Abstract
Insulin resistance (IR) is a harbinger of type 2 diabetes (T2D) and partly determined by genetic factors. However, genetically regulated mechanisms of IR remain poorly understood. Using gene expression, genotype, and insulin sensitivity data from the African American Genetics of Metabolism and Expression (AAGMEx) cohort, we performed transcript-wide correlation and expression quantitative trait loci (eQTL) analyses to identify IR-correlated cis-regulated transcripts (cis-eGenes) in adipose tissue. These IR-correlated cis-eGenes were tested in the European ancestry individuals in the Metabolic Syndrome in Men (METSIM) cohort for trans-ethnic replication. Comparison of Matsuda index–correlated transcripts in AAGMEx with the METSIM study identified significant correlation of 3,849 transcripts, with concordant direction of effect for 97.5% of the transcripts. cis-eQTL for 587 Matsuda index–correlated genes were identified in both cohorts. Enoyl-CoA hydratase domain-containing 3 (ECHDC3) was the top-ranked Matsuda index–correlated cis-eGene. Expression levels of ECHDC3 were positively correlated with Matsuda index, and regulated by cis-eQTL, rs34844369 being the top cis-eSNP in AAGMEx. Silencing of ECHDC3 in adipocytes significantly reduced insulin-stimulated glucose uptake and Akt Ser473 phosphorylation. RNA sequencing analysis identified 691 differentially expressed genes in ECHDC3-knockdown adipocytes, which were enriched in γ-linolenate biosynthesis, and known IR genes. Thus, our studies elucidated genetic regulatory mechanisms of IR and identified genes and pathways in adipose tissue that are mechanistically involved in IR.
Introduction
The deterioration of systemic insulin responses related to glucose (and other metabolite) handling is referred to as insulin resistance (IR) (1). Reduced insulin sensitivity, or IR, promotes glucose intolerance and is a forerunner of type 2 diabetes (T2D) (1). Longitudinal studies in high-risk individuals suggest that IR occurs years before glucose intolerance or β-cell failure (2). Thus, reduced insulin sensitivity is an intermediate phenotype and early marker of T2D risk. Diet, lifestyle, and other environmental factors play important roles in the etiology of IR. However, several lines of evidence, including differences in the prevalence of IR among ethnic groups, suggest that genetic factors are important influences on IR (3). Deciphering the underlying molecular defects and genetic regulatory mechanisms of insulin sensitivity is an unmet need to develop novel and safe therapeutic options to prevent IR and slow or halt progression to T2D and its devastating complications.
Previous efforts to understand the role of genetic factors in determining IR were primarily focused on identifying susceptibility loci (e.g., single nucleotide polymorphisms [SNPs]) by genetic association studies in human populations. Large-scale genetic association studies provide valuable insights into the genetic architecture of phenotypes such as fasting glucose and insulin (4–7); IR, whether derived from oral glucose tolerance tests (OGTTs), frequently sampled intravenous glucose tolerance tests, or euglycemic clamp studies (8–10); and T2D. Functional interpretation of those association signals remains challenging, and recent efforts were only nominally successful in defining genes specifically modulated by these SNPs (11,12). Alternative strategies are required for a more precise understanding of genetic regulatory mechanisms of IR.
Adipose tissue integrates various physiologic pathways, including glucose and fatty acid homeostasis (13). The failure of adipocytes to sequester excess fuel during nutritional abundance results in ectopic fat accumulation and is proposed as a trigger for systemic IR (1,14,15). Transcriptomic analyses by us and others suggest that IR results from a derangement in gene expression in tissues involved in glucose homeostasis, including adipose tissue (16–18). Interactions among genetic factors and dietary components likely determine variability in adipose tissue remodeling and expansion by modulating gene expression, leading to adipose tissue dysfunction and onset of IR. Genetic variants such as SNPs may determine tissue gene transcript levels. Thus, to identify genetic IR regulatory mechanisms, we focused on adipose tissue IR–correlated transcripts modulated by expression regulatory loci (expression quantitative trait loci [eQTL]). We hypothesized that expression of a substantial subset of IR-correlated genes, in part, is genetically driven and causally linked to altered insulin sensitivity. Further, we hypothesized that focused interrogation of these genes would help to identify novel susceptibility loci within a functional context.
Similar to most complex diseases, IR is a heterogeneous mix of molecular phenotypes caused by perturbations of multiple genes contributing to altered insulin sensitivity. To identify genetic IR regulatory mechanisms that are common in both African and European ancestry populations, we implemented a systems biological approach. Using gene expression, genotype, and insulin sensitivity data from an African American cohort, we identified insulin sensitivity–correlated cis-regulated transcripts (cis-eGenes) in adipose tissue. Data from a European ancestry cohort replicated 587 insulin sensitivity–correlated cis-eGenes. These analyses suggested that genetically regulated expression of adipose tissue enoyl-CoA hydratase domain-containing 3 (ECHDC3) may play an important role in determining insulin sensitivity. In vitro genetic silencing studies in a human adipocyte model were successful in elucidating cellular and molecular mechanisms for the role of ECHDC3 in IR.
Research Design and Methods
Human Subjects
Glucometabolic phenotype, gene expression, and genotype data available from 256 unrelated and individuals without diabetes in the African American Genetics of Metabolism and Expression (AAGMEx) cohort (18,19) were used to identify genetically regulated adipose tissue insulin sensitivity–correlated transcripts. Cohort participants were healthy, self-reported African American men and women residing in North Carolina, aged 18–60 years, with a BMI between 18 and 42 kg/m2. All participants provided written informed consent under protocols approved by the institutional review boards at Wake Forest School of Medicine. A standard 75-g OGTT was used to evaluate insulin sensitivity and exclude individuals with diabetes. Fasting blood samples were drawn for DNA isolation and biochemical analyses. Subcutaneous adipose tissue biopsies were collected by Bergstrom needle from participants after an overnight fast. Clinical, anthropometric, and physiological characteristics of the AAGMEx cohort have previously been described (18).
We sought to replicate our findings in AAGMEx cohort using adipose tissue gene expression and genotype data from the European Ancestry individuals in Metabolic Syndrome in Men (METSIM) cohort (Finland [N = 770]) (20). In AAGMEx, insulin sensitivity was measured by both OGTT and frequently sampled intravenous glucose tolerance tests. However, the Matsuda index, a measure of insulin sensitivity derived from OGTT, was available for both the AAGMEx and METSIM cohorts and was used in this study as the primary phenotype. Laboratory, statistical, and bioinformatic methods are described briefly below.
Laboratory Measures and Physiologic Phenotypes
Details of clinical laboratory measures have previously been described (18,20). In brief, for the AAGMEx cohort, plasma glucose levels were analyzed at a Clinical Laboratory Improvement Amendments–certified commercial laboratory (LabCorp). Plasma insulin was measured using an immunochemiluminometric assay (Invitron Limited, Monmouth, U.K.). Plasma glucose and insulin data from five OGTT time points (0, 30, 60, 90, and 120 min) were used to calculate the Matsuda insulin sensitivity index (http://mmatsuda.diabetes-smc.jp/MIndex.html). Insulin sensitivity in the METSIM cohort was evaluated by calculating Matsuda index from three OGTT data points (0, 30, and 120 min).
