Abstract
OBJECTIVE
Observational studies show that higher hemoglobin A1c (A1C) predicts coronary artery disease (CAD). It remains unclear whether this association is driven entirely by glycemia. We used Mendelian randomization (MR) to test whether A1C is causally associated with CAD through glycemic and/or nonglycemic factors.
RESEARCH DESIGN AND METHODS
To examine the association of A1C with CAD, we selected 50 A1C-associated variants (log10 Bayes factor ≥6) from an A1C genome-wide association study (GWAS; n = 159,940) and performed an inverse-variance weighted average of variant-specific causal estimates from CAD GWAS data (CARDIoGRAMplusC4D; 60,801 CAD case subjects/123,504 control subjects). We then replicated results in UK Biobank (18,915 CAD case subjects/455,971 control subjects) and meta-analyzed all results. Next, we conducted analyses using two subsets of variants, 16 variants associated with glycemic measures (fasting or 2-h glucose) and 20 variants associated with erythrocyte indices (e.g., hemoglobin [Hb]) but not glycemic measures. In additional MR analyses, we tested the association of Hb with A1C and CAD.
RESULTS
Genetically increased A1C was associated with higher CAD risk (odds ratio [OR] 1.61 [95% CI 1.40, 1.84] per %-unit, P = 6.9 × 10−12). Higher A1C was associated with increased CAD risk when using only glycemic variants (OR 2.23 [1.73, 2.89], P = 1.0 × 10−9) and when using only erythrocytic variants (OR 1.30 [1.08, 1.57], P = 0.006). Genetically decreased Hb, with concomitantly decreased mean corpuscular volume, was associated with higher A1C (0.30 [0.27, 0.33] %-unit, P = 2.9 × 10−6) per g/dL and higher CAD risk (OR 1.19 [1.04, 1.37], P = 0.02).
CONCLUSIONS
Genetic evidence supports a causal link between higher A1C and higher CAD risk. This relationship is driven not only by glycemic but also by erythrocytic, glycemia-independent factors.
Introduction
Hemoglobin A1c (A1C) is a convenient test used to diagnose diabetes, monitor glycemic control, and assess risk of diabetes-related complications, including coronary artery disease (CAD). Epidemiologic studies have shown in people without diabetes that A1C is strongly associated with CAD risk, even after accounting for fasting glucose and other clinical risk factors (1,2). Mendelian randomization (MR) can be used to assess the causal association between A1C and CAD risk through the application of an instrumental variable analysis with A1C-associated genetic variants. As genetic variants are randomized at birth and have effects that are potentially lifelong, MR studies are less vulnerable to unmeasured and residual confounding than traditional epidemiologic studies where biomarker measures may not be reliably measured or only reflect a brief snapshot in time (3).
Previous MR studies have shown that genetically increased A1C is associated with higher CAD risk (4–6). However, a major challenge has been the examination of the underlying mechanisms linking A1C and CAD risk. As A1C is not a direct measurement of glycemia but rather a measure of the proportion of glycated hemoglobin in the blood, nonglycemic determinants of A1C that are intrinsic to the erythrocyte (7–10) may also influence CAD risk independently of glycemia.
Here, we undertook a series of MR analyses to determine whether the association between A1C and CAD risk was driven by glycemic or erythrocytic factors, or both, through subsets of genetic variants previously classified by their probable biological categories (glycemic or erythrocytic). As some genetic determinants of A1C may act through changes in hemoglobin (Hb) levels, we performed additional MR analyses to test whether Hb and other erythrocytic traits were also associated with A1C and CAD risk.
