hydrophobic binding surfaces suitable for the interaction with MED25 (Malik and Roeder, 2010; Fondell, 2013; Poss et al., 2013; Allen and Taatjes, 2015). The COI1–JAZ–MYC2 module in JA signaling resembles the metazoan NR system with respect to its interactions with MED25. JA-Ile-triggered COI1-dependent degradation of JAZ proteins releases the interaction surface of MYC2 with MED25, which resembles the hormone-triggered conformational changes in NRs (An et al., 2017; Zhai et al., 2017a). These studies reveal that plants and animals have evolved distinct, but largely similar, mechanisms for NR activation at the level of transcriptional regulation.
MED25 cooperates with HAC1 to activate MYC2-mediated transcription
During the activation stage, MED25 directly interacts with and recruits HISTONE ACETYLTRANSFERASE1 (HAC1), an evolutionarily conserved histone modification enzyme, to MYC2 target promoters (An et al., 2017). MED25 interacts with HAC1 via its ACID, which also is required for the MED25–MYC2 interaction (Chen et al., 2012; An et al., 2017), suggesting that the ACID is important for the MED25 activation function (Fig. 2). HAC1 selectively regulates H3K9ac at MYC2 target promoters, which favors gene activation (An et al., 2017). This action mode of MED25 also is conserved between Arabidopsis and human. Human MED25 cooperates with CREB-binding protein, which is the human ortholog of the Arabidopsis HAC1, in the activation of retinoic acid receptor-mediated gene expression (Lee et al., 2007).
These combined results reveal that MED25 is involved in the assembly of a MYC2–MED25 functional transcription complex, which acts as an integrative hub that coordinates the actions of multiple regulators during hormone-triggered MYC2 activation (Fig. 3).
The MYC2–MED25 functional complex regulates the termination of JA signaling
Our recent study identified an additional and unexpected function of the MYC2–MED25 complex in regulating the termination of JA signaling in tomato (Liu et al., 2019). MYC2 and MED25 activate the expression of three bHLH TFs, MYC2-TARGETED BHLH 1 (MTB1), MTB2, and MTB3. MTB proteins negatively regulate JA signaling via their antagonistic effects on the functionality of the MYC2–MED25 transcriptional activation complex. MTB proteins impair the formation of the MYC2–MED25 complex and compete with MYC2 in binding to its target gene promoters. Therefore, MYC2–MED25 and MTB proteins form an autoregulatory negative feedback circuit to terminate JA signaling in a highly organized manner (Liu et al., 2019). Considering that the MYC2–MED25 functional complex has a central role in the initiation and amplification of JA-mediated transcriptional responses (Chen et al., 2012; An et al., 2017; Du et al., 2017), the formation of the MYC2–MTB feedback loop is already pre-programmed during the induction phase of JA signaling. Thus, in addition to controlling the initiation and amplification of JA-mediated transcriptional responses, the MYC2–MED25 functional complex also executes intrinsic termination of JA-mediated defense responses.
Phylogenetic analysis revealed that MTB proteins are homologs of the Arabidopsis JASMONATE-ASSOCIATED MYC2-LIKE (JAM) proteins (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014; Goossens et al., 2017; Liu et al., 2019). Similarly, JAM proteins play a negative role in JA-mediated defense responses by competing with the DNA-binding capacity of Arabidopsis MYC2 (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014). However, in tomato, MTB1 physically interacts with MYC2 and competitively inhibits the interaction of MYC2 and MED25; because the MYC2–MED25 complex is essential for MYC2-dependent PIC formation, the interference of MTB1 determines the transcriptional output of the MYC2–MED25 transcriptional activation complex (Chen et al., 2012; An et al., 2017; Du et al., 2017; Liu et al., 2019). In this regard, the action mechanism of MTB proteins is different from the Arabidopsis JAM proteins, which did not show physical interaction with AtMYC2 in yeast two-hybrid and coimmunoprecipitation assays (Song et al., 2013; Fonseca et al., 2014).