Gene Expression Analyses and Genotyping Methods
Details of adipose tissue gene expression analyses, genotyping, and data quality-control methods for both cohorts have previously been published (18–20). Genome-wide expression data in AAGMEx were generated using HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA) whole-genome gene expression arrays. Data were submitted to the Gene Expression Omnibus (GEO) (identification no. GSE95674). In the METSIM cohort, subcutaneous adipose tissue expression data were generated by Affymetrix U219 arrays and submitted to GEO (identification no. GSE70353). Infinium HumanOmni5Exome-4 v1.1 DNA Analysis BeadChips (Illumina) were used to genotype DNA samples from the AAGMEx cohort based on the manufacturer’s instructions. METSIM samples were genotyped using the Illumina HumanOmniExpress BeadChip array and the Illumina HumanCoreExome array.
Bioinformatics and Statistical Analyses
As described previously (18), in the AAGMEx data set, genome-wide gene expression data (probe level) were extracted using Illumina GenomeStudio V2011.1. Expression levels were log2 transformed, robust multiarray average normalized (includes quantile normalization), and batch corrected using ComBat (https://www.bu.edu/jlab/wp-assets/ComBat/). Details of the gene expression data processing for METSIM have previously been published (20).
In AAGMEx, genotype data were examined to verify sample and SNP quality. Genotype assays of 4,210,443 SNPs passed technical quality filters. Genotypes from 2,296,925 autosomal SNP assays (representing 2,210,735 unique high-quality genotyped SNPs with minor allele frequency [MAF] >0.01 and Hardy-Weinberg equilibrium P value >1 × 10−6) were used in our initial eQTL analyses (19). We imputed these genotyped SNPs to the 1000 Genomes data set (1KGP, phase 3 cosmopolitan reference panel) using the genotype imputation program Minimac3, implemented on the Michigan Imputation Server (https://imputationserver.sph.umich.edu/). The combined set of 14,502,313 autosomal genotyped and imputed SNPs was used for expanded eQTL analyses for selected Matsuda index–correlated transcripts. After quality control and genotype imputation of the 681,789 directly genotyped variants, the METSIM study used 7,677,146 variants (MAF ≥0.01) for eQTL analysis (20).
To test for associations between insulin sensitivity and expression level in the AAGMEx cohort, we computed a linear regression model using R(glm) software with the Matsuda index (natural log transformed) as the outcome and expression level (log2) as the predictor. Models included age, sex, and African ancestry proportion (admixture estimates were computed using the ADMIXTURE program [http://software.genetics.ucla.edu/admixture/index.html]) as covariates (Supplementary Fig. 1). A secondary analysis included BMI as an additional covariate. We computed P values adjusted for Benjamini-Hochberg false discovery rate (FDR) (q value). Expression of a transcript probe correlated with Matsuda index at q < 0.01 was considered significant, excluding probes with a SNP within the probe sequence. We also correlated the Matsuda index with each transcript using Spearman semipartial correlation coefficients adjusted for age, sex, and ancestry proportion (admixture). We used the DAVID functional annotation tool (v6.9) (https://david.ncifcrf.gov/) and Ingenuity Pathway Analysis (IPA) (Build version: 470319M, Content version: 43605602) (https://apps.ingenuity.com/) to identify enrichment of Matsuda index–correlated genes in biological pathways. In the METSIM data set, regression analyses were computed to evaluate correlations between adipose tissue gene expression and cardiometabolic-related traits, including the Matsuda index (20). We compared results from the METSIM and AAGMEx cohorts. Expression of a transcript correlated with Matsuda index at q < 0.05 in both cohorts and showing the same direction of effect was considered statistically replicated.
We also conducted cis-eQTL analyses (i.e., within ± 500 Kb 5′ and 3′ of respective transcript). For each transcript probe, we computed linear regression with the log2-transformed expression value as the outcome and an additive genetic model for the SNP as implemented in the R package MatrixEQTL, with age, sex, and African ancestry proportion as covariates in AAGMEx. Cis-eQTL with P < 2.96 × 10−5 corresponding to a q value <0.01 (or <1.0%) were considered significant (19). A secondary eQTL analysis included BMI as an additional covariate. Results from factored spectrally transformed linear mixed model (FaST-LMM) eQTL analysis in the METSIM cohort (https://systems.genetics.ucla.edu/) were used for comparison (20). Considering the difference in sample sizes for eQTL analyses, transcripts with cis-eQTL P value < 2.46 × 10−4 in METSIM (corresponding to 1% FDR) (20) and P ≤ 1.75 × 10−4 (corresponding to q < 0.04) in AAGMEx were considered statistically replicated cis-eGenes.
To test for an association between a SNP and Matsuda index (natural log transformed) in AAGMEx, a linear regression model (SNPLASH) (https://www.phs.wakehealth.edu/public/home.cfm) was computed that included age, sex, and African ancestry proportion as covariates. The primary inference was based on the additive genetic model. Similar linear regression model analysis adjusting for age was implemented in R to test association between cis-expression regulatory single nucleotide polymorphisms (cis-eSNPs) in selected loci and Matsuda index in METSIM.
Studies in Cultured Adipocyte Models
Gene expression analysis in adipose tissue from human cohorts identified enoyl-CoA hydratase domain-containing 3 (ECHDC3) as the top-ranked Matsuda index–correlated cis-eGene (see results). Thus, we studied the expression of ECHDC3 at different stages of differentiation in human adipose stroma-derived stem cells (hADSCs). The hADSCs (AG17928 and AG172929) derived from abdominal subcutaneous tissue samples donated by two different women were obtained from Coriell Cell Repositories (Camden, NJ). To identify the role of ECHDC3 in adipocytes by knocking down its expression, we used Simpson-Golabi-Behmel syndrome (SGBS) preadipocytes, a well-characterized human adipocyte cell model (21) that is more amenable to genetic manipulation than hADSCs and has an expression profile after differentiation that closely mimics mature adipocytes. We grew hADSC and SGBS cells under standard culture condition in DMEM/Ham’s F-12 (1:1 v/v) adipocyte basal medium (BM-1; ZenBio, Inc.; Research Triangle Park, NC) supplemented with 10% FBS and antibiotics. Cells were differentiated to adipocytes using adipocyte differentiation medium (DM2; ZenBio) for 7 days and maintained for an additional 7 days in adipocyte maintenance medium (AM-1; ZenBio) for complete maturation of adipocytes following the vendor-recommended protocol.
