Abstract
The composition of the gastrointestinal microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine, are elevated in high-fat diet–induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon-like peptide 1. Obese Fpr1 knockout mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. Overall, we describe a new mechanism by which the gut microbiota can modulate glucose metabolism, providing a potential approach for the treatment of metabolic disease.
Introduction
Chronic inflammation in obesity can induce insulin resistance leading to type 2 diabetes (T2D) (1). The commensal microbiota of the gastrointestinal (GI) tract serve as a key link between diet and metabolism, producing numerous metabolites that influence host physiology (2,3). Changes in gut microbiota, termed dysbiosis, are associated with obesity as well as other disease states. Interestingly, obesity-associated dysbiosis has been implicated in the development of insulin resistance and T2D (4). As of yet, most studies that have linked dysbiosis with obesity or T2D involve associations and correlations without detailed mechanisms. As rates of obesity and T2D continue to rise across the globe (5), the identification of the mechanisms by which gut microbiota interact with the host to modulate metabolism is necessary before therapeutic interventions can be developed.
The effects of the microbiome and microbiota-derived byproducts extend beyond the local intestinal epithelium or gut immune cells. In mice fed a high-fat diet (HFD), gut dysbiosis preceded elevations in circulating inflammatory cytokines, suggesting an early role of the microbiome in the induction of inflammation (6). As well, an “obese” microbiome signature confers an increased capacity to harvest energy from the diet, which could contribute to the obese state (7). Obesity is strongly associated with an increased microbiome content of the Firmicutes phylum and a decreased representation of the Bacteroidetes phylum (8). The importance of the microbiome was further highlighted in a study where germ-free (GF) mice that received microbiota transplants from an obese human twin gained 10% more fat mass compared with mice receiving a microbiota transplant from the lean twin (9). However, the mechanisms connecting microbiome changes and insulin resistance or obesity remain to be elucidated, and are likely to involve both immune cells and enteroendocrine cells (10).
Incretins are a class of metabolic hormones released from enteroendocrine cells in the gut (11). After eating, glucagon-like peptide 1 (GLP-1) is released from L cells, which act on β-cells to potentiate glucose-stimulated insulin secretion (11). Indeed, antagonism of the GLP-1 receptor (Glp1r) lowers insulin secretion and increases blood glucose concentrations (12). In T2D humans, postprandial GLP-1 secretion is generally impaired (13), although the mechanism has not been established (14). The microbiome, including metabolites and byproducts, may play a role in gut hormone secretion since enteroendocrine cells contain receptors to these products and can be influenced by them (10).
Metabolites and byproducts of the microbiome have been shown to orchestrate a variety of immune responses in the mammalian host. Among these different products, N-formyl peptides are derived from foreign bacteria or destroyed host cells (mitochondria) and activate G-protein–coupled N-formyl peptide receptors (FPRs) (15). N-formyl peptides traditionally define a large group of oligopeptides that contain a formyl-containing methionine (16). These motifs are part of a larger group known as pathogen- or danger-associated molecular patterns because they stimulate immune responses and inflammation (17). N-formyl peptides can stimulate chemotaxis of FPR-expressing immune cells (18), which leads to a signaling cascade that can cause phagocytosis (19) or apoptosis (20). Intestinal epithelial cells also express the Fpr1 receptor and, in response to an infection, can affect epithelial cell restitution (21) or migration (22). However, a role for Fpr1 in metabolism remains unexplored (23).
In the current study, we investigated whether formyl-methionyl-leucyl-phenylalanine (fMLF) levels are altered in conditions of obesity and the associated dysbiosis, and reveal a mechanism whereby this bacterial product influences glucose metabolism.
Research Design and Methods
Animal Care and Use
Frp1 knockout (Frp1-KO) and Glp1r-KO mice were generated as previously described (24,25). Only male mice were used in this study, fed ad libitum and maintained on a 12-h light/dark cycle. At 8 weeks of age, mice were placed on a 60% HFD (Research Diets) or normal chow diet (NCD) (13.5% fat; LabDiet) for 8–10 weeks before analyses. For broad-spectrum antibiotic treatment, mice were subject to daily oral gavage (OG) for 2 weeks with water alone or containing a mixture of antibiotics (Sigma-Aldrich): 0.5 mg/mL vancomycin HCl, 1 mg/mL ampicillin sodium salt, 1 mg/mL neomycin sulfate, 1 mg/mL metronidazole, and 1 mg/mL gentamycin sulfate. Mice were treated with the Glp1r antagonist exendin 9-39 (Sigma-Aldrich) or saline vehicle in a 0.2 mg/kg dose 20 min before metabolic studies. fMLF (Sigma-Aldrich) was administered at 1 mg/kg in saline via OG 1 h prior to analyses. Cinnamoyl-phenylalanyl-(D)leucine-phenylalanyl-(D)leucine-phenylalanine (cFLFLF) was provided daily at 0.5 mg/kg by OG for 2 weeks prior to analyses. For all drug treatments, body weight and food intake were monitored prior to and during the treatment period. GF C57BL/6 mice maintained in the UC San Diego Gnotobiotic Mouse Facility. GF mice were fed either an autoclaved chow diet (2019S Teklad Global 19% Protein Extruded Rodent Diet; Harlan Laboratories) or an irradiated sterilized 60% HFD (06414 Teklad), as previously described (26). For fecal transplantation, 8- to 10-week-old GF mice were gavaged with fecal suspensions from either WT HFD or Fpr1-KO HFD animals (50 mg/mL). Animal housing and procedures were performed according to UC San Diego and Institutional Animal Care and Use Committee–approved protocols, and conformed to the Guide for Care and Use of Laboratory Animals of the National Institutes of Health (NIH).
fMLF Detection and Analysis
Tissues, feces, and plasma were collected, homogenized and lyophilized, if necessary. Metabolites were extracted by adding PBS containing 4 nmol/L isotopically labeled fMLF (N-formyl-L-methionyl-L-leucyl [13C-6, 15N]-L-phenylalanine) as an internal standard (New England Peptide). The mixture was extracted using a Waters Oasis HLB SPE cartridge (30 mg, 1 mL) then eluted. The eluent was dried in vacuo using a Thermo Savant vacuum concentrator (Thermo Fisher Scientific) with the resulting dried pellet resuspended in MeOH:H2O (40:60, v/v) for liquid chromatography-mass spectrometry (MS) analysis.
Liquid chromatography-MS analysis was performed on Agilent 1290 Infinity II UPLC system (Agilent Technologies) coupled to an Agilent 6495 triple-quadrupole mass spectrometer equipped with an electrospray ionization source. The acquired data were analyzed by Agilent MassHunter workstation software (version B.07.01), and the concentration of fMLF was determined using calibration curves of isotopically labeled standard spiked into pooled plasma or stool at 1, 2.5, 5, 10, 25, and 50 nmol/L.
Bacterial DNA Measurement From Feces
The PowerViral DNA/RNA Isolation Kit (MoBio) was used to isolate DNA from feces, followed by quantitative PCR (qPCR) for the bacterial 16S rRNA gene V2 region. Bacterial DNA concentration was estimated using a standard curve of bacterial DNA.
Cell Culture and siRNA Transfection
Intraperitoneal macrophages were elicited from mice by injection of 3% thioglycollate and harvested in PBS after 3 days. mGLUTag cells were cultured as previously described (27). Forty-eight hours after plating cells, we transfected pooled siRNA libraries against Fpr1 or Fpr2, or a scrambled negative control (ON-TARGETplus SMARTpool; Dharmacon), using GenMute Transfection Reagent (SignaGen Laboratories).