Research Design and Methods
A1C Candidate Instrument Selection and Classification as Either Glycemic or Erythrocytic
Genome-wide association studies (GWAS) data sets used in the MR analyses are summarized in Table 1. We extracted association summary statistics from a large-scale transethnic meta-analysis GWAS on A1C in individuals without diabetes (the Meta-Analyses of Glucose and Insulin-Related Traits Consortium [MAGIC]) (11). As GWAS data sets were overwhelmingly European, we drew a set of distinct variants reaching genome-wide (GW) significance in the transethnic analysis (log10 Bayes factor ≥6; n = 159,940, European-ancestry linkage disequilibrium r2 < 0.05) and excluded those with P > 5 × 10−5 in European samples (n = 123,665) (Supplementary Fig. 1). Previous work has classified these variants as “glycemic” or “erythrocytic” (Supplementary Table 1) (11). We performed separate MR analyses restricting instruments to A1C variants classified as “glycemic” or “erythrocytic” to test the hypothesis that variation in A1C altered CAD risk through glycemic or erythrocytic factors, respectively. A1C variants that were not associated with glycemic or erythrocytic traits were excluded from these subanalyses.
Table 1.
Data sets of GWAS used in the MR analysis to estimate the causal effect of A1C on CAD risk
| MAGIC | CARDIoGRAMplusC4D | UKBB | |
|---|---|---|---|
| Population | Multinational | Multinational | U.K. |
| Number of cohorts/studies | 82 | 48 | 1 |
| Sample size, n | 159,940 participants without diabetes* | 60,801 CAD case subjects and 123,504 control subjects | 18,915 CAD case subjects and 455,971 control subjects |
| Ethnicities | European 77% | European 77% | European 84% |
| East Asian 13% | South Asian 13% | Non-European 16% | |
| African American 5% | East Asian 6% | ||
| South Asian 5% | Hispanic or African American 4% | ||
| Imputation reference panel | Phase 2 of the International HapMap Project | Phase 1 version 3 of 1000 Genomes | Haplotype Reference Consortium |
| Phenotype | Measured A1C by NGSP percent | CAD: myocardial infarction, acute coronary syndrome, chronic stable angina, or coronary stenosis >50% | CAD: myocardial infarction, percutaneous coronary angioplasty, coronary artery bypass graft, or triple heart bypass |
| Model | Transethnic meta-analysis; linear regression, additive model adjusted for study-specific covariates, age, sex, and genomic control | Transethnic meta-analysis; logistic regression, additive model, adjusted for study-specific covariates, age, sex, and genomic control | Linear mixed, additive model adjusted for the first five principal components |
| Publication | Wheeler et al. (11) | Nikpay et al. (12) | Methods adapted from Nelson et al. (15) |
*European-only effect estimates for A1C were used for this MR analysis (n = 123,665).
MR Analysis Using CAD GWAS Summary Data
To estimate the causal association of A1C on CAD, we performed a two-sample MR using summary data from the Coronary Artery Disease Genome-Wide Replication and Meta-analysis Plus Coronary Artery Disease Genetics (CARDIoGRAMplusC4D; n = 60,801 case subjects/123,504 control subjects) GWAS (12). The causal effect was evaluated by the inverse-variance weighted (IVW) method, whereby genetic variant outcome coefficients were modeled as a function of genetic variant exposure coefficients weighted by the inverse of the squared genetic variant outcome SE (13).
Replication of A1C-CAD MR Analysis in UK Biobank
UK Biobank (UKBB) recruited half a million participants and followed their health through questionnaires, National Health Service records, and national death and hospital registries. Of the 487,409 participants in the full release of imputed genotypes to the European Haplotype Reference Consortium panel (14), we excluded 373 with mismatched sex and 9 without kinship calculation. We defined “hard” CAD case subjects as having a history of myocardial infarction, percutaneous coronary angioplasty, coronary artery bypass graft, or triple heart bypass (n = 18,915) and excluded “soft” CAD case subjects with a history of angina or chronic ischemic heart disease but none of the above-mentioned cardiac conditions (n = 12,141) (15). The rest of the participants were considered control subjects (n = 455,971). We conducted a “hard” CAD GWAS on ∼20 million variants (imputation INFO >0.4; minor allele frequency >0.001) using the first five principal components using BOLT-LMM version 2.3 (16). After correcting for genomic inflation, effect estimates were transformed from the linear to log-odds scale (17). Next, we performed two-sample MR analysis to estimate the causal effect of A1C on CAD risk and combined MR estimates from UKBB and CARDIoGRAMplusC4D by an inverse-variance weighted fixed-effects meta-analysis. We used P < 0.05 to declare statistical significance.