MED25 interacts with intermediate TFs of JA signaling
MYC2 is a master regulator of JA signaling and targets groups of intermediate TFs, which directly regulate the JA-induced transcription of late defense genes (Bu et al., 2008; Zheng et al., 2012; Zhai et al., 2013; Du et al., 2017). MED25 also interacts with these intermediate TFs, such as the AP2/ERF TFs OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59) and ERF1 (Ou et al., 2011; Çevik et al., 2012). The MED25 ACID and the transcription activation domain (TAD) of these ERFs are necessary for their interactions (Çevik et al., 2012). Transcriptional activation experiments indicate that ORA59- and ERF1-dependent activation of pathogen defense genes such as PLANT DEFENSIN1.2 (PDF1.2) requires a functional MED25. These studies implicate an extensive function of MED25 in regulating JA-mediated transcription. First, MYC2 recruits MED25 to activate the expression of the intermediate TFs; further, the intermediate TFs also recruit MED25 to activate the expression of downstream plant defense genes. MED25 acts as a major player in the hierarchical transcriptional cascade that initiates and amplifies transcriptional output. MED25 uses its ACID to interact with the MID (or TAD) of these different TFs, suggesting that interacting with MED25 is an important part of TF-mediated transcriptional activation in JA signaling.
Other Mediator subunits in JA signaling
Several other Mediator subunits have been reported to participate in JA-mediated plant immunity, including MED8, MED16, MED18, and CDK8 (Kidd et al., 2009; Wathugala et al., 2012; Zhang et al., 2012; Lai et al., 2014; Zhu et al., 2014; Li et al., 2018).
A mutation in the Arabidopsis MED8 gene results in susceptibility to Alternaria brassicicola but resistance to Fusarium oxysporum, suggesting a role for MED8 in JA-dependent plant immunity (Kidd et al., 2009). The med8 med25 double mutant exhibits an additive effect in the phenotypic deficiency relative to the single mutants, suggesting that MED8 and MED25 probably affect JA-mediated plant immunity via independent and additive mechanisms (Kidd et al., 2009). A recent study reported that MED8 regulates plant immunity to Botrytis cinerea by interacting with the FAMA TF, which was previously shown to control the final proliferation/differentiation switch during stomatal development (Li et al., 2018). The fama mutant exhibits increased susceptibility to B. cinerea infection and reduced defense-responsive gene expression. Genetic analyses of MED8 and FAMA suggest that FAMA-regulated plant defense against B. cinerea depends on MED8 function. FAMA recruits MED8 to the ORA59 promoter and activates ORA59 expression, which in turn regulates the expression of downstream plant defense genes (Li et al., 2018).
The role of Arabidopsis MED16 in regulating JA/ethylene (ET)-mediated plant immunity has been reported recently (Wathugala et al., 2012; Zhang et al., 2012; Wang et al., 2015). Mutants of MED16 are more susceptible to Sclerotinia sclerotiorum than mutants of 13 other Mediator subunits, and med16 is also more susceptible to S. sclerotiorum than coi1-1, which is the most susceptible mutant reported so far (Wang et al., 2015). Hence, MED16 is an important regulator of basal resistance against S. sclerotiorum. Furthermore, ET-induced suppression of JA-activated wound responses is compromised in med16, suggesting a role for MED16 in JA–ET cross talk (Wang et al., 2015). MED16 also physically associated with TF WRKY33 to regulate the transcription of PDF1.2 and ORA59, indicating that MED16 regulates resistance to S. sclerotiorum by governing both JA/ET-mediated and WRKY33-activated defense signaling in Arabidopsis (Wang et al., 2015). MED16 not only positively regulates the expression of JA-responsive genes but also positively regulates the expression of salicylic acid (SA)-responsive genes (Wathugala et al., 2012; Zhang et al., 2012). It is generally believed that the JA and SA signaling pathways antagonize each other (Kunkel and Brooks, 2002). Recently, it has been found that MED16 and the other two-tail module subunits, MED14 and MED15, are required for both SA- and ET-promoted inhibition of JA-mediated wound signaling. These results indicate that MED16 and some other Mediator subunits not only relay defense signaling from the SA and JA/ET pathways to the Pol II transcription machinery, but also fine-tune defense-related transcriptional changes (Wang et al., 2016). Another interesting point is that MED25 could interact with MED16 via its vWF-A domain (Çevik et al., 2012). Thus, it will be crucial to elucidate the functional relevance of these two subunits in regulating JA signaling or other signal transduction pathways.