In Vitro Chemotaxis Assay
After siRNA transfection, 1 × 105 cells were placed in the upper chamber of a Transwell plate (8 μm polycarbonate filter; Corning), whereas RPMI medium containing fMLF (250 nmol/L) (Sigma-Aldrich) or MCP-1 (100 ng/mL) (Sigma-Aldrich) was placed in the lower chamber, and incubated for 4 h. Cells that migrated to the other side of the Transwell filter were stained with DAPI, images captured on an Olympus MVX10 MacroView Microscope and cells counted using ImageJ software (NIH).
Gene Expression Analysis
RNA was isolated with the DirectZol RNA isolation kit (Zymo Research), and cDNA synthesized using the Applied Biosystems High Capacity Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed using iTaq SYBR Green Supermix (BIO-RAD) and the Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). Relative gene expression was calculated using the ΔΔCt method with GAPDH as an internal control. Inferred expression of intestinal epithelial cell Fpr1 was calculated by subtracting lamina propria gene expression from the total ileum gene expression. Primer sequences are listed in Supplementary Table 1.
Intracellular cAMP Analysis
Intracellular cAMP levels in intraperitoneal macrophages were measured using the Bridge-It cAMP Designer Fluorescence Assay Kit (Mediomics).
Metabolic Studies
Mice were fasted for 6 h prior to conducting insulin tolerance tests and glucose tolerance tests (GTTs), which have been described previously (28).
GLP-1 Detection
Blood serum was collected from mice fasted for 6 h, immediately before (basal) and 10–15 min after dextrose gavage, in the presence of DPP-4 (dipeptidyl peptidase 4) inhibitor (EMD Millipore). Postprandial blood collection was performed 10 min after gavage in NCD mice and 15 min after gavage in HFD mice. Analysis of GLP-1 was conducted by High Sensitivity GLP-1 Active ELISA Kit, Chemiluminescent (Millipore Sigma).
Glucose-Stimulated Insulin Secretion Assay
Primary murine islets were isolated as previously described (29). Secreted insulin was normalized to the corresponding intracellular insulin levels in the cell pellet of each sample.
Glucose-Stimulated GLP-1 Secretion Assay
mGLUTag cells were stimulated with 3 mmol/L glucose in the presence or absence of Fpr1 agonists (fMLF) for 4 h.
Plasma Protein Measurements
Fasting insulin, glucagon, total GLP-1, and active GLP-1 levels were measured by ELISA (Alpco, Mercodia, and EMD Millipore, respectively). Other fasting plasma proteins were measured by MILLIPLEX Multiplex Assay (EMD Millipore).
Histological Analyses
Tissue sections of pancreas, epididymal white adipose tissue (eWAT), small intestine, or colon were immunostained with appropriate antibodies, which are listed in Supplementary Table 1 and were analyzed as described previously (30).
RNA Fluorescence In Situ Hybridization
RNA fluorescence in situ hybridization was performed on frozen sections of the small intestine and colon using an RNAscope Fluorescent Multiplex Kit with RNAscope probes designed against mouse Fpr1 (catalog #319251) and mouse Proglucagon (catalog #400601), according to the manufacturer instructions (Advanced Cell Diagnostics, Inc.). Slides were counterstained with DAPI, and images were captured with an Olympus FV1000 Spectral Confocal Microscope. For quantification, 15–30 randomly selected Proglucagon+ L cells on each slide were observed for the Fpr1 probe signal, and images were analyzed for Fpr1 probe fluorescence intensity using ImageJ software.
Immunoblot Analysis
Proteins from tissue lysates loaded onto an SDS-PAGE gel then transferred to a polyvinylidene fluoride membrane. Membranes were blocked then incubated with antibodies, which are listed in Supplementary Table 2 and were analyzed as previously described (21).
Flow Cytometry Analysis
Lamina propria leukocytes were isolated from the small intestine and colon as previously described, followed by staining with fluorescence-tagged antibodies to detect cell lineages (see table in Goodyear et al. [31]) (Supplementary Table 2). Data were collected with a BD FACSCanto flow cytometer (BD Biosciences) and analyzed using FlowJo software.
Fecal Albumin Analysis
Fecal pellets were collected prior to and 8–10 weeks after placing animals on an HFD and were frozen at −80°C. Feces were resuspended at 10 mg/mL in PBS, and the albumin concentration determined by ELISA (Bethyl Laboratories, Inc).
In Vivo FITC-Dextran Assay
Analysis was performed as previously described (32). The concentration of FITC-dextran was determined by fluorescence spectroscopy relative to a linear standard curve of FITC-dextran prepared in plasma from untreated mice.
Bacterial 16S rRNA Gene Sample Processing and Sequencing
Bacterial DNA was isolated from feces using the PowerFecal DNA Isolation Kit (MoBio), according to the manufacturer guidelines. Pools of PCR amplification using bacterial 16S V4 rDNA fusion primers (515f and 806r) were loaded into an Illumina MiSeq and sequenced for 250 cycles for paired-end sequencing (33).
Sequencing Data Analyses
Sequenced paired-end reads were merged, quality filtered, and dereplicated with USEARCH (34). Representative sequences from the de novo operational taxonomic units (OTUs) were assigned taxonomic classification via mothur’s Bayesian classifier at 80% confidence; the classifier was trained against the GREENGENES reference database of 16S rRNA gene sequences clustered at 99% (35). The α- and β-diversity metrics were calculated as previously described (36,37).
Inference of Metagenomes
Metagenome prediction was conducted from 16S rRNA sequencing data using the Piphillin algorithm, using the reference genome databases KEGG 70.1 and BioCyc 18.0 (38).
Synthesis and Purification of cFLFLF
The synthetic peptide cFLFLF was synthesized using a standard solid-phase Wang resin supported Fmoc (fluorenylmethyloxycarbonyl) peptide synthesis protocol using an auto synthesizer (CSBio), similar to previous publications (39).
Significance Testing
Sample sizes necessary for animal studies were estimated using Russ Lenth’s power calculator (University of Iowa, Iowa City, IA [http://homepage.divms.uiowa.edu/∼rlenth/Power/]). Whole microbiome significance testing was performed by permutational ANOVA (PERMANOVA, version 3.2.2 [www.r-project.org]) (40). Taxon significance, or the univariate differential abundance of OTUs, was tested using the DESeq2 package, and q values were calculated with the Benjamini-Hochberg procedure to correct P values, controlling for false discovery rates (41,42). All other data were analyzed for significance testing using GraphPad Prism software.