Accounting for Pleiotropy in the A1C-CAD Association
An assumption of MR is that instruments do not influence the outcome independently of the risk factor of interest (i.e., nonmediated pleiotropy). We tested this assumption in two sensitivity analyses. First, we used the weighted median estimator (WME) (18), which requires ≥50% of the contribution to the causal estimate to be from valid instruments; if so, its causal estimate is stable. Second, we used MR-Egger regression (19) whereby a linear regression of variant outcome on variant exposure coefficients was performed without constraining the intercept to the origin. The slope of the regression line provides the corrected causal estimates even when none of the instruments are valid (19). Causal estimates were calculated using the R package “MendelianRandomization” version 0.2.2 (20).
Causal Effects of Hb and Mean Corpuscular Volume on A1C, LDL, and CAD Risk
We posited that Hb may partly explain the genetic relationship between A1C and CAD risk. To identify genetic instruments for Hb, we selected genetic variants that reached GW significance (P < 1 × 10−8) with one or more erythrocytic traits (e.g., Hb, mean corpuscular volume [MCV], mean corpuscular Hb, and mean corpuscular Hb concentration) that were also associated with Hb at P < 5 × 10−5 in a large meta-analysis GWAS for erythrocytic traits by the HaemGen consortium (21). As Hb-associated genetic variants have been shown to be associated with lipid levels (22,23), a known causal risk factor for CAD (24), we tested for causal associations between decreased Hb and A1C, LDL, and CAD risk using GWAS summary data from MAGIC (11), Global Lipids Genetics Consortium (GLGC) (25), and CARIoGRAMplusC4D (12) and conducted bidirectional MR analyses for Hb, A1C, and LDL to clarify the directionality of associations (3). To identify genetic instruments for LDL, we selected genetic variants that reached GW significance (P < 5 × 10−8) in GLGC (25) (Supplementary Fig. 1).
MCV is measured in clinical practice to assist in the morphological classification of anemia (low Hb), specifically microcytic (low MCV), macrocytic (high MCV), and normocytic (normal MCV) anemia. To identify which forms of anemia underlie the genetic relationship between A1C and CAD risk, we repeated the analysis using three subsets of Hb-lowering genetic variants: those associated with MCV in the same direction of effect as Hb (P < 0.05), in the opposite direction of effect to Hb (P < 0.05), or not associated with MCV (P ≥ 0.05) in the GWAS of erythrocytic traits (21).
Results
Selection of A1C Genetic Instruments
Of the 60 genome-wide significant variants from the transethnic A1C GWAS, 57 were polymorphic in European ancestry (11). We excluded four because they were not associated with A1C in Europeans (rs2073285, rs12132919, rs2237896, and rs17256082) and three that were in linkage disequilibrium (r2 > 0.05) with other candidate variants with stronger P values (rs3824065, rs13387347, and rs10823343). The remaining 50 variants served as instruments in the MR analysis (Supplementary Table 1).