For stable RNA interference, the ECHDC3 gene was silenced by infecting the SGBS preadipocytes with lentiviral particles to deliver gene-specific shRNA expression vectors (sc-90758-V; Santa Cruz Biotechnology, Santa Cruz, CA) (a pool of transduction-ready viral particles containing three target-specific constructs that encode shRNA) in the presence of polybrene (sc-134220 [8 μg/mL]; Santa Cruz Biotechnology) according to the manufacturer’s protocol. A control shRNA, lentiviral Particles-A (sc-108080; Santa Cruz Biotechnology) (encodes a scrambled shRNA sequence that will not lead to the specific degradation of any known cellular mRNA), was used as a negative control. Cells successfully transduced and stably expressing shRNA were selected using 2 μg/mL puromycin (A1113803; Gibco, Thermo Fisher Scientific).
Total RNA was isolated using an RNAeasy kit (Qiagen) from three sets of cells: 1) undifferentiated hADSC and SGBS cells, 2) hADSC and SGBS cells at different stages of differentiation, and 3) ECHDC3 shRNA and control shRNA–expressing SGBS cells at the 14th day of differentiation. RNA samples were reverse transcribed using a QuantiTect reverse transcription kit (Qiagen) based on the manufacturer’s protocol. For determination of expression levels at different stages of adipocyte differentiation and for confirmation of shRNA-mediated downregulation, ECHDC3 expression was measured in cDNA by quantitative real-time PCR (qRT-PCR) using Power SYBR green chemistry (Applied Biosystems, Inc., Foster City, CA). Oligonucleotide primers used to amplify ECHDC3 were 5′-ACGGCATAAGGAACATCGTC-3′ (forward) and 5′-AAAACACAGGCCCCTCAG-3′ (reverse). The expression of target genes was normalized to the expression of an endogenous control gene, 36B4/RPLP0. Two independent experiments with three biological replicates for each treatment condition were performed for confirmation of successful ECHDC3 knockdown by gene-specific shRNA.
Glucose Uptake Assay
Similar to a published study (22), on day 14 of differentiation, the control shRNA and ECHDC3 shRNA (ECHDC3-KD) SGBS cells grown in biological triplicates in sixwell plates were washed with Dulbecco’s PBS and preincubated with HEPES-buffered Krebs-Ringer solution (Alfa Aesar J67795) for 6 h at 37°C. After preincubation, the cells were incubated with or without 1 μmol/L insulin in Krebs-Ringer solution for 20 min. Next, 0.5 µCi/mL labeled 2-deoxy-d-[3H]glucose and 0.25 mmol/L d-glucose were added for an additional 20 min at 37°C. Reactions were terminated by placing the cells onto the ice and washing three times with ice-cold Dulbecco’s PBS. The cells were solubilized with 1 mL of 0.2 N NaOH per well and incubated overnight at room temperature with constant shaking. Radioactivity and protein content in cell lysates were measured using liquid scintillation spectroscopy and bicinchoninic acid assays, respectively. Cellular radiolabeled glucose uptake was normalized to cell protein content.
Western Blots to Determine Insulin-Stimulated Akt Phosphorylation
On the 14th day of differentiation, control shRNA and ECHDC3 shRNA (ECHDC3-KD) SGBS cells (in sixwell plates) were washed once with 0.1% BSA-PBS and serum starved in 0.1% BSA-PBS for 30 min. Cells were then stimulated with or without 100 nmol/L insulin for 15 min (23). Cells were harvested using 200 μL RIPA buffer (no SDS) per well of a sixwell plate. Cell lysates were sonicated (3 pulses for 3 s) on ice and centrifuged (12,000 rcf for 15 min), and protein concentrations in clear lysates were measured by bicinchoninic acid assays. Molecular weight markers and heat-denatured cell lysates containing 10 µg protein were loaded in each well for electrophoretic separation on a 10% SDS-PAGE gel (30 min at 80 V and 90 min at 110 V). Protein bands were transferred to polyvinylidene difluoride membranes (3.5–4 h at 65 V or 400 mA). Membranes were blocked with 5% nonfat milk and 0.1% Tween 20 in Tris-buffered saline (0.1% TBST) for 1 h at room temperature. Membranes were washed three times (5 min each) in 0.1% TBST. Blocked and washed membranes were then incubated overnight at 4°C in 0.1% TBST buffer containing 5% fatty acid–free BSA and primary phosphorylated Akt (pAkt) at Ser473 (1:1,000, cat. no. 4060; Cell Signaling Technologies), total Akt (1:1,000, cat. no. 4691; Cell Signaling Technologies), or GAPDH (1:2,000, cat. no. sc-32233; Santa Cruz Biotechnology) antibody. Membranes were then washed three times in 0.1% TBST (5 min each) and incubated with conjugated secondary antibody (1:5,000, cat. no. A9169, goat anti-rabbit IgG–horseradish peroxidase, Sigma-Aldrich, and cat no. sc-2031, goat anti-mouse IgG–horseradish peroxidase, Santa Cruz Biotechnology) in 0.1% TBST buffer containing 5% nonfat milk at room temperature for 1–2 h. Finally, membranes were washed (three washes of 0.1% TBST for 5 min each) and images were acquired using darkroom development techniques or cold camera for chemiluminescence.
RNA Sequencing and Bioinformatic Analyses
We performed genome-wide expression profiling of control and ECHDC3 shRNA SGBS cells in triplicate at day 14 of differentiation by RNA sequencing (RNA-seq). RNA-seq experiments were performed at Beijing Genome Institute (BGI, Shenzhen, China). In brief, high-quality total RNA samples (RNA integrity number [RIN] > 9.0) isolated from SGBS cells were treated with DNase I to remove DNA contamination, and then the mRNA was enriched by using oligo (dT) magnetic beads. Sequencing adaptor-ligated cDNA libraries were prepared from mRNA templates and were used with BGISEQ-500 RNA-seq technology to obtain >20 million single-end 50 base pair (bp) reads per sample. On average, 24,135,731 raw sequencing reads were generated per sample, and after filtering low-quality reads, on average 23,841,269 clean reads were obtained. Clean reads were mapped to the human reference genome using the HISAT/Bowtie2 tool (24,25). The average mapping ratio with the reference gene was 84.41% and average genome mapping ratio was 96.51%. Gene expression levels were quantified as fragments per kilobase million (FPKM) values using RSEM software (26). Differentially expressed genes (DEGs) between groups were screened using NOISeq (27). DEGs with Bayes posterior probability >70% in NOISeq and average log2 fold change ± 0.58 were considered significant. IPA (https://apps.ingenuity.com/) (Build version: 481437M, Content version: 39480507) was used to identify enrichment of DEGs in biological pathways.