Results
fMLF Is Increased in HFD/Obese Mice and Requires Fpr1 to Induce Chemotaxis
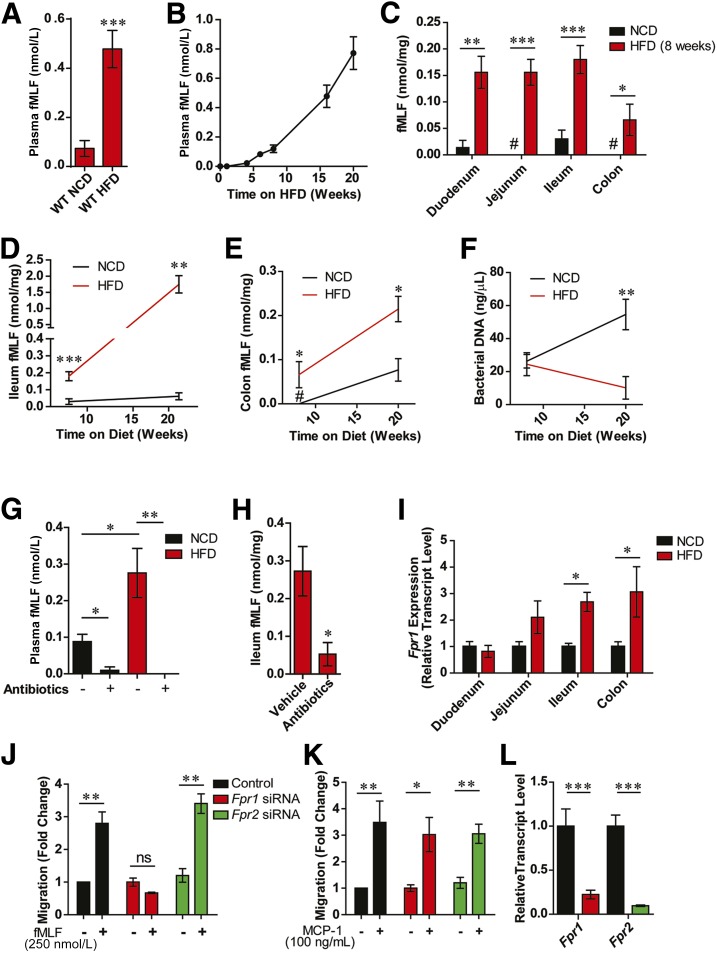
To determine whether levels of N-formyl peptides are altered by HFD-induced obesity, we conducted MS analysis of blood plasma and GI tissues from wild-type (WT) mice fed either a NCD or a 60% HFD. We found that blood levels of the N-formyl peptide fMLF are several fold higher in obese/HFD animals compared with lean/NCD animals, and increase over time with animals being fed an HFD (Fig. 1A and B). GI levels of fMLF across all gut regions are even more dramatically increased while being fed an HFD and are in a much higher range compared with those found in the circulation (Fig. 1C and E). These elevated fMLF levels are particularly dramatic considering the reduction in total bacterial load that occurs over time while on an HFD (Fig. 1F). Consistent with a microbiota-derived origin, treatment of NCD or HFD mice with broad-spectrum antibiotics for 2 weeks led to an ∼80–100% reduction in blood and intestinal fMLF levels (Fig. 1G and H). Additionally, GF animals also had barely detectable intestinal and blood levels of fMLF on NCD (Supplementary Fig. 1A and B). One of the receptors for fMLF is the N-formyl receptor, Fpr1, which we found expressed in all parts of the intestine and is increased in the ileum and colon under HFD feeding (Fig. 1I). Intestinal lamia propria showed the highest expression of Fpr1 (Supplementary Fig. 1C). However, Fpr1 expression was ∼50% reduced in the small intestinal lamina propria of HFD animals; therefore, the increased Fpr1 expression in the total intestinal tissue in these mice resides in the epithelial compartment (Supplementary Fig. 1D). Although mammals have several FPRs, in vitro macrophage chemotaxis experiments revealed that fMLF acts through the Gi-coupled Fpr1, rather than Fpr2, and also reduces forskolin-induced cAMP (Fig. 1J–L and Supplementary Fig. 1E and F) (43). In addition, qPCR expression analyses confirmed that Fpr1 is expressed throughout the intestine, where fMLF levels are highest, and is increased in HFD animals compared with NCD animals (Supplementary Fig. 1G and H). These findings, together with the marked differences between blood and intestinal fMLF levels, led us to suggest that Fpr1 is stimulated by fMLF present in the intestinal lumen, which is consistent with reports that Fpr1 is expressed on the apical surface of intestinal epithelial cells (44).
Figure 1.
fMLF is increased in HFD/obese mice and requires Fpr1 for chemotaxis. A: Blood plasma fMLF levels in lean/NCD vs. obese/HFD mice after 16 weeks on an HFD (n = 8–12 mice/group). B: Timecourse of fMLF levels after placement on an HFD (n = 6–8 mice per time point). C: fMLF levels in various intestinal tissues in WT NCD and HFD animals after 8 weeks on diet (n = 8 mice/group). Ileum (D) and colon (E) fMLF levels after 8 and 20 weeks of HFD or NCD (n = 6–8 mice/group). F: Bacterial DNA levels in colon fecal samples, determined by 16S rRNA gene expression and presented as a measure of bacterial load (n = 8 mice/group). G: Blood plasma fMLF levels in NCD and HFD mice (after 8 weeks on diet) treated with or without antibiotics (n = 6–8 mice/group). H: Ileum fMLF levels in HFD antibiotic-treated animals, after 8 weeks on diet (n = 4 mice/group). I: Relative Fpr1 transcript levels in various intestinal tissues of NCD and HFD WT animals after 8 weeks on diet (n = 8 mice/group). J: Chemotaxis assays of thioglycollate-elicited intraperitoneal macrophages (IP-Macs) in response to fMLF treatment (250 nmol/L), and after siRNA knockdown of Fpr1 or Fpr2 (n = 3). K: Chemotaxis assays in response to MCP-1 treatment (100 ng/mL) demonstrate that knockdown of Fpr1 does not inhibit macrophage activation by other chemokines. L: mRNA transcript levels of Fpr1 and Fpr2 in IP-Macs after siRNA knockdown. #Below detection limit. Data are mean ± SEM. ns, not significant. *P < 0.05, **P < 0.001, ***P < 0.0001 by one-way ANOVA with Bonferroni post-test (D–G) or two-tailed t test comparing the indicated groups. In D–F, statistical significance is indicated by comparing groups at each respective time point.
Loss of Fpr1 in HFD/Obese Mice Improves Glucose Tolerance and Is GLP-1 Dependent
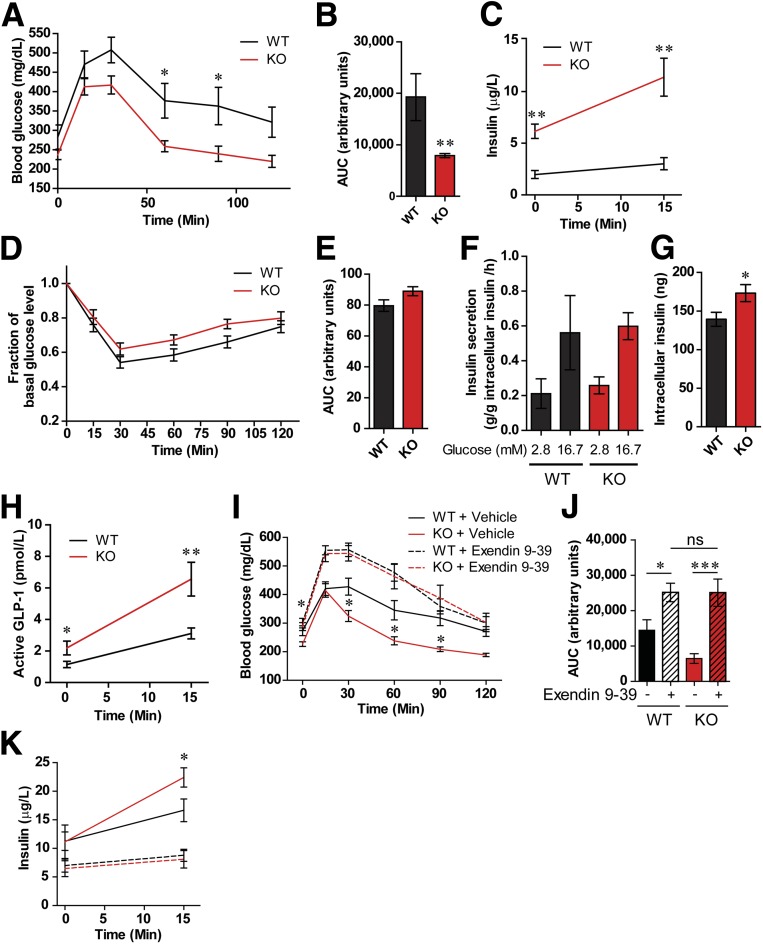
To assess the metabolic effects of Fpr1, we placed WT and Fpr1-KO littermates on an HFD for 10 weeks and measured glucose and insulin tolerance. OG-GTTs revealed a significant improvement in glucose tolerance in Fpr1-KO versus WT mice, with no change in body weight, organ weight, or overall food intake (Fig. 2A and B and Supplementary Fig. 2A–I). In addition, Fpr1-KO mice displayed increased plasma insulin levels in both the basal state and in response to glucose (Fig. 2C), while insulin tolerance tests revealed no improvement in insulin sensitivity (Fig. 2D and E). In contrast, the loss of Fpr1 did not lead to a significant improvement in glucose or insulin tolerance on NCD (Supplementary Fig. 3). Overall, this is consistent with the idea that elevated fMLF levels in the HFD state activates Fpr1 leading to glucose intolerance.