Causal Effect of Increased A1C on CAD Risk
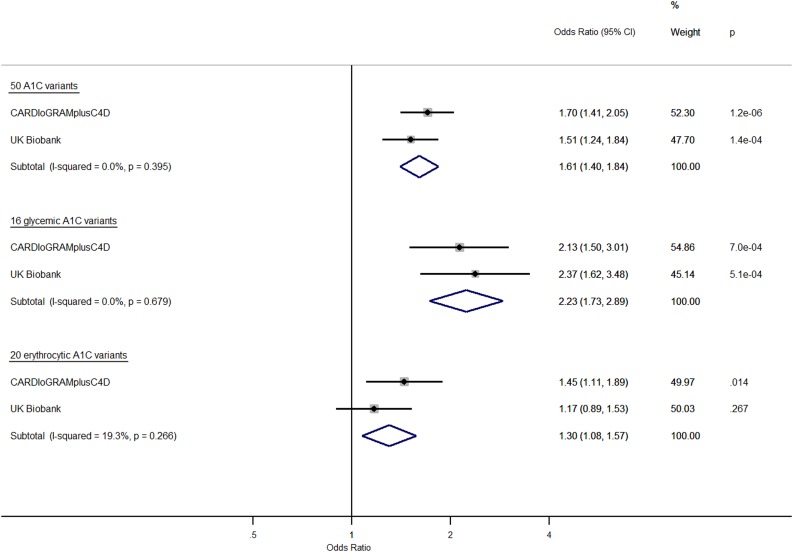
Genetically increased A1C was associated with higher CAD risk in CARDIoGRAMplusC4D (odds ratio [OR] 1.70 [95% CI 1.41, 2.05]) per %-unit in A1C, in UKBB (OR 1.51 [1.24, 1.85]) per %-unit, and in a meta-analysis of both data sets (OR 1.61 [1.40, 1.84] per %-unit, P = 6.9 × 10−12) (Fig. 1). WME results (meta-analysis: OR 1.56 [1.24, 1.95] per %-unit, P = 0.0001) (Supplementary Fig. 2A) were consistent with IVW. MR estimates from MR-Egger were consistent with those from IVW and WME in CARDIoGRAMplusC4D but not in UKBB, where the association was null (Supplementary Fig. 2B).
Figure 1.
Causal effect on CAD risk in CARDIoGRAMplusC4D and UKBB of increased A1C instrumented by all A1C-associated genetic variants, glycemic-only A1C variants, and erythrocytic-only A1C variants. MR analyses were performed by the IVW method. Effect estimates are OR of CAD per %-unit increase in A1C.
Causal Effect of A1C on CAD Risk by Glycemic Versus Erythrocytic Factors
Genetically higher A1C was associated with higher CAD risk when restricting instruments to glycemic A1C variants in CARDIoGRAMplusC4D and UKBB (meta-analysis: OR 2.23 [1.73, 2.89] per %-unit, P = 1.0 × 10−9) and when restricting instruments to erythrocytic A1C variants (meta-analysis: OR 1.30 [1.08, 1.57] per %-unit, P = 0.006) (Fig. 1). Causal estimates instrumented by glycemic A1C variants were attenuated using WME compared with IVW, and null with wide CI using MR-Egger (Supplementary Fig. 2A and B). Conversely, causal estimates instrumented by erythrocytic A1C variants were quite consistent using IVW, WME (meta-analysis: OR 1.36 [1.02, 1.81], P = 0.04), and MR-Egger (meta-analysis: OR 1.56 [1.13, 2.16], P = 0.006), even when excluding variants associated with BMI, LDL, triglycerides, and systolic blood pressure (25–28) in CARDIoGRAMplusC4D (Supplementary Fig. 3).
Causal Effect of Hb and MCV on A1C, LDL, and CAD Risk
Genetically decreased Hb was associated with higher A1C, higher LDL, and higher CAD risk when restricting instruments to Hb-lowering genetic variants that also decreased MCV, but not when restricting to those that increased MCV or were not associated with MCV (Table 2). In reverse MR analyses, increased LDL, instrumented by 29 LDL-associated genetic variants, was associated with lower Hb (P = 0.002), suggesting that the genetic relationship between Hb and LDL was bidirectional. Increased A1C, instrumented by erythrocytic variants, was associated with higher LDL (P = 2.3 × 10−24) but not in the reverse MR analysis (P = 0.40), indicating that the A1C-LDL relationship was unidirectional. Figure 2 summarizes the interrelationship of these causal associations.
Table 2.