Arabidopsis MED18 is another subunit that contributes to plant immunity against necrotrophic pathogens (Lai et al., 2014). Mutants of MED18 exhibit enhanced susceptibility to B. cinerea, suggesting that MED18 positively regulates B. cinerea-induced plant immunity. However, the expression of PDF1.2 induced by B. cinerea or MeJA was increased rather than decreased in the med18 mutant, indicating that MED18 might function in a JA-independent manner (Lai et al., 2014). MED18 interacts with the YIN YANG1 (YY1) TF, which acts as a transcriptional repressor. MED18 positively regulates the function of YY1 to suppress the transcription of thioredoxin (TRX-h5, LIV1) and glutaredoxin (GRXS13, GRX480). Genetic data indicate that elevated expression of GRX and TRX contributes to the enhanced susceptibility of med18 and yy1 mutants to fungal infection (Lai et al., 2014). Another study reported that MED18 and MED20 form a subdomain within Mediator that controls the balance of JA and SA signaling pathways (Fallath et al., 2017). In that study, MED18 and MED20 conferred susceptibility to F. oxysporum. In F. oxysporum-infected med18 and med20, the expression of JA-responsive genes was significantly reduced, whereas the expression of SA-associated genes and several genes associated with reactive oxygen species (ROS) production was upregulated. It is possible that both JA and SA signaling is channeled through the Mediator complex via MED18 and MED20; alternatively, misregulation of the JA pathway in med18 and med20 mutants may lead to defects in ROS production and tolerance (Fallath et al., 2017). Consistent with the role of MED18 in regulating the balance of JA and SA signaling pathways, MED18 is recruited by the histone acetyltransferase HOOKLESS1 to the WRKY33 promoter, thereby increasing WRKY33 expression (Liao et al., 2016). It is believed that WRKY33 has an important role in regulating JA–SA crosstalk and redox homeostasis (Birkenbihl et al., 2012). These studies indicate that MED18 regulates plant immunity in both JA-independent and JA-dependent manners.
The CDK8 module is involved in JA-mediated plant immunity (Zhu et al., 2014). The cdk8 mutant exhibits enhanced resistance to B. cinerea infection but susceptibility to A. brassicicola infection. However, both B. cinerea- and A. brassicicola-induced PDF1.2 expression is reduced in the cdk8 mutant, which is similar to the MeJA-induced expression levels of PDF1.2 in the cdk8 mutant. CDK8 interacts with MED25 and is required for ERF1- and ORA59-dependent activation of PDF1.2 expression, indicating that CDK8 regulates plant immunity through a JA-dependent pathway (Zhu et al., 2014). CDK8 regulates resistance to A. brassicicola through direct transcriptional regulation of AGMATINE COUMAROYLTRANSFERASE (AACT1), which is critical for the biosynthesis of hydroxycinnamic acid amides, secondary metabolites that function in fungal resistance (Zhu et al., 2014). CDK8 negatively regulates B. cinerea-induced plant immunity by interacting with WAX INDUCER1 (WIN1), an ERF family protein that regulates cuticular wax biosynthesis. Genetic evidence supports the notion that the resistance to B. cinerea in the cdk8 mutant is due to the altered cuticle profile linked to the loss of CDK8 functions (Zhu et al., 2014). The kinase activity is necessary for CDK8 regulation of A. brassicicola-induced plant immunity, but not for regulation of B. cinerea-induced plant immunity (Zhu et al., 2014). Two other kinase module mutants, med12 and med13, exhibit disease responses and increased cuticle permeability similar to that of the cdk8 mutant, revealing the common functions and structural conservation of the kinase module (Zhu et al., 2014).