Figure 2.
Loss of Fpr1 in HFD/obese mice improves glucose tolerance and is GLP-1 dependent. A and B: OG-GTT after 10 weeks on an HFD, and area under the curve (AUC) (n = 7–10 mice/group). C: Plasma insulin levels during OG-GTT. D and E: Intraperitoneal insulin tolerance test after 12 weeks on an HFD and AUC (n = 7–10 mice/group). F and G: Glucose-stimulated insulin secretion and intracellular insulin levels in pancreatic islets from HFD WT and Fpr1-KO mice (n = 4 mice/per group, 20 islets/incubation). H: Active GLP-1 levels during the OG-GTT in A (n = 24–25 mice/group). I and J: OG-GTT and corresponding AUC of WT and Fpr1-KO HFD mice after 10 weeks on an HFD and treatment with or without the Glp1r antagonist exendin 9-39 (n = 10–11 mice/group). K: Plasma insulin levels during the OG-GTT in I (n = 10–11 mice/group). Error bars indicate the SEM. ns, not significant. *P < 0.05, **P < 0.001, ***P < 0.0001 by two-way ANOVA and Bonferroni post-test (A and I), one-way ANOVA and Bonferroni post-test (J) or two-tailed t test. Statistical significance is indicated comparing groups at each respective time point. In I and K, significance is indicated for WT vs. Fpr1-KO animals treated with vehicle.
To examine how loss of Fpr1 leads to increased insulin levels, we measured ex vivo glucose-stimulated insulin secretion using pancreatic islets isolated from HFD WT and Fpr1-KO mice. Surprisingly, we found no change in insulin secretion (Fig. 2F). In addition, islet size and total mass in Fpr1-KO mice was unchanged (Supplementary Fig. 2J and K), although intracellular levels of insulin were elevated (Fig. 2G). This suggests that Fpr1 deficiency increases insulin levels through an extrinsic, or islet-nonautonomous, manner. This is also consistent with findings that Fpr1 is expressed only at very low levels in WT pancreatic islets and is reduced further in HFD conditions (Supplementary Fig. 1D). Interestingly, circulating GLP-1 levels were higher in HFD Fpr1-KO mice compared with WT mice (Fig. 2H). No significant changes were detected in levels of other circulating gastric hormones or glucagon (Supplementary Fig. 4), suggesting that GLP-1 may drive the increased insulin secretion in Fpr1-KO animals. We also found that antibiotic-treated HFD mice displayed improved glucose tolerance and increased GLP-1 levels, consistent with a connection between the gut microbiota and GLP-1 modulation (Supplementary Fig. 5). To determine whether the glucose-tolerant phenotype of HFD Fpr1-KO mice is dependent upon GLP-1, we pretreated WT and Fpr1-KO HFD mice with the Glp1r antagonist exendin 9-39 (45). This antagonist prevented the improved glucose tolerance in Fpr1-KO mice compared with vehicle-treated littermates (Fig. 2I and J). Similarly, the elevated insulin levels in Fpr1-KO animals were suppressed by Glp1r inhibition (Fig. 2K). Thus, the beneficial effects of Fpr1 deficiency are dependent upon GLP-1 signaling.
GLP-1 Secretion Is Dependent on Fpr-1 Expression
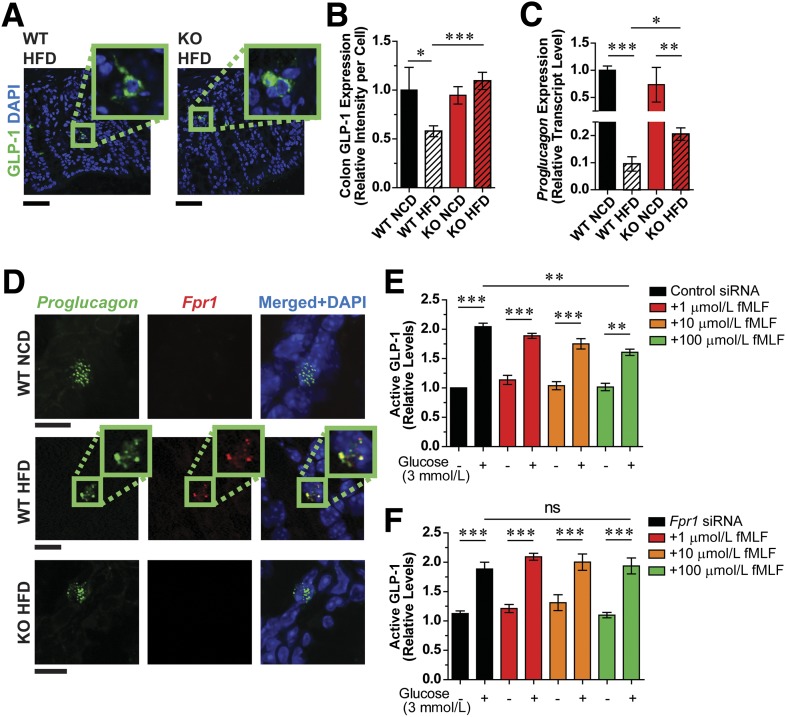
Consistent with elevated GLP-1 levels in HFD Fpr1-KO mice, immunofluorescence analysis revealed increased GLP-1 expression in enteroendocrine L cells of the ileum and colon in these mice compared with reduced levels observed in WT HFD mice (Fig. 3A and B and Supplementary Fig. 6A). mRNA transcript levels of Proglucagon, the precursor of GLP-1, were also increased in Fpr1-KO mice relative to WT mice on an HFD (Fig. 3C and Supplementary Fig. 6B and C), indicating a potential transcriptional basis for increased GLP-1 levels. We also demonstrate Fpr1 expression in L cells through RNA fluorescence in situ hybridization and immunofluorescence analyses (Fig. 3D and Supplementary Fig. 6D and E). Interestingly, whereas Fpr1 expression was quite low and difficult to detect in NCD L cells, we found increased Fpr1 expression in HFD L cells. In addition, Fpr1 expression was increased in the colon of HFD mice, as measured by qPCR and Western blot analyses (Fig. 1I and Supplementary Figs. 1G and 6D–G). This provides a reasonable explanation for the more dramatic phenotypic difference in Fpr1-KO mice on an HFD. GLP-1 can also be produced in pancreatic α-cells, but our studies show that Fpr1 expression is very low in isolated murine islets, with even lower expression in HFD (Supplementary Fig. 1C and D). This suggests that the actions of Fpr1 to modulate GLP-1 levels are primarily through L cells in the intestine.