Causal effect of decreased Hb on LDL, A1C, and CAD risk instrumented by all Hb-associated genetic variants and subsets of Hb-associated genetic variants based on their association with MCV
| No. of variants | A1C (%-unit) change per 1 g/dL decrease in Hb |
LDL (SD) change per 1 g/dL decrease in Hb |
CAD odds per 1 g/dL decrease in Hb |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | P | β | 95% CI | P | OR | 95% CI | P | ||
| All Hb genetic variants | 27 | 0.16 | 0.14, 0.18 | 1.7 × 10−16 | 0.21 | 0.17, 0.26 | 2.7 × 10−10 | 1.05 | 0.97, 1.13 | 0.20 |
| Effect on MCV | ||||||||||
| Lowering | 6 | 0.30 | 0.27, 0.33 | 2.9 × 10−6 | 0.21 | 0.13, 0.29 | 0.001 | 1.19 | 1.04. 1.37 | 0.02 |
| Raising | 8 | 0.07 | 0.02, 0.12 | 0.009 | 0.40 | 0.27, 0.52 | 1.3 × 10−4 | 1.12 | 0.93, 1.36 | 0.19 |
| No effect | 13 | 0.04 | 0.01, 0.07 | 0.018 | 0.12 | 0.04, 0.21 | 0.006 | 0.88 | 0.77, 0.99 | 0.04 |
Outcome GWAS data sets were obtained from MAGIC (A1C), GLGC (LDL), and CARDIoGRAMplusC4D and UKBB (CAD). Effect estimates for CAD were combined by a fixed-effects IVW meta-analysis. As the number of Hb variants used as instruments was especially small when using subsets based on their effect on MCV, the t distribution was used. Genetic variants used as instruments were aligned to the alleles associated with lower Hb relative to the alternate alleles. The signs of the MR estimates were flipped so that the interpretation of the causal estimate would be the change in outcome measure per 1 g/dL decrease in Hb. β, causal estimate.
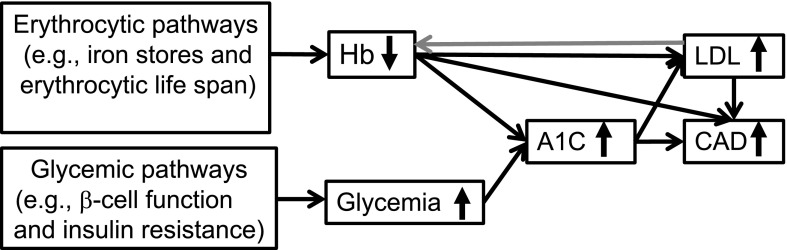
Figure 2.
MR diagram of glycemic and erythrocytic factors underlying the genetic relationship between A1C and CAD risk. Genetically decreased Hb with concomitantly decreased MCV was associated with higher A1C and higher odds of CAD (Table 2). Hb and LDL had bidirectional associations: increased LDL was associated with 0.06 g/dL decrease in Hb per SD change in LDL (P = 0.002), and decreased Hb was associated with 0.21 SD increase in LDL (P = 2.7 × 10−10). Generally, increased A1C when instrumented by all 50 A1C genetic variants was associated with higher LDL by 0.49 SD (P = 5.4 × 10−36) per 1%-unit in A1C and higher odds of CAD (OR 1.61, P = 6.9 × 10−12) per 1%-unit in A1C. Increased A1C when instrumented by 20 erythrocytic A1C variants was associated with higher LDL by 0.57 SD (P = 2.3 × 10−24) per 1%-unit in A1C and higher odds of CAD (OR 1.30, P = 0.004) per 1%-unit in A1C. The causal association of higher LDL with higher CAD risk has been shown in the literature (22,23) and so the MR analysis was not performed.