Conclusions and perspectives
Multiple Mediator subunits are important for JA-dependent plant immunity. MED25 is the most well-studied Mediator subunit; it has critical roles in different stages of JA signaling. MED25 acts as a master coordinator to integrate the actions of the hormone receptor COI1, the master TF, MYC2, and its coactivator, HAC1, into a concerted transcriptional program. Future structural studies will help to fully identify the mechanism by which MED25 dynamically changes its interaction with different regulators to spatiotemporally regulate the transcriptional output. In addition to JA, the main receptors for several other plant hormones are localized in the nucleus and directly linked to hormone-regulated gene transcription. An important future direction will be to determine whether the action mode of MED25 in regulating JA signaling can be extended to other hormones whose receptors are localized in the nucleus. If so, it will be crucial to identify the mechanisms that regulate the general and context-specific functions of Mediator, and especially to determine how a single multiprotein complex can perform so many diverse tasks.
Several other Mediator subunits, including MED8, MED16, MED18, and CDK8, have been identified as important players in JA-dependent plant immunity. However, compared with MED25, the mechanisms of these subunits in regulating JA signaling remain elusive. Considering that MED8, MED16, and CDK8 physically interact with MED25, it is possible that these Mediator subunits execute their functions in JA signaling by cooperating with MED25. Alternatively, these Mediator subunits could regulate JA-mediated plant immunity by cooperating with new players in JA signaling.
Plant Mediator is a large complex that currently comprises more than 33 subunits. A more comprehensive screen could identify additional Mediator subunits that function in the JA signaling pathway. Future studies of the intact plant Mediator complex will shed new light on our understanding of plant Mediator and its role in regulating the JA signaling pathway.
Acknowledgements
We acknowledge the support of Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB11030200; National Natural Science Foundation of China Grant 31730010; the Chinese Academy of Sciences Youth Innovation Promotion Association Grant 2014082.
References
- Allen BL, Taatjes DJ. 2015. The Mediator complex: a central integrator of transcription. Nature Reviews. Molecular Cell Biology 16, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Li L, Zhai Q, You Y, Deng L, Wu F, Chen R, Jiang H, Wang H, Chen Q, Li C. 2017. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proceedings of the National Academy of Sciences, USA 114, E8930–E8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Mou Z. 2013. The function of the Mediator complex in plant immunity. Plant Signaling & Behavior 8, e23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. 1999. Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985–987. [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. 2002. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. The EMBO Journal 21, 6036–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. 2007. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Molecular Cell 26, 717–729. [DOI] [PubMed] [Google Scholar]
- Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. 2011. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biology 9, e1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE. 2012. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiology 159, 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek E, Mittler G, Meisterernst M. 2005. The mediator of RNA polymerase II. Chromosoma 113, 399–408. [DOI] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M. 2010. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. The Journal of Biological Chemistry 285, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S. 2004. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes & Development 18, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM. 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Research 36, 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. 2009. Jasmonate passes muster: a receptor and targets for the defense hormone. Annual Review of Plant Biology 60, 183–205. [DOI] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. 2008. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research 18, 756–767. [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. 2003. Regulation of flowering time by light quality. Nature 423, 881–885. [DOI] [PubMed] [Google Scholar]
- Cevher MA, Shi Y, Li D, Chait BT, Malik S, Roeder RG. 2014. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nature Structural & Molecular Biology 21, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, et al. 2012. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiology 160, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, et al. 2012. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. The Plant Cell 24, 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Li T, Rahl PB, et al. 2012. Functional association of Gdown1 with RNA polymerase II poised on human genes. Molecular Cell 45, 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. 2011. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Molecular Plant 4, 279–288. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends in Biochemical Sciences 30, 250–255. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. 1997. Biosynthesis and action of jasmonates in plants. Annual Review of Plant Physiology and Plant Molecular Biology 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Davis JA, Takagi Y, Kornberg RD, Asturias FA. 2002. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Molecular Cell 10, 409–415. [DOI] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. 2002. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. The Plant Journal 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Dolan WL, Chapple C. 2017. Conservation and divergence of mediator structure and function: insights from plants. Plant & Cell Physiology 58, 4–21. [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature Structural & Molecular Biology 17, 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. 2000. Structural organization of yeast and mammalian mediator complexes. Proceedings of the National Academy of Sciences, USA 97, 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Zhao J, Tzeng DTW, et al. 2017. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. The Plant Cell 29, 1883–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N, Davoine C, Benlloch R, et al. 2011. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proceedings of the National Academy of Sciences, USA 108, 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallath T, Kidd BN, Stiller J, Davoine C, Björklund S, Manners JM, Kazan K, Schenk PM. 2017. MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana. PLoS One 12, e0176022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Browse J. 2015. Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. The Plant Journal 81, 849–860. [DOI] [PubMed] [Google Scholar]
- Fondell JD. 2013. The Mediator complex in thyroid hormone receptor action. Biochimica et Biophysica Acta 1830, 3867–3875. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proceedings of the National Academy of Sciences, USA 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nature Chemical Biology 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Fonseca S, Fernández-Calvo P, Fernández GM, et al. 2014. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PloS One 9, e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, et al. 2013. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Solano R. 2015. Novel players fine-tune plant trade-offs. Essays in Biochemistry 58, 83–100. [DOI] [PubMed] [Google Scholar]
- Goossens J, Mertens J, Goossens A. 2017. Role and functioning of bHLH transcription factors in jasmonate signalling. Journal of Experimental Botany 68, 1333–1347. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Young RA. 2017. New insights into genome structure: genes of a feather stick together. Molecular Cell 67, 730–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annual Review of Plant Biology 59, 41–66. [DOI] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. 2016. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proceedings of the National Academy of Sciences, USA 113, 6562–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. 2017. The Mediator complex: At the nexus of RNA polymerase II transcription. Trends in Cell Biology 27, 765–783. [DOI] [PubMed] [Google Scholar]
- Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG. 2012. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Molecular Cell 45, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2013. MYC2: the master in action. Molecular Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Keaveney M, Berkenstam A, Feigenbutz M, Vriend G, Stunnenberg HG. 1993. Residues in the TATA-binding protein required to mediate a transcriptional response to retinoic acid in EC cells. Nature 365, 562–566. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ 3rd, Flanagan PM, Kornberg RD. 1990. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61, 1209–1215. [DOI] [PubMed] [Google Scholar]
- Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K. 2011. Diverse roles of the Mediator complex in plants. Seminars in Cell & Developmental Biology 22, 741–748. [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. 2009. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. The Plant Cell 21, 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Chen X. 2011. The plant mediator and its role in noncoding RNA production. Frontiers in Biology 6, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. 2009. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes & Development 23, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Meisterernst M, Roeder RG. 1993. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proceedings of the National Academy of Sciences, USA 90, 11508–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. 2002. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun DJ, Mengiste T. 2014. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nature Communications 5, 3064. [DOI] [PubMed] [Google Scholar]
- Lee HK, Park UH, Kim EJ, Um SJ. 2007. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. The EMBO Journal 26, 3545–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. 2004. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. The Plant Cell 16, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang R, Chen H. 2018. The Arabidopsis thaliana Mediator subunit MED8 regulates plant immunity to Botrytis cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS One 13, e0193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CJ, Lai Z, Lee S, Yun DJ, Mengiste T. 2016. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. The Plant Cell 28, 1662–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q. 2019. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. The Plant Cell 31, 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature Reviews. Genetics 11, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK. 2011. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiology 157, 1609–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. 1993. DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365, 227–232. [DOI] [PubMed] [Google Scholar]