Figure 3.
GLP-1 secretion is dependent on Fpr-1 expression. A: Immunofluorescence labeling of GLP-1 in L cells of WT and Fpr1-KO HFD mouse colon sections (representative images; scale bars = 50 µm; 16 weeks on an HFD). B: Quantification of A, including NCD controls (n = 14–20/group). C: Proglucagon mRNA levels in the ileum of WT and Fpr1-KO mice, relative to WT NCD (n = 8, 16 weeks on an HFD). D: RNA fluorescence in situ hybridization of Proglucagon and Fpr1 in colonic L cells (representative images; scale bars = 10 µm) (n = 6/group). E and F: Glucose-stimulated GLP-1 secretion in mGLUTag cells treated with scrambled control siRNA (E) or Fpr1 siRNA (F) and different concentrations of fMLF (n = 5). Error bars indicate SEM. ns, not significant. *P < 0.05, **P < 0.001, ***P < 0.0001 by one-way ANOVA and Bonferroni post-test, comparing the indicated groups.
Given these results, we postulated that fMLF acts upon L cells to suppress glucose-stimulated GLP-1 secretion. To test this, we measured glucose-stimulated GLP-1 secretion in murine mGLUTag L cells, treated with or without fMLF. Treatment with fMLF suppressed GLP-1 secretion in a dose-dependent fashion (Fig. 3E). Furthermore, siRNA-mediated Fpr1 knockdown abrogated the effect of fMLF to reduce GLP-1 secretion at any of the concentrations tested (Fig. 3F and Supplementary Fig. 6F). Whereas Fpr1 was well expressed, we could not detect the low-affinity Fpr2 in these cells (data not shown). Treatment of mGLUTag cells with palmitate to mimic HFD conditions also led to increased expression of Fpr1, similar to that observed in vivo (Supplementary Fig. 6E). Overall, we conclude that fMLF can signal through FPR1 in L cells to inhibit GLP-1 secretion.
Flow cytometric analysis of the small intestinal lamina propria confirmed previously reported immune cell population differences in HFD versus NCD animals, including reduced eosinophil and increased macrophage numbers (Supplementary Fig. 7A–H) (32). However, loss of Fpr1 did not change immune cell composition in either diet condition. qPCR analysis of these cells revealed little change in proinflammatory gene expression, although anti-inflammatory interleukin-10 expression was increased in Fpr1-KO HFD mice (Supplementary Fig. 7I and J). Similarly, elevated interleukin-10 levels were detected in the blood of these mice (Supplementary Fig. 7K). Analysis of inflammation in eWAT also showed no significant change in macrophage numbers and only a modest improvement in inflammatory gene expression (Supplementary Fig. 8).
Pharmacological Control of Fpr1 Activity Modulates GLP-1 Levels and Glucose Tolerance
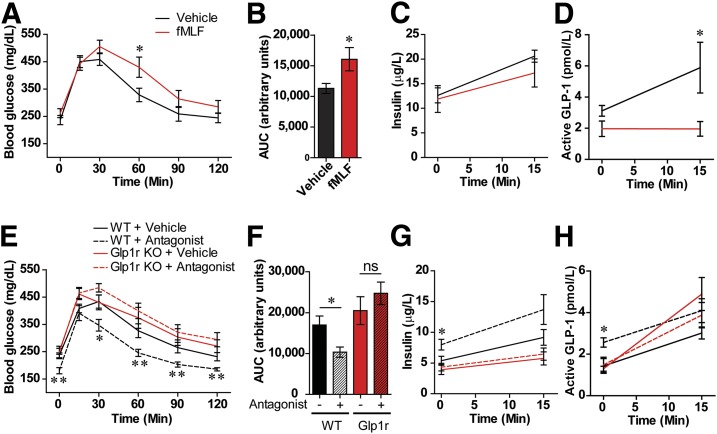
To confirm that the Fpr agonist fMLF detrimentally impacts glucose metabolism in vivo, we acutely treated WT HFD mice with fMLF. Treatment reduced glucose tolerance (Fig. 4A–C) and GLP-1 levels (Fig. 4D). In contrast, fMLF had no significant effect in NCD mice, likely due to the low expression of GI L-cell Fpr1 in these animals compared with HFD animals (Supplementary Fig. 9A). In addition, we determined whether pharmacological inhibition of Fpr1 could provide benefits similar to those seen in Fpr1-KO mice. In contrast to agonist administration, the treatment of WT HFD mice with the Fpr1 antagonist cFLFLF (Supplementary Fig. 9B) improved glucose tolerance (Fig. 4E and F) and increased plasma insulin and GLP-1 levels, without changing body weight or food intake (Fig. 4G and H and Supplementary Fig. 9C–H). The improved glucose tolerance induced by the Fpr1 antagonist is dependent upon the Glp1r, as treatment had no effect in HFD Glp1r-KO littermates (Fig. 4E and F). These results suggest that GLP-1 dependency is the dominant mechanism for the Fpr1-KO phenotype.
Figure 4.
Pharmacological control of Fpr1 activity modulates GLP-1 levels and glucose tolerance. A: OG-GTT of WT HFD mice after acute OG with the Fpr1 agonist fMLF (1 mg/kg) (n = 6 mice/group, 10 weeks on HFD). B–D: Area under the curve (AUC), insulin levels, and active GLP-1 levels measured during the OG-GTT. The information for A also applies to C and D. E: OG-GTT of WT and Glp1r-KO mice after 10 weeks on an HFD and daily OG of the Fpr1 antagonist cFLFLF for 2 weeks (0.5 g/kg) (n = 8 mice/group). F–H: AUC, insulin levels, and active GLP-1 levels measured during the OG-GTT. Information for E also applies to G and H. Data are mean ± SEM. ns, not significant. *P < 0.05, **P < 0.001 by two-way ANOVA and Bonferroni post-test (A and E), one-way ANOVA and Bonferroni post-test (F) or two-tailed t test. Statistical significance is indicated comparing groups at each respective time point. In E, G, and H, significance is indicated for WT animals treated with vehicle vs. WT animals treated with antagonist.