Conclusions
Using exposure and outcome variables from large samples of people of primarily European descent, we provided strong genetic evidence for A1C as a causal predictor of CAD risk, supporting other MR studies that tested this hypothesis (4–6). The results supported a causal interpretation of longitudinal epidemiologic studies showing that higher A1C was associated with higher CAD incidence (1,2). We additionally showed that higher A1C was associated with higher CAD risk even when restricting to genetic variants that were associated with erythrocytic traits, indicating that mediating or pleiotropic effects on erythrocytic traits may partly explain glycemia-independent associations of A1C with CAD risk and its ability to better predict CAD events compared with fasting glucose. Causal estimates were consistent even after excluding variants associated with BMI, LDL, triglycerides, and systolic blood pressure in CARDIoGRAMplusC4D, suggesting that this association with CAD attributed to erythrocytic factors may even be independent of this metabolic cluster of clinical risk factors.
As genetic determinants of A1C could exert their effects through Hb, we examined whether Hb partly explained the genetic relationship underlying A1C and CAD risk. We showed that the causal effect of Hb on A1C and CAD risk was strongest when using Hb-lowering genetic variants that concomitantly decreased MCV, indicating that a genetic predisposition to microcytic anemia may partly underlie this relationship. Epidemiologic studies have shown that iron deficiency, a common cause of microcytic anemia, is associated with artificially raised A1C, although underlying mechanisms remain poorly understood (8). Our genetic findings are consistent with a previous MR study (29) and a meta-analysis of prospective studies showing an association between higher iron status and lower CAD risk (30). Three of the five genetic variants used to instrument decreased Hb and MCV reside in or near genes implicated in iron metabolism, HFE-HIST1H2A (two variants) and TMPRSS6 (one variant). Conversely, hereditary hemochromatosis, a genetic condition characterized by iron overload, is associated with higher diabetes risk (31,32) and higher CAD risk (33), suggesting a detrimental effect, and not a protective effect, of excess iron stores.
Bidirectional MR analyses revealed that Hb and LDL had bidirectional relationships in keeping with results from a previous study that identified 10 genetic variants associated with both erythrocytic and lipid traits (24). As erythrocytic membranes have structural regions that are highly enriched with cholesterol (34), this genetic overlap may imply that genetic variation that influences erythrocytic volume or erythrocyte count also determines circulating lipid levels.
Hematologic conditions that reduce erythrocytic life span also lower A1C. A previous study showed that between-person differences in mean erythrocytic age explain nonglycemic variation in A1C (35). Common variants in genes implicated in hematologic diseases characterized by hemolytic anemia, e.g., G6PD (G6PD deficiency), HBB (sickle cell disease), SPTA1, ANK1 (spherocytosis/elliptocytosis), HK1 (hexokinase deficiency), and PEIZO1 (stomatocytosis), have been identified to be associated with A1C through GWAS (11). A previous MR study showed that higher reticulocyte count, a marker of shortened erythrocyte life span, was associated with higher CAD risk (36). Without direct measures of erythrocytic age, it remains unknown to what degree erythrocytic life span explains the glycemia-independent association between A1C and CAD risk.
Our study had limitations. There was partial overlap in the MAGIC GWAS and CARIoGRAMplusC4D GWAS samples, which could inflate MR estimates (37), but minimal overlap in MAGIC samples and UKBB. As effect estimates were consistent from both analyses, sample overlap likely did not create a spurious finding. The variance explained in A1C by genetic variants was modest but in a range typical for complex quantitative traits, especially when using subsets of them (4.7% by all variants, 2.0% by the glycemic A1C variants, and 1.6% by the erythrocytic A1C variants). Weak instruments tend to bias results toward the null, so our positive findings may underestimate true effect sizes (13). Whereas the causal association of A1C with CAD risk was consistent in IVW and WME, results from MR-Egger were less compelling, suggesting that IVW estimates may be biased by pleiotropy or other confounding factors. However, we note that MR-Egger is a less efficient estimator than WME and IVW (18) and is generally considered as only one of several sensitivity analyses used to assess the plausibility of MR findings. Our study subjects were without clinical diabetes. So, although our causal inferences apply to effects of A1C in the nondiabetic range, no conclusion can be made about genetic effects on A1C in people with diabetes, and we suggest caution extrapolating our findings to the extremes of the A1C distribution. Finally, well-powered studies in people of other ancestral origins are needed to determine whether the A1C-CAD relationship is similar to that in this European ancestry sample.