- Mittler G, Stühler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, Berti L, Meisterernst M. 2003. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. The EMBO Journal 22, 6494–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. 1999. Mediator protein mutations that selectively abolish activated transcription. Proceedings of the National Academy of Sciences, USA 96, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, Kim Y, Myers LC. 2005. Mediator and TFIIH govern carboxyl-terminal domain-dependent transcription in yeast extracts. The Journal of Biological Chemistry 280, 33739–33748. [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. 2013. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. The Plant Cell 25, 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. 2011. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. Journal of Experimental Botany 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B, Yin KQ, Liu SN, et al. 2011. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Molecular Plant 4, 546–555. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Larivière L, Wenzeck L, et al. 2015. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380. [DOI] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ. 2013. The Mediator complex and transcription regulation. Critical Reviews in Biochemistry and Molecular Biology 48, 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D. 2015. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. The Plant Cell 27, 1620–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli HG, Martin GB. 2015. Functional genomics of tomato for the study of plant immunity. Briefings in Functional Genomics 14, 291–301. [DOI] [PubMed] [Google Scholar]
- Ryan CA. 2000. The systemin signaling pathway: differential activation of plant defensive genes. Biochimica et Biophysica Acta 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Samanta S, Thakur JK. 2015. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Frontiers in Plant Science 6, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. 2013. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiology 163, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. 2005. Systemic signaling in the wound response. Current Opinion in Plant Biology 8, 369–377. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. 2010. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. 2013. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genetics 9, e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J. 2018. Transcription regulation by the Mediator complex. Nature Reviews. Molecular Cell Biology 19, 262–274. [DOI] [PubMed] [Google Scholar]
- Sun JQ, Jiang HL, Li CY. 2011. Systemin/jasmonate-mediated systemic defense signaling in tomato. Molecular Plant 4, 607–615. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. 2013. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nature Structural & Molecular Biology 20, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Yu X, Gopalan S, et al. 2017. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. 2002. The jasmonate signal pathway. The Plant Cell 14(Suppl), S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Du X, Mou Z. 2016. The mediator complex subunits MED14, MED15, and MED16 are involved in defense signaling crosstalk in Arabidopsis. Frontiers in Plant Science 7, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z. 2015. The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiology 169, 856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Q, Ding Z, Ji J, Wang J, Kong X, Yang J, Cai G. 2014. Redefining the modular organization of the core Mediator complex. Cell Research 24, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala DL, Hemsley PA, Moffat CS, Cremelie P, Knight MR, Knight H. 2012. The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytologist 195, 217–230. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. 2014. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Molecular Cell 54, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YM, Chang JW, Wang CH, Lin YC, Wu PL, Huang SH, Chang CC, Hu X, Gnatt A, Chang WH. 2012. Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-EM. The EMBO Journal 31, 3575–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. 2002. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. The Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, et al. 2009. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. 2007. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li L, Qu LJ. 2016. Plant Mediator complex and its critical functions in transcription regulation. Journal of Integrative Plant Biology 58, 106–118. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ. 2014. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. The Plant Journal 77, 838–851. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Li L, An C, Li C. 2017a Conserved function of mediator in regulating nuclear hormone receptor activation between plants and animals. Plant Signaling & Behavior 13, e1403709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Yan C, Li L, Xie D, Li C. 2017b Jasmonates . In: Smith MS, Li C, Li J, eds. Hormone metabolism and signaling in plants. London: Elsevier/Academic Press, 243–272. [Google Scholar]
- Zhai Q, Yan L, Tan D, Chen R, Sun J, Gao L, Dong MQ, Wang Y, Li C. 2013. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genetics 9, e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Yao J, Ke J, et al. 2015. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z. 2012. The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. The Plant Cell 24, 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhai Q, Sun J, et al. 2006. Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiology 141, 1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. 2012. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host & Microbe 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Schluttenhoffer CM, Wang P, Fu F, Thimmapuram J, Zhu JK, Lee SY, Yun DJ, Mengiste T. 2014. CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. The Plant Cell 26, 4149–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]