Loss of Fpr1 Alters Intestinal Microbiome Composition in HFD/Obese Mice
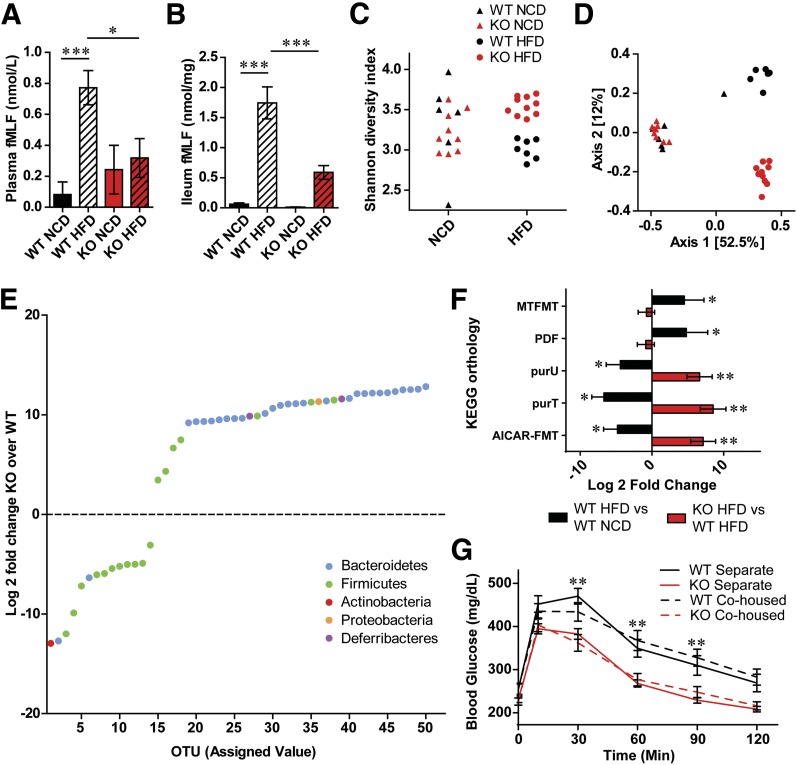
We next assessed the effects of diet and genotype on the microbiome. Levels of fMLF in the blood and intestine of HFD Fpr1-KO mice were reduced compared with WT mice (Fig. 5A and B). Measurement of GI permeability as assessed by fecal albumin content and analysis of blood levels of orally administered FITC-dextran confirmed increased permeability in HFD animals, consistent with the increased plasma fMLF levels (Supplementary Fig. 10A and B) (32). However, we found no change in GI permeability in Fpr1-KO mice compared with WT mice, suggesting that the microbiota composition in the Fpr1-KO mice might be responsible for decreased fMLF. To investigate this, we conducted fecal 16S rRNA gene sequencing of WT mice and Fpr1-KO littermates fed a NCD or HFD. Sequences were clustered based on similarity into OTUs and compared against a reference 16S rRNA database to identify their bacterial classifications (Supplementary Fig. 10C) (35). There was a modest change in α-diversity between lean/NCD and HFD animals as measured by either the Shannon diversity index or OTU richness (Fig. 5C), while the loss of Fpr1 led to a marked increase in diversity compared with WT mice on an HFD, although not on a NCD (Fig. 5C and Supplementary Fig. 10D and E). We next examined compositional differences between samples, termed β-diversity, based on dissimilarity of whole microbiome abundance profiles, using Bray-Curtis and principal component analyses. Diet was the strongest determinant of differences, with HFD and NCD samples highly dissimilar regardless of genotype (Fig. 5D). Interestingly, the Fpr1-KO significantly affected microbiome composition, but only in HFD animals, revealing that HFD interacts with the Fpr1-KO genotype to uncover an otherwise masked phenotype.
Figure 5.
Loss of Fpr1 alters intestinal microbiome composition in obese/HFD mice. A: fMLF levels in blood plasma (A) and ileum (B) of Fpr1-KO vs. WT mice after 20 weeks on an HFD (n = 6–11 mice/group). C: Shannon diversity index in fecal 16S rRNA gene-sequencing data from WT and Fpr1-KO mice on a NCD and an HFD (n = 6–10 mice/group). The information for C also applies to D. D: Analysis of dissimilarity between samples or β-diversity. The pairwise Bray-Curtis dissimilarity between samples is represented by a two-dimensional principal coordinate analysis ordination plot. Samples separate into three clusters based on diet (Axis 1) and genotype (Axis 2). The percentage of sample variation due to each axis is indicated. E: Log2 fold change of OTU abundances in Fpr1-KO vs. WT mice on an HFD, showing the 50 most significantly changed taxa, color coded by phylum. F: Log2 fold change of bacterial enzymes predicted altered by inferred metagenomics content analysis, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (n = 6–10 mice/group). AICAR-FMT, phosphoribosylaminoimidazolecarboxamide-formyltransferase; PDF, peptide deformylase; purU, formyltetrahydrofolate deformylase; purT, phosphoribosylglycinamide formyltransferase. G: OG-GTT of WT and Fpr1-KO mice after 10 weeks on an HFD, comparing mice housed separately based on genotype or cohoused. Data are mean ± SEM. ns, not significant. *P < 0.05, **P < 0.001, ***P < 0.0001 by one-way ANOVA with Bonferroni post-test (A and B), two-way ANOVA and Bonferroni post-test (G) or PERMANOVA and DESEq. 2 (C–F). In G, significance is indicated for WT cohoused vs. Fpr1-KO cohoused animals at each respective time point.
Comparison between Fpr1-KO and WT microbiome abundances on an HFD revealed differences in >360 OTUs (Fig. 5E and Supplementary Table 3). The mean relative abundances of several obesity-associated families within these phyla, including Lachnospiraceae and Rikenellaceae (Supplementary Fig. 10F–H) (46), were altered by diet and genotype. To illuminate what functional bacterial pathways and enzymes might be altered, we performed Piphillin inferred metagenomic content analysis, which predicts metagenomes from 16S rRNA sequencing data by comparison with reference genome databases (47). This analysis predicted numerous pathways and enzymes altered by diet or genotype (Fig. 5F, Supplementary Fig. 10I, and Supplementary Tables 4 and 5).
As changes in host metabolism can influence gut microbiota composition (4), we tested whether the differences in microbiota observed in HFD Fpr1-KO mice might be the result or a cause of improved glucose tolerance. Given the coprophagic nature of mice, one would predict that a microbiota-dependent effect would be transferrable through cohousing. We found that even when cohoused, differences in glucose tolerance were readily observed between genotypes, arguing against microbiome-transferable effects (Fig. 5G). Analysis of whole microbiome abundance profiles from multiple cohorts containing cohoused or separated mice showed that, whereas WT and Fpr1-KO mice raised in separate cages display differences in microbiota composition, those that are cohoused are more similar or the same (Supplementary Fig. 10J).
Fecal Microbiota Transfer From Fpr1-KO HFD Donor Mice to GF Recipient Mice Does Not Alter Glucose Tolerance Compared With Controls
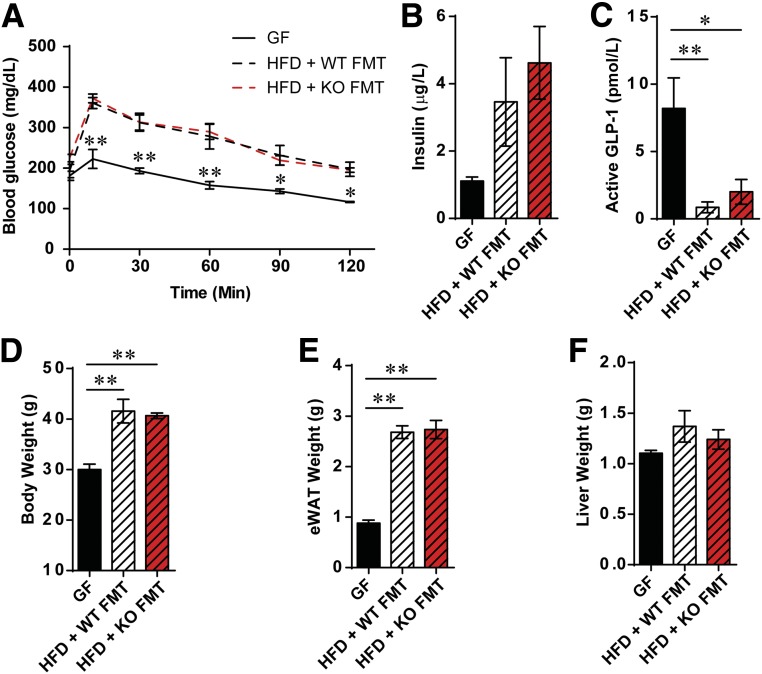
To further elucidate the role of the microbiome, we performed fecal microbiota transfer experiments, in which we transferred microbiota by gavage from either WT or Fpr1-KO HFD donor mice to GF recipient animals. After 8 weeks on an HFD, no differences in glucose tolerance were found between animals receiving microbiota from WT or Fpr1-KO mice (Fig. 6A). Insulin and GLP-1 levels were also unchanged, suggesting that the altered microbiome of Fpr1-KO mice does not transfer the improved metabolic phenotype (Fig. 6B and C). Rather, the enhanced glucose tolerance in Fpr1-KO mice appears to be genotype related. Body weight, epididymal fat, and liver weight were also unchanged between mice that received WT or Fpr1-KO HFD microbiome transplants (Fig. 6D–F).