A1C is a valuable diabetes biomarker used worldwide. We found genetic evidence that A1C is a causal CAD risk factor representing two distinct etiologic pathways, glycemic and erythrocytic. The ability for A1C to predict CAD risk likely extends beyond its reflection of ambient glycemia and points to the importance of further research on the biology underlying the A1C-CAD relationship. Our study also suggests the importance of adequate iron stores and correcting anemia, which could lower CAD risk and improve the accuracy of A1C in estimating glycemia. In addition to suggesting new biology, these results illuminate the clinical utility of A1C as a biomarker in disease prediction and show how modern large-scale genotype-phenotype data can be used in MR frameworks to test biological hypotheses supporting epidemiologic associations with broad clinical implications.
Supplementary Material
Article Information
Acknowledgments. Part of this research has been conducted using the UKBB resource (application number 3913).
Funding. J.M. is supported by an individual fellowship funded by the Horizon 2020 Marie Skłodowska-Curie Actions of the European Commission (H2020-MSCA-IF-2015-703787). J.I.R. is supported in part by the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1-TR-001881, and the National Institute of Diabetes and Digestive and Kidney Diseases Diabetes Research Center grant DK-063491 to the Southern California Diabetes Research Center. J.C.F. is a Massachusetts General Hospital Research Scholar and is supported by K24-DK-110550. I.B. acknowledges funding from Wellcome (WT206194). J.B.M. is supported by R01-DK-078616, U01-DK-078616, K24-DK-080140, and the American Diabetes Association Mentored Scientist Career Development Award.
None of the funding sources were involved in the study design, data collection, data analysis, data interpretation, writing of the manuscript, or the decision to submit the manuscript for publication. None of the authors have been paid to write this article by a pharmaceutical company or other agency.
Duality of Interest. J.B.M. consults as an Academic Associate with Quest Diagnostics, Inc. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. A.L. performed the literature search, conceived the study, developed the study design, generated the figures and tables, contributed to data analysis and data interpretation, and wrote the initial version of the manuscript. J.C. contributed to the study design, data analysis, data interpretation, generation of figures and tables, and writing of the initial version of the manuscript. E.W., M.-F.H., C.-T.L., J.M., J.D., ES.T., J.I.R., J.C.F., I.B., and J.B.M. contributed to the study design and data interpretation and revised the entire manuscript critically. All authors approved the final version of the manuscript. A.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. An oral abstract for this study was presented at the 76th Scientific Sessions of the American Diabetes Association, New Orleans, LA, 10–14 June 2016.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc18-1712/-/DC1.
References
- 1.Di Angelantonio E, Gao P, Khan H, et al.; Emerging Risk Factors Collaboration . Glycated hemoglobin measurement and prediction of cardiovascular disease. JAMA 2014;311:1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selvin E, Steffes MW, Zhu H, et al. Glycated hemoglobin, diabetes, and cardiovascular risk in nondiabetic adults. N Engl J Med 2010;362:800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad OS, Morris JA, Mujammami M, et al. A Mendelian randomization study of the effect of type-2 diabetes on coronary heart disease. Nat Commun 2015;6:7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross S, Gerstein HC, Eikelboom J, Anand SS, Yusuf S, Paré G. Mendelian randomization analysis supports the causal role of dysglycaemia and diabetes in the risk of coronary artery disease. Eur Heart J 2015;36:1454–1462 [DOI] [PubMed] [Google Scholar]
- 6.Au Yeung SL, Luo S, Schooling CM. The impact of glycated hemoglobin (HbA1c) on cardiovascular disease risk: a Mendelian randomization study using UK Biobank. Diabetes Care 2018;41:1991–1997 [DOI] [PubMed] [Google Scholar]
- 7.Panzer S, Kronik G, Lechner K, Bettelheim P, Neumann E, Dudczak R. Glycosylated hemoglobins (GHb): an index of red cell survival. Blood 1982;59:1348–1350 [PubMed] [Google Scholar]
- 8.Coban E, Ozdogan M, Timuragaoglu A. Effect of iron deficiency anemia on the levels of hemoglobin A1c in nondiabetic patients. Acta Haematol 2004;112:126–128 [DOI] [PubMed] [Google Scholar]