Results
Expression of Subcutaneous Adipose Tissue Transcripts Enriched for Salient Biological Pathways Is Correlated With Matsuda Index of Insulin Sensitivity in African Americans
Individuals in the AAGMEx cohort had a broad range of insulin sensitivity as measured by OGTT-derived Matsuda index (mean ± SD 6.2 ± 6.7). High-quality adipose tissue gene expression and Matsuda index data (N = 246 subjects) were used to determine correlations between insulin sensitivity and adipose tissue transcript expression levels. Expression of 5,102 transcripts (probes representing 4,273 Entrez Gene IDs) in adipose tissue was significantly correlated (q value <0.01) with Matsuda index (Supplementary Table 1). Matsuda index was positively and negatively correlated, respectively, with the expression of 2,645 and 2,457 adipose transcripts. Among these transcripts, 83% (4,239 probes) remain significantly associated (P < 0.05) with Matsuda index even after additional adjustment for BMI. Membrane-spanning 4-domains subfamily-A member 6A (MS4A6A) was the most significant negatively (inversely) correlated transcript (ILMN_1721035, β = −1.06, P = 4.27 × 10−24), whereas zinc-binding α-2-glycoprotein 1 (AZGP1) was the most significant positively correlated transcript (ILMN_1797154, β = 1.33, P = 2.96 × 10−23) with the Matsuda index (Table 1). Multiple probes putatively representing isoforms of 707 Entrez ID genes were correlated with the Matsuda index, including probes for 691 Entrez ID genes (e.g., three probes for MS4A6A) showing the same effect direction. However, probes for 16 Entrez ID genes (e.g., two probes for clusterin [CLU]) showed discordant directions of effect, suggesting isoform-specific roles of these genes in insulin sensitivity.
Table 1.
Twenty most significantly correlated expression levels of subcutaneous adipose tissue transcripts with Matsuda insulin sensitivity index in AAGMEx
| Probe ID | Symbol | Entrez Gene ID | β | SE of β | Adjusted R2 | P value | q value |
|---|---|---|---|---|---|---|---|
| ILMN_1721035 | MS4A6A | 64231 | −1.06 | 0.093 | 0.365 | 4.27E-24 | 2.02E-19 |
| ILMN_2397721 | GLB1 | 2720 | −1.94 | 0.174 | 0.361 | 1.01E-23 | 2.39E-19 |
| ILMN_1796409 | C1QB | 713 | −0.94 | 0.087 | 0.342 | 3.44E-22 | 2.71E-18 |
| ILMN_1722622 | CD163 | 9332 | −1.01 | 0.095 | 0.338 | 6.67E-22 | 4.51E-18 |
| ILMN_1805992 | KIAA1598 | 57698 | −1.51 | 0.144 | 0.333 | 1.91E-21 | 1.13E-17 |
| ILMN_1745963 | FOLR2 | 2350 | −1.50 | 0.145 | 0.326 | 6.27E-21 | 2.97E-17 |
| ILMN_1792473 | AIF1 | 199 | −1.19 | 0.117 | 0.319 | 2.16E-20 | 7.86E-17 |
| ILMN_1740015 | CD14 | 929 | −1.20 | 0.118 | 0.317 | 2.97E-20 | 9.06E-17 |
| ILMN_1757387 | UCHL1 | 7345 | −0.66 | 0.066 | 0.315 | 4.36E-20 | 1.15E-16 |
| ILMN_1780533 | RNASE6 | 6039 | −2.42 | 0.241 | 0.314 | 5.50E-20 | 1.30E-16 |
| ILMN_1740024 | NAALAD2 | 10003 | 1.06 | 0.111 | 0.296 | 1.18E-18 | 1.22E-15 |
| ILMN_1757882 | PPP1R16A | 84988 | 1.51 | 0.157 | 0.296 | 1.20E-18 | 1.22E-15 |
| ILMN_3285959 | LOC645515 | 645515 | 2.54 | 0.265 | 0.296 | 1.22E-18 | 1.22E-15 |
| ILMN_1785284 | ALDH6A1 | 4329 | 1.22 | 0.124 | 0.305 | 2.79E-19 | 4.41E-16 |
| ILMN_1778104 | ACADM | 34 | 1.44 | 0.144 | 0.311 | 9.30E-20 | 2.00E-16 |
| ILMN_1694106 | GPD1L | 23171 | 1.37 | 0.136 | 0.317 | 3.06E-20 | 9.06E-17 |
| ILMN_1718924 | ETFA | 2108 | 1.76 | 0.175 | 0.317 | 3.02E-20 | 9.06E-17 |
| ILMN_1690040 | TM7SF2 | 7108 | 1.14 | 0.112 | 0.320 | 1.75E-20 | 6.89E-17 |
| ILMN_1678323 | AASS | 10157 | 2.84 | 0.279 | 0.322 | 1.42E-20 | 6.12E-17 |
| ILMN_1797154 | AZGP1 | 563 | 1.33 | 0.120 | 0.355 | 2.96E-23 | 4.64E-19 |
Top 10 positively and top 10 inversely correlated genes are shown. Results for all significant probes are shown in Supplementary Table 1. β, β-coefficient value; P value, significance level of correlation of transcript level with Matsuda index; adjusted R2, proportion of the variation in Matsuda index (outcome) explained by expression levels (predictor) in linear regression analysis adjusted for age, sex, and ancestry proportion/admixture; q value, FDR-corrected P value.
We found significant enrichment (Benjamini-Hochberg–corrected P < 0.05) of 205 canonical pathways (in IPA knowledgebase) among these Matsuda index–correlated transcripts in adipose tissue (Supplementary Table 2A). Gene expression profiles also suggested significant activation (activation z score >2) or repression (z < −2) of 41 pathways enriched in adipose tissue of insulin-sensitive individuals. Among the enriched pathways, expression profile of genes in oxidative phosphorylation (Benjamini-Hochberg–corrected P = 1.58 × 10−12, z = 7.216), EIF2 signaling (P = 6.17 × 10−8, z = 3.111), and valine degradation (P = 7.24 × 10−7, z = 3.357) pathways suggested significant activation, whereas genes in integrin signaling (P = 4.47 × 10−9, z = −3.801), IL-8 signaling (P = 1.70 × 10−6, z = −3.204), and Fcγ receptor–mediated phagocytosis in macrophage and monocyte (P = 2.88 × 10−6, z = −3.904) pathways suggested significant repression in insulin-sensitive individuals. Expression levels of 55 genes in oxidative phosphorylation pathways were positively, and of 33 genes in Fcγ receptor–mediated phagocytosis in macrophage and monocyte pathway were inversely, correlated with Matsuda index. DAVID functional annotation analysis using KEGG and GO annotation also validated the enrichment of multiple pathways (Supplementary Table 2B).
Correlation of Adipose Tissue Transcript Levels With Matsuda Index in African Americans Replicates in an Independent European Ancestry Cohort
Using adipose tissue expression data from METSIM and European ancestry cohorts, we sought replication of Matsuda index–correlated transcripts identified in African Africans. Individuals in the METSIM cohort also had a broad range of Matsuda index values (mean ± SD 7.3 ± 4.3). We compared the most significant Matsuda index–correlated probe for each gene in METSIM with transcripts significantly correlated with Matsuda index and matched for gene symbol annotation in AAGMEx. A total of 3,849 transcripts were significantly correlated (q < 0.05) with Matsuda index in both cohorts (Supplementary Table 3). Matsuda index was positively and inversely correlated with the expression of 2,047 and 1,708 adipose transcripts in both cohorts, respectively. (Supplementary Fig. 2A). Thus, our results suggest a strong replication of correlation and direction of effect for 97.5% of Matsuda index–correlated adipose tissue transcripts in independent cohorts of African and European ancestry individuals.
Matsuda Index–Correlated Transcripts Are Regulated by eQTL in African and European Ancestry Individuals
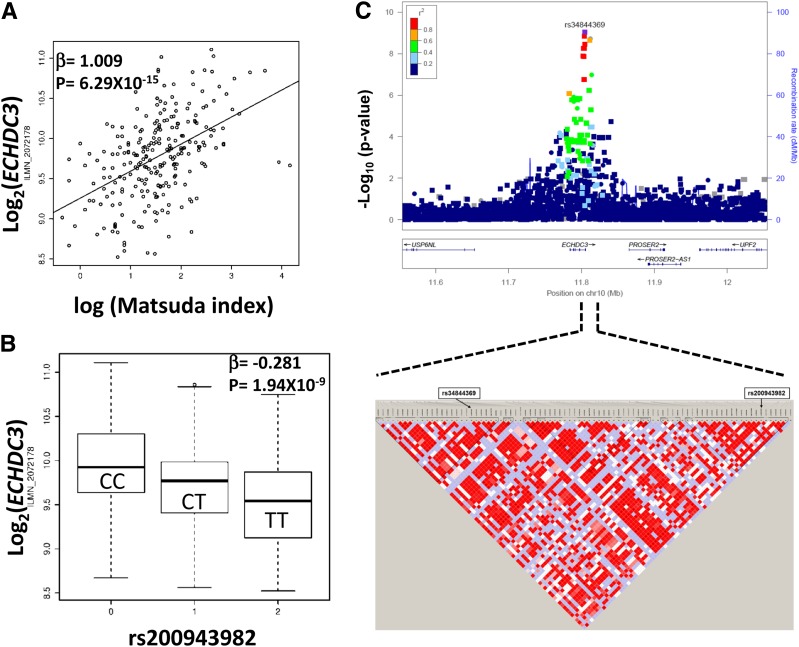
Our analyses in these cohorts suggested deranged expression of thousands of adipose tissue genes in those with IR. However, it is difficult to differentiate causal effects from reactive effects based on transcriptomic data. Genotype variations, including SNPs in regulatory regions of the genome or eQTL, may determine tissue transcript levels. The eQTL analyses integrate SNP genotypes with transcript expression profiles and provide evidence for genotype-dependent variations in transcript abundance. eQTL analyses identified significant cis-eSNPs for 587 Matsuda index–correlated transcripts in adipose tissue of AAGMEx and METSIM participants (FDR 4% and FDR 1%, respectively) (Supplementary Table 4). Top cis-eSNPs in one cohort showed high concordance in effect direction and strong correlation of effect sizes in the other cohort (Supplementary Table 4A and B and Supplementary Fig. 3). In the AAGMEx cohort, we also compared the eQTL analysis with and without adjustment for BMI. Of the 587 Matsuda index–correlated cis-eGenes that we report, 474 are significant in BMI-corrected eQTL analysis (FDR <4% [in Supplementary Table 4C]), and results from both analysis (β and –log10 P value) were highly correlated (r2 > 0.9). Thus, adipose tissue expression levels of these 587 transcripts are correlated with insulin sensitivity and are genetically regulated in both ethnicities. These transcripts were ranked based on their statistical significance of correlation with Matsuda index and association with genotype in eQTL analysis in AAGMEx and METSIM cohorts. The top 10 Matsuda index–correlated cis-eGenes are shown in Table 2. Based on average ranking, the enoyl-CoA hydratase domain-containing 3 (ECHDC3) gene was the top-ranked Matsuda index–correlated cis-eGene. Its expression was positively correlated with Matsuda index (ILMN_2072178, β = 1.009, P = 6.29 × 10−15 in AAGMEx [Fig. 1A] and 11728810_a_at, β = 0.34, P = 2.35 × 10−22, in METSIM [Supplementary Fig. 2B]). Adipose tissue ECHDC3 expression was strongly regulated by cis-eQTL (P = 1.94 × 10−9 in AAGMEx and P = 8.26 × 10−52 in METSIM [Fig. 1B and Supplementary Fig. 2C and D]).
Table 2.
Top 10 ranked Matsuda insulin sensitivity–correlated cis-eGenes in adipose tissue from African American (AAGMEx cohort) and European ancestry (METSIM) individuals
| Gene symbol | Chr | AAGMEx cohort |
METSIM cohort |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matsuda insulin sensitivity index |
eQTL analysis |
Matsuda insulin sensitivity index |
eQTL analysis |
||||||||||||||
| ρ | P | Probe ID | Top cis-eSNP | A1 | MAF | β* | eQTL_ P value | β | P value | Probeset ID | Top cis-eSNP | alt_ allele | MAF | β* | eQTL_ P value | ||
| ECHDC3 | 10 | 0.471 | 5.33E-15 | ILMN_2072178 | rs200943982 | T | 0.401 | −0.281 | 1.94E-09 | 0.342 | 2.35E-22 | 11728810_a_at | rs3814627 | G | 0.312 | −0.755 | 8.3E-52 |
| MIF | 22 | −0.390 | 2.26E-10 | ILMN_1807074 | rs4822455 | T | 0.435 | −0.188 | 4.56E-13 | −0.257 | 5.64E-13 | 11744470_x_at | rs4822443 | A | 0.225 | −0.989 | 1.3E-78 |
| CENPV | 17 | 0.411 | 1.86E-11 | ILMN_3246608 | rs3112521 | T | 0.427 | 0.107 | 5.83E-09 | 0.288 | 4.85E-16 | 11722721_a_at | rs3112526 | C | 0.485 | 0.751 | 2.3E-53 |
| DDT | 22 | 0.362 | 4.71E-09 | ILMN_1690982 | rs79966373 | G | 0.178 | −0.302 | 2.81E-14 | 0.277 | 5.30E-15 | 11731089_a_at | chr22:24334948 | C | 0.353 | −1.055 | 1E-125 |
| APOB | 2 | 0.401 | 6.50E-11 | ILMN_1664024 | rs1429974 | T | 0.241 | 0.293 | 5.28E-09 | 0.256 | 7.13E-13 | 11758033_s_at | rs11693870 | C | 0.265 | 0.950 | 9E-72 |
| ORMDL3 | 17 | 0.445 | 2.24E-13 | ILMN_1662174 | rs28820390 | A | 0.091 | −0.355 | 3.37E-07 | 0.392 | 1.52E-29 | 11736188_a_at | rs2872516 | T | 0.495 | −0.565 | 1.2E-29 |
| GLIPR1 | 12 | −0.458 | 3.55E-14 | ILMN_1769245 | rs117251563 | A | 0.013 | 0.705 | 8.07E-07 | −0.217 | 1.30E-09 | 11721839_at | rs35619460 | C | 0.194 | −1.445 | 1E-158 |
| ACSS3 | 12 | 0.449 | 1.32E-13 | ILMN_1659885 | rs4842386 | G | 0.167 | −0.111 | 6.97E-06 | 0.332 | 2.99E-21 | 11759049_at | rs3794325 | A | 0.332 | −0.919 | 5.9E-81 |
| ITIH5 | 10 | −0.434 | 9.54E-13 | ILMN_1731862 | rs201694044 | G | 0.375 | −0.154 | 5.05E-07 | −0.325 | 2.66E-20 | 11734677_x_at | rs867490 | G | 0.312 | 0.610 | 6.8E-33 |
| DMRT2 | 9 | 0.401 | 6.05E-11 | ILMN_1751785 | rs756145 | G | 0.315 | 0.282 | 2.77E-08 | 0.300 | 1.80E-17 | 11727728_a_at | rs10959032 | T | 0.271 | −0.699 | 9.4E-37 |
Matsuda insulin sensitivity–correlated transcripts of Entrez ID genes in adipose tissue associated with a SNP (q value <0.04) within ±500 Kb of the 5′ and 3′ end of the transcript in AAGMEx are shown. Data for most significantly associated genotyped cis-eSNP in AAGMEx are presented. Data on European ancestry males are from the METSIM cohort (Civelek et al. [20]). In AAGMEx: ρ, Spearman partial correlation coefficient; P value, significance level of correlation of transcript level with Matsuda index in Spearman partial correlation analysis; A1, minor allele; β, effect size of minor allele (A1); eQTL_P value, significance in additive model (in MatrixEQTL analysis). In METSIM: β, β value; P value, significance level of correlation of transcript level with Matsuda index in linear regression analysis; alt_allele, alternative allele; β*, effect size for alternative allele of best cis-eSNP in FaST-LMM eQTL analysis. Results for all significant probes are shown in Supplementary Table 4. Top cis-eSNPs are not the same in both cohorts, but comparison of their effects is shown in Supplementary Table 4A and B and in Supplementary Fig. 3. Chr, chromosome.
Figure 1.
ECHDC3 transcript expression in adipose tissue is correlated with Matsuda index of insulin sensitivity and is genetically regulated in African Americans. The scatter plot shows correlation of ECHDC3 transcript expression (ILMN_2072178) in adipose tissue with Matsuda index in AAGMEx (A). The box plot shows association of ECHDC3 transcript expression (ILMN_2072178) in adipose tissue with genotype of the cis-eSNP rs200943982 (B). LocusZoom plots show regional association of ECHDC3 cis-eQTL region SNPs (genotyped and imputed) with transcript expression (C). Significance level (−log10 P values) of genotyped SNPs are indicated as circles and imputed SNPs are indicated as squares in the LocusZoom plot. Linkage disequilibrium plot below the LocusZoom plot shows linkage disequilibrium relationship (r2) between SNPs in the marked region and indicates the location of top imputed and genotyped cis-eSNP for ECHDC3, rs34844369 and rs200943982, respectively.
Ten genotyped SNPs (within ±500 Kb of transcript and MAF >0.01) showed significant association (q < 0.04) with ECHDC3 transcript levels in adipose tissue of African Americans in the AAGMEx cohort, with rs200943982_T being the strongest genotyped cis-eSNP (MAF 0.393, β = −0.281, P = 1.94 × 10−9 [Fig. 1B]). Expanded eQTL analysis using imputed SNPs identified 53 cis-eSNPs (P < 1 × 10−4) in AAGMEx; rs34844369_A, an imputed SNP, was the strongest cis-eSNP (MAF 0.438, β = −0.283, P = 9.37 × 10−10 [Fig. 1C]) for ECHDC3 (Supplementary Table 5). ECHDC3 expression was positively correlated with Matsuda index in all genotype groups (Supplementary Fig. 4). The rs34844369 SNP is located in the intron of ECHDC3 and rs200943982 is located downstream (∼6.2 Kb 3′ of ECHDC3). These two cis-SNPs are 7,717 bp apart on chromosome 10 but showed moderate linkage disequilibrium (D′ = 0.89, r2 = 0.67). Neither SNP was significantly associated with ECHDC3 expression with adjustment for the other. Stepwise regression analysis identified no other significant eSNPs after adjustment for rs200943982 and rs34844369. Haplotype analyses of rs34844369 and rs200943982 suggested significant associations with ECHDC3 expression level (global haplotype test P = 1.32 × 10−13), with the A-T haplotype (rs34844369_A-rs200943982_T) associated with reduced ECHDC3 expression (β = −0.2976, P = 2.75 × 10−15, adjusted for age, sex, admixture). In the METSIM cohort, rs34844369 was a significant ECHDC3 cis-eSNP (P = 1.41 × 10−32 [Supplementary Table 6]). However, in METSIM, the strongest ECHDC3 cis-eQTL signal was identified at SNP rs3814627_G (MAF 0.312, β = −0.755, P = 8.26 × 10−52 [Supplementary Fig. 1D]), located in the putative ECHDC3 promoter region (−482 bp upstream of the transcription start site). The SNP rs3814627 was not a significant cis-eSNP in AAGMEx (MAF 0.122, P = 0.68); however, another SNP near this putative promoter region (rs10906007_G, P = 8.44 × 10–7, −1,157 bp upstream of the transcription start site) was significantly associated, suggesting the presence of common and ethnicity-specific genetic regulation for ECHDC3 expression in adipose tissue. Epigenetic regulatory annotation of SNPs in ECHDC3 cis-eQTL identified in the AAGMEx and METSIM cohorts, and the GTEx study by HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), suggested the presence of promoter or enhancer histone marks and altered transcription factor–binding motifs for many of these cis-eSNPs (Supplementary Table 7A). Bioinformatic analysis using RegulomeDb-v1.1 (http://regulomedb.org/) and SNP2TFBS (https://ccg.vital-it.ch/snp2tfbs/) also supported the putative regulatory role and ability to alter transcription factor binding of these SNPs in ECHDC3 cis-eQTL (Supplementary Table B and C). Despite a strong effect of regulatory genetic variants on proximal molecular phenotypes, i.e., transcript expression, these cis-eSNPs individually showed no statistically significant association with Matsuda index in either the AAGMEx or the METSIM cohort (Supplementary Tables 5 and 6). However, large genome-wide association study data sets curated in the Accelerating Medicines Partnership Type 2 Diabetes Knowledge Portal (http://www.type2diabetesgenetics.org/) for glycemic traits showed significant association (below genome-wide threshold) of top ECHDC3 cis-eSNPs, and allele effect direction on traits was consistent with their effect on gene expression in our eQTL analysis. For example, rs3814627_G allele that reduces ECHDC3 expression was associated with reduced Stumvoll insulin sensitivity index (β = −0.023, P = 0.0448) and increased HbA1c (β = 0.027, P = 0.0309) in MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) genome-wide association study data sets.