Figure 6.
Fecal microbiota transfer (FMT) from WT and Fpr1-KO HFD donor mice to GF recipient mice leads to glucose intolerance. A: OG-GTT of GF mice fed a NCD or placed on an HFD and gavaged with fecal preparations from WT or Fpr1-KO mice fed an HFD (n = 3–4 mice/group; 8 weeks on an HFD). B: Basal insulin levels taken immediately prior to OG-GTTs. C: Active GLP-1 levels. D: Body weight at the time of OG-GTTs. E: Weight of eWAT. F: Weight of liver. *P < 0.05, **P < 0.001 by two-way ANOVA and Bonferroni post-test (A) or one-way ANOVA and Bonferroni post-test, comparing the indicated groups. In A, significance is indicated for GF vs. HFD + WT FMT animals at each respective time point.
Discussion
In these studies, we identify a bacterial product and host receptor signaling mechanism that modulates GLP-1 to influence metabolism in diet-induced obesity. As such, fMLF is among the first identified microbiota-produced compounds increased in obesity, which can directly impair glucose homeostasis. These data support a model whereby lean animals have a healthy microbiota producing lower levels of fMLF, which, together with low Fpr1 expression in L cells, allows for abundant postprandial GLP-1 secretion. In contrast, obesity-induced dysbiosis leads to elevated fMLF levels and increased Fpr1 expression, culminating in suppressed GLP-1 secretion and reduced glucose tolerance (Supplementary Fig. 11).
Incretins are important hormones that stimulate β-cell glucose-stimulated insulin secretion (11). In particular, GI L cells produce GLP-1 under the influence of dietary glucose as well as other regulatory inputs. Recent studies have also demonstrated that glucagon-producing α-cells in the pancreatic islets also produce GLP-1 and that this may play an important role in stimulating insulin secretion in neighboring β-cells (48). In addition, with respect to α-cells versus L cells, our studies found low expression of Fpr1 in α-cells compared with robust expression in L cells, along with high local levels of fMLF in the GI tract (Supplementary Fig. 1C and D). This suggests that the major physiologic effects of fMLF are on GI L cells. It is also well known that GLP-1 has robust effects to improve glucose homeostasis in an insulin-independent manner (49). Thus, GLP-1 causes decreased α-cell glucagon secretion, delayed gastric emptying, and neuronal signaling from the gut to the brain that may contribute to modulation of metabolic homeostasis (50). With this paradigm, the effects of GLP-1 from either α-cells or L cells on the stimulation of insulin secretion would represent only one arm of GLP-1 regulation of glucose homeostasis.
Our studies show that feeding mice an HFD had a strong effect that caused microbiome changes, commonly termed dysbiosis. In addition to the diet-induced dysbiosis, we found that Fpr1-KO also led to alterations in the composition of the microbiota. Interestingly, predicted levels of methionyl-tRNA formyltransferase (MTFMT), the enzyme that produces formylmethionine, were increased in WT HFD versus NCD mice, consistent with increased fMLF production (51). Furthermore, in Fpr1-KO mice versus WT mice on an HFD, predicted levels of purine production enzymes were elevated, potentially decreasing substrate availability for MTFMT and contributing to the reduced fMLF levels observed in Fpr1-KO animals (52). Interestingly, in the current study, whereas Fpr1-KO mice demonstrated an altered microbiome, when we did fecal transplant studies from Fpr1-KO mice into GF WT animals, we found that the microbiome transplant did not confer any changes in metabolic status. These data suggest that the microbiome alterations we observed in the Fpr1-KO mice are secondary to their improved metabolic status.
Another important finding from this study is that HFD mice display elevated fMLF levels, which are associated with glucose intolerance and reduced GLP-1 secretion. This increased fMLF level is likely derived from the microbiome, as antibiotic treatment of mice abolished blood levels of fMLF and GF animals also displayed barely detectable intestinal and blood fMLF levels (Fig. 1G and H and Supplementary Fig. 1A and B). Although these data suggest that most of the fMLF present in HFD mice appears to be microbiome derived, there may be a small contribution from host mitochondria, which remains to be determined. Since the treatment of WT and Fpr1-KO mice with the GLP1 antagonist (9–39) blocked the effects of Fpr1-KO to improve glucose tolerance, this suggests that the major aspects of the metabolic phenotype in the KO animals can be traced back to GLP-1/GLP1R interactions. Furthermore, treating mice with exogenous fMLF by OG led to worsening of glucose tolerance, whereas the treatment of these mice with the Fpr1 antagonist cFLFLF markedly improved glucose tolerance with enhanced insulin secretion. G-protein–coupled receptors have been established as important regulators of L-cell GLP-1 secretion through the elevation of intracellular cAMP levels leading to a rise in intracellular Ca2+ with membrane depolarization (53). Since Fpr1 is a Gαi-coupled G-protein–coupled receptor, our data are consistent with the proposal that fMLF signaling through Fpr1 can lower intracellular cAMP levels by activating Gαi (Supplementary Fig. 1F). These lowered levels of cAMP would impair membrane depolarization and reduce GLP-1 secretion. This raises the possibility that Fpr1 antagonists could eventually be used in the context of antidiabetic therapy.
In summary, our findings illustrate a mechanism whereby the gut microbiota influences metabolism in obesity (Supplementary Fig. 11). We demonstrate that fMLF levels are significantly increased in HFD mice and that Fpr1 KO improves glucose tolerance and promotes GLP-1 and insulin secretion. GLP-1 antagonism reverses the beneficial metabolic effects of Fpr1 KO, indicating a primary role of incretins in the phenotype. We also found that the treatment of HFD mice with fMLF can exacerbate glucose intolerance, although antagonizing fMLF with cFLFLF significantly improves glucose tolerance. Finally, microbiome changes occur with an HFD and Fpr1 KO, which highlight a dynamic host-microbiome relationship. The antidiabetic effects of fMLF antagonism should be further explored in order to maximize the incretin effect in patients who have uncontrolled hyperglycemia.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Ji Wang and Dr. Phillip Murphy at the NIH for providing Fpr1-KO mice; Dr. Daniel Drucker at the University of Toronto for providing Glp1r-KO mice; Dr. Patricia McDonald at Scripps Research Institute of Florida for providing the mGLUTag cell line; and Dr. Lars Eckmann and Dr. Yukiko Miyamoto at the UC San Diego Gnotobiotic Mouse Facility for providing GF mice and fecal transplantation. The authors also thank Angela Tyler (University of California, San Diego) for administrative support.
Funding. This study was funded in part by grants from the NIH Office of the Director (F32-DK-105686 and T32-DK-007494) and National Institute of Diabetes and Digestive and Kidney Diseases (DK-033651, DK-063491, DK-074868, and DK-101395). Imaging was conducted at the UC San Diego Neuroscience Microscopy Facility (NIH grant P30-NS-047101). The synthesis of cFLFLF was supported by a private donation to D.P. (147807-101-DR02793-41200).