- 9.Little RR, Roberts WL. A review of variant hemoglobins interfering with hemoglobin A1c measurement. J Diabetes Sci Technol 2009;3:446–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bry L, Chen PC, Sacks DB. Effects of hemoglobin variants and chemically modified derivatives on assays for glycohemoglobin. Clin Chem 2001;47:153–163 [PubMed] [Google Scholar]
- 11.Wheeler E, Leong A, Liu CT, et al.; EPIC-CVD Consortium; EPIC-InterAct Consortium; Lifelines Cohort Study . Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med 2017;14:e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikpay M, Goel A, Won HH, et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG; EPIC-InterAct Consortium . Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015;30:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton A, Brooks LD, Durbin RM, et al.; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature 2015;526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson CP, Goel A, Butterworth AS, et al.; EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD Working Group . Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–1391 [DOI] [PubMed] [Google Scholar]
- 16.Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for Biobank-scale datasets. Nat Genet 2018;50:906–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Jones LR, Robinson MR, Yang J, Visscher PM. Transformation of summary statistics from linear mixed model association on all-or-none traits to odds ratio. Genetics 2018;208:1397–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 2017;46:1734–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature 2012;492:369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do R, Willer CJ, Schmidt EM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet 2013;45:1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess S, Freitag DF, Khan H, Gorman DN, Thompson SG. Using multivariable Mendelian randomization to disentangle the causal effects of lipid fractions. PLoS One 2014;9:e108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chami N, Chen MH, Slater AJ, et al. Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am J Hum Genet 2016;99:8–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Schmidt EM, Sengupta S, et al.; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotsapas C, Voight BF, Rossin E, et al.; FOCiS Network of Consortia . Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 2011;7:e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehret GB, Munroe PB, Rice KM, et al.; International Consortium for Blood Pressure Genome-Wide Association Studies . Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011;478:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill D, Del Greco M F, Walker AP, Srai SKS, Laffan MA, Minelli C. The effect of iron status on risk of coronary artery disease: a Mendelian randomization study—brief report. Arterioscler Thromb Vasc Biol 2017;37:1788–1792 [DOI] [PubMed] [Google Scholar]
- 30.Das De S, Krishna S, Jethwa A. Iron status and its association with coronary heart disease: systematic review and meta-analysis of prospective studies. Atherosclerosis 2015;238:296–303 [DOI] [PubMed] [Google Scholar]
- 31.Qi L, Meigs J, Manson JE, et al. HFE genetic variability, body iron stores, and the risk of type 2 diabetes in U.S. women. Diabetes 2005;54:3567–3572 [DOI] [PubMed] [Google Scholar]
- 32.Raffield LM, Louie T, Sofer T, et al. Genome-wide association study of iron traits and relation to diabetes in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL): potential genomic intersection of iron and glucose regulation? Hum Mol Genet 2017;26:1966–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen ML, Folsom AR, Catellier DJ, Tsai MY, Garg U, Eckfeldt JH. A prospective study of coronary heart disease and the hemochromatosis gene (HFE) C282Y mutation: the Atherosclerosis Risk in Communities (ARIC) study. Atherosclerosis 2001;154:739–746 [DOI] [PubMed] [Google Scholar]
- 34.Himbert S, Alsop RJ, Rose M, et al. The molecular structure of human red blood cell membranes from highly oriented, solid supported multi-lamellar membranes. Sci Rep 2017;7:39661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malka R, Nathan DM, Higgins JM. Mechanistic modeling of hemoglobin glycation and red blood cell kinetics enables personalized diabetes monitoring. Sci Transl Med 2016;8:359ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Astle WJ, Elding H, Jiang T, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 2016;167:1415–1429.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.