ECHDC3 Is Primarily Expressed in Adipocytes
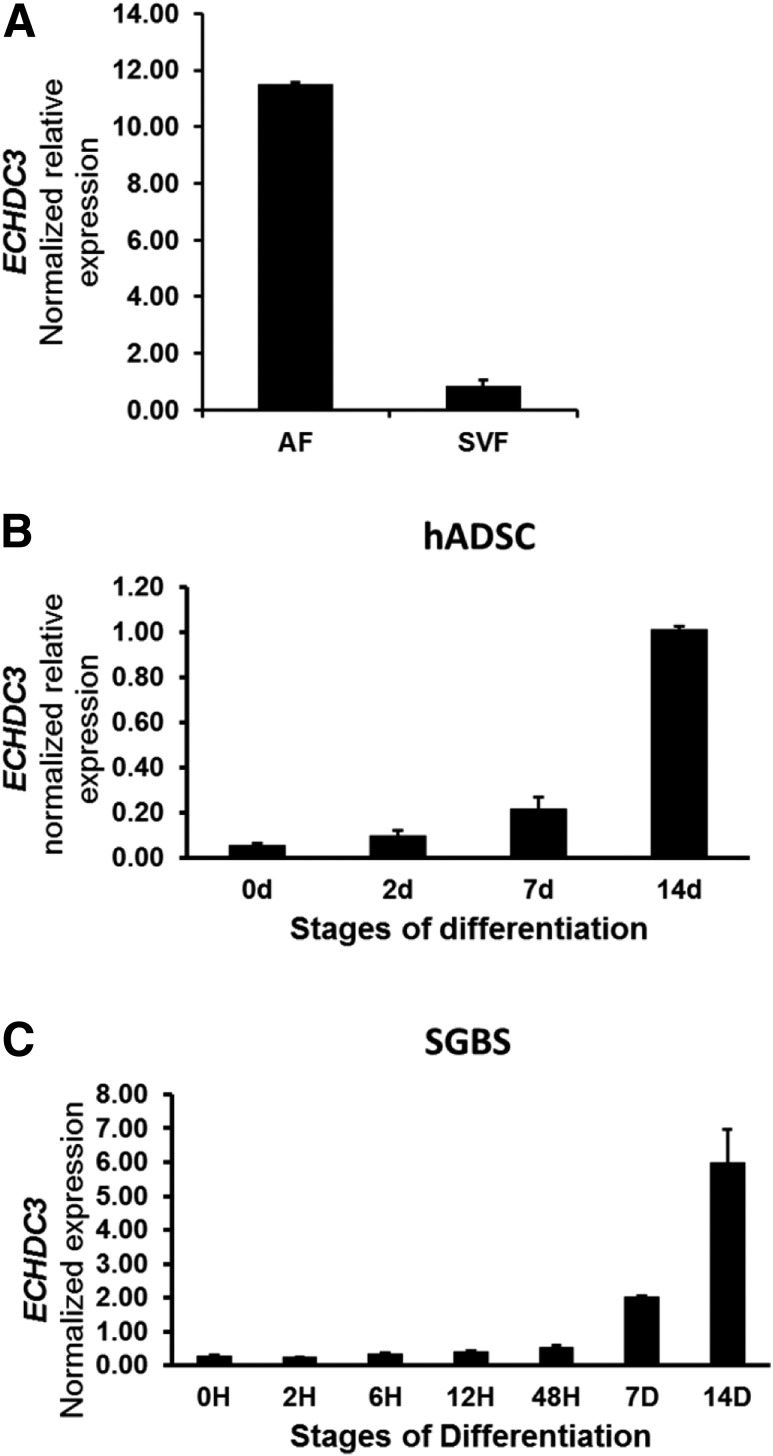
Human cohort analyses suggested that genetically regulated expression of ECHDC3 in adipose tissue may be important in determining insulin sensitivity. Defining the contribution of adipose tissue cell types in ECHDC3 expression levels may help identify cell-specific roles in correlations between ECHDC3 expression and the Matsuda index. Thus, we fractionated fresh subcutaneous adipose tissue into adipocytes and adipose stromal vascular cells as previously described (28). Total RNA extracted from five to six African American individuals in the AAGMEx cohort was used to prepare separate pools of adipocyte fraction and stromal vascular fraction cDNA. In a qRT-PCR assay, ECHDC3 expression was 13.6-fold higher in the adipocyte fraction (relative expression mean ± SD 11.53 ± 0.04) than the stromal vascular fraction (relative expression 0.85 ± 0.22) (Fig. 2A), suggesting that ECHDC3 expression is primarily attributable to adipocytes.
Figure 2.
In subcutaneous adipose tissue, ECHDC3 is primarily expressed in adipocytes and ECHDC3 expression is induced with adipocyte differentiation. Results from the qRT-PCR analysis of expression of ECHDC3 mRNA (normalized to RPLP0 [36B4] endogenous control gene) are shown. A: Relative expression of ECHDC3 in pooled RNA samples from stromal vascular fractions (SVFs) compared with the adipocyte fraction (AF) of subcutaneous adipose tissue in African Americans (data for two technical replicates of the pooled RNA samples from 5–6 individual donors are shown). Expression of ECHDC3 at different stages of in vitro differentiation of hADSCs (B) and SGBS preadipocytes (C). Data are shown for independent biological triplicates at each stage of differentiation. d and D, days; H, hours.
Adipocyte Differentiation Induces ECHDC3 Expression
We analyzed the expression of ECHDC3 transcripts at different stages of in vitro differentiation of hADSCs and SGBS cells. ECHDC3 mRNA expression was low in undifferentiated hADSCs (relative expression 0.05 ± 0.01), began to rise in cells after 7 days in differentiation media, and was strongly induced at 14 days (relative expression 1.01 ± 0.02) (Fig. 2B). Similarly, ECHDC3 mRNA expression at 14 days was 23-fold higher in SGBS cells compared with undifferentiated cells (Fig. 2C).
ECHDC3 Silencing in SGBS Adipocytes Decreases Insulin Sensitivity
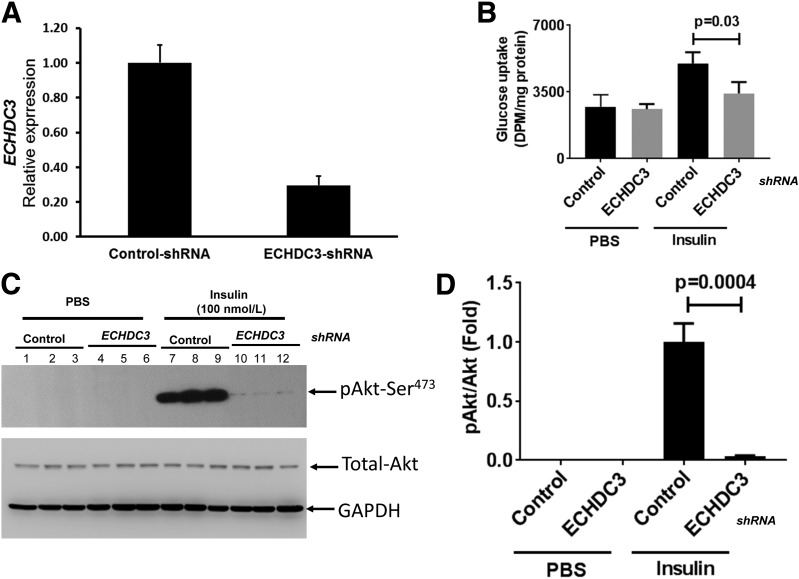
Individuals with low mRNA abundance of ECHDC3 in adipose tissue had low insulin sensitivity. Thus, we next determined whether decreasing ECHDC3 expression in cultured human adipocytes reduced insulin sensitivity. SGBS cells stably expressing ECHDC3-specific shRNA (ECHDC3-KD) showed 70% downregulation (P = 1.20 × 10−5) of ECHDC3 mRNA (Fig. 3A) compared with cells stably expressing control shRNA at 14 days after the induction of differentiation. Insulin (1 μmol/L) stimulated glucose uptake by 84% in control cells (P = 0.009) relative to PBS controls, but only 32% in ECHDC3-KD cells, demonstrating that insulin-stimulated glucose uptake was significantly (P = 0.03) decreased in ECHDC3-shRNA SGBS cells (ECHDC3-KD) compared with controls (Fig. 3B). As shown in a representative Western blot in Fig. 3C, pAkt at Ser473 after insulin stimulation was significantly decreased (P = 0.0004) (Fig. 3D) in ECHDC3-KD cells compared with controls.
Figure 3.
Knocking down of ECHDC3 gene in SGBS adipocytes affects insulin sensitivity. A: Lentiviral particle-mediated delivery of gene-specific shRNA expression vector stably knocks down ECHDC3 mRNA levels in SGBS adipocytes. Results from the qRT-PCR analysis of expression of ECHDC3 mRNA (normalized to RPLP0 [36B4] endogenous control gene) in SGBS cells stably expressing control shRNA or ECHDC3 shRNA at 14 days post–initiation of differentiation are shown. Bar graph indicates the mean ± SD of six biological replicates for each condition from two independent experiments. B: Basal and insulin-stimulated glucose uptake in control shRNA and ECHDC3 shRNA (ECHDC3-KD) SGBS cells at 14 days of differentiation. Bar graph indicates the mean ± SD of biological triplicates for each condition. Western blot analysis at day 14 of differentiated control shRNA and ECHDC3-KD SGBS cells using antibodies specific for pAKt, total Akt, and GAPDH at basal and insulin-stimulated (100 nmol/L for 15min) conditions (C) and quantified data (mean ± SD of biological triplicates) from scanning of a representative (selected from two independent experiments) Western blot (D) are shown. DPM, disintegrations per minute.
ECHDC3 Silencing in Adipocytes Modulates Many Pathways and Genes Correlated With IR and Metabolism of Fatty Acid and Triacylglycerol
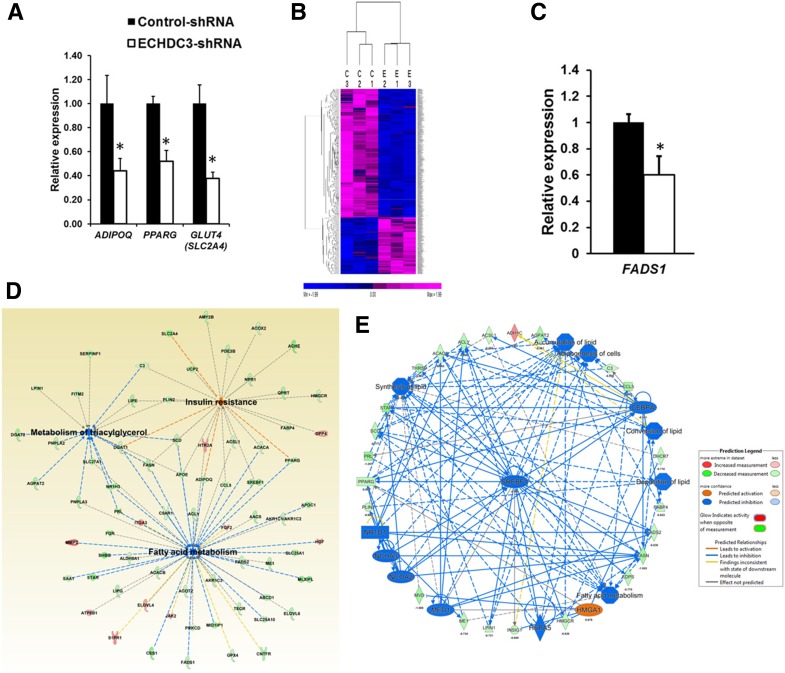
The functional role of ECHDC3 in adipocytes or other cells is unknown. Our qRT-PCR analyses suggested significant downregulation of three genes important in IR—adiponectin (ADIPOQ) (P = 1.15 × 10−3), facilitated glucose transporter-4 (GLUT4/SLC2A4) (P = 3.36 × 10−6), and peroxisome proliferator–activated receptor γ (PPARG) P = 2.51 × 10−6)—in ECHDC3-shRNA SGBS cells (ECHDC3-KD) compared with controls (Fig. 4A). To identify genes modulated by ECHDC3 and to define its biological role, we compared triplicate cultures of ECHDC3-shRNA SGBS cells (ECHDC3-KD) with control-shRNA SGBS cells at day 14 of differentiation by RNA-seq analysis. This genome-wide analysis identified 691 DEGs (at Bayes posterior probability >70% and average log2 fold change ±0.58) in ECHDC3-KD cells compared with control cells (Fig. 4B and Supplementary Table 8), including 479 downregulated and 212 upregulated genes. The IPA result suggested an enrichment of 63 canonical pathways (P < 0.05) among DEGs, with the γ-linolenate biosynthesis pathway as the most enriched (P = 3.8 × 10−8) (Supplementary Table 9). Eight genes in this pathway (e.g., FADS1, FADS2, ACSL1, ACSL5) were downregulated in ECHDC3-KD cells. Downregulation of FADS1 (P = 9.44 × 10−5) in ECHDC3-KD cells was validated by qRT-PCR in independent experiments (Fig. 4C). Canonical pathways for fatty acid, triglycerides, and cholesterol biosynthesis were also enriched among DEGs. Among the enriched pathways, expression profile of genes suggested significant inactivation of an LXR/RXR activation