Duality of Interest. This study was funded in part by grants from Merck and Johnson & Johnson (UCSD 2016-1729). No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.W., M.R., Y.-J.X., A.M.F.J., J.M.O., W.Y., D.E.O., Y.S.L., and J.D.W. performed in vivo and in vitro studies. J.W., M.R., Y.-J.X., A.M.F.J., J.M.O., W.Y., D.E.O., Y.S.L., J.D.W., and M.J. analyzed the data. J.W. and J.M.O. designed the studies and wrote the manuscript. M.R., Y.-J.X., A.M.F.J., J.M.O., W.Y., D.E.O., L.S.C., A.W.H., N.A.M., C.N.R., Y.S.L., J.D.W., M.D.C., D.P., and M.J. gave input to the design of the studies and the writing of the manuscript. L.S.C., A.W.H., N.A.M., and C.N.R. conducted and analyzed 16S rRNA sequencing. M.D.C. and D.P. synthesized and purified cFLFLF. J.W. and J.M.O. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1307/-/DC1.
References
- 1.Esser N, Legrand-Poels S, Piette J, Scheen AJ, Paquot N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res Clin Pract 2014;105:141–150 [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102:11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell 2013;152:673–684 [DOI] [PubMed] [Google Scholar]
- 5.Shamseddeen H, Getty JZ, Hamdallah IN, Ali MR. Epidemiology and economic impact of obesity and type 2 diabetes. Surg Clin North Am 2011;91:1163–1172, vii [DOI] [PubMed] [Google Scholar]
- 6.Guo X, Li J, Tang R, et al. High fat diet alters gut microbiota and the expression of paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators Inflamm 2017;2017:9474896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 8.Castaner O, Goday A, Park YM, et al. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol 2018;2018:4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu VB, Gribble FM, Reimann F. Free fatty acid receptors in enteroendocrine cells. Endocrinology 2018;159:2826–2835 [DOI] [PubMed] [Google Scholar]
- 11.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 2006;368:1696–1705 [DOI] [PubMed] [Google Scholar]
- 12.Miyawaki K, Yamada Y, Yano H, et al. Glucose intolerance caused by a defect in the entero-insular axis: a study in gastric inhibitory polypeptide receptor knockout mice. Proc Natl Acad Sci U S A 1999;96:14843–14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toft-Nielsen MB, Damholt MB, Madsbad S, et al. Determinants of the impaired secretion of glucagon-like peptide-1 in type 2 diabetic patients. J Clin Endocrinol Metab 2001;86:3717–3723 [DOI] [PubMed] [Google Scholar]
- 14.Larsen MP, Torekov SS. Glucagon-like peptide 1: a predictor of type 2 diabetes? J Diabetes Res 2017;2017:7583506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts EC, Hobson CH, Anderson RP, Chadwick VS. Radio-immunoassay for formyl methionyl leucyl phenylalanine. II. Demonstration of an enterohepatic circulation of immunoreactive bacterial chemotactic peptides in man. J Gastroenterol Hepatol 1990;5:38–43 [DOI] [PubMed] [Google Scholar]
- 16.Prevete N, Liotti F, Marone G, Melillo RM, de Paulis A. Formyl peptide receptors at the interface of inflammation, angiogenesis and tumor growth. Pharmacol Res 2015;102:184–191 [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010;464:104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prossnitz ER, Ye RD. The N-formyl peptide receptor: a model for the study of chemoattractant receptor structure and function. Pharmacol Ther 1997;74:73–102 [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Chen K, Chen J, et al. The G-protein-coupled formylpeptide receptor FPR confers a more invasive phenotype on human glioblastoma cells. Br J Cancer 2010;102:1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagener BM, Marjon NA, Prossnitz ER. Regulation of N-formyl peptide receptor signaling and trafficking by arrestin-src kinase interaction. PLoS One 2016;11:e0147442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babbin BA, Jesaitis AJ, Ivanov AI, et al. Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of Rac1 and Cdc42. J Immunol 2007;179:8112–8121 [DOI] [PubMed] [Google Scholar]
- 22.Leoni G, Alam A, Neumann PA, et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest 2013;123:443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molloy MJ, Grainger JR, Bouladoux N, et al. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 2013;14:318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao JL, Lee EJ, Murphy PM. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J Exp Med 1999;189:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scrocchi LA, Brown TJ, MaClusky N, et al. Glucose intolerance but normal satiety in mice with a null mutation in the glucagon-like peptide 1 receptor gene. Nat Med 1996;2:1254–1258 [DOI] [PubMed] [Google Scholar]
- 26.Chen P, Miyamoto Y, Mazagova M, Lee KC, Eckmann L, Schnabl B. Microbiota protects mice against acute alcohol-induced liver injury. Alcohol Clin Exp Res 2015;39:2313–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drucker DJ, Jin T, Asa SL, Young TA, Brubaker PL. Activation of proglucagon gene transcription by protein kinase-A in a novel mouse enteroendocrine cell line. Mol Endocrinol 1994;8:1646–1655 [DOI] [PubMed] [Google Scholar]
- 28.Riopel M, Seo JB, Bandyopadhyay GK, et al. Chronic fractalkine administration improves glucose tolerance and pancreatic endocrine function. J Clin Invest 2018;128:1458–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Morinaga H, Kim JJ, et al. The fractalkine/CX3CR1 system regulates β cell function and insulin secretion. Cell 2013;153:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cinti S, Mitchell G, Barbatelli G, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 2005;46:2347–2355 [DOI] [PubMed] [Google Scholar]
- 31.Goodyear AW, Kumar A, Dow S, Ryan EP. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J Immunol Methods 2014;405:97–108 [DOI] [PubMed] [Google Scholar]
- 32.Johnson AM, Costanzo A, Gareau MG, et al. High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PLoS One 2015;10:e0122195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996–998 [DOI] [PubMed] [Google Scholar]
- 35.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon CE. A mathematical theory of communication. Bell Syst Tech J 1948;27:623–656 [Google Scholar]
- 37.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 1957;27:326–349 [Google Scholar]
- 38.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353–D361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Kundu B, Zhong M, et al. PET imaging detection of macrophages with a formyl peptide receptor antagonist. Nucl Med Biol 2015;42:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol 2001;26:32–46 [Google Scholar]
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye RD, Boulay F, Wang JM, et al. International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol Rev 2009;61:119–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol 2010;177:2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards CM, Todd JF, Mahmoudi M, et al. Glucagon-like peptide 1 has a physiological role in the control of postprandial glucose in humans: studies with the antagonist exendin 9-39. Diabetes 1999;48:86–93 [DOI] [PubMed] [Google Scholar]
- 46.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwai S, Weinmaier T, Schmidt BL, et al. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 2016;11:e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fava GE, Dong EW, Wu H. Intra-islet glucagon-like peptide 1. J Diabetes Complications 2016;30:1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers AP, Sorrell JE, Haller A, et al. The role of pancreatic preproglucagon in glucose homeostasis in mice. Cell Metab 2017;25:927–934.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah M, Vella A. Effects of GLP-1 on appetite and weight. Rev Endocr Metab Disord 2014;15:181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meinnel T, Mechulam Y, Blanquet S. Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie 1993;75:1061–1075 [DOI] [PubMed] [Google Scholar]
- 52.Nagy PL, Marolewski A, Benkovic SJ, Zalkin H. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J Bacteriol 1995;177:1292–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson AK, Ward PS, Wong KY, et al. Cyclic AMP triggers glucagon-like peptide-1 secretion from the GLUTag enteroendocrine cell line. Diabetologia 2007;50:2181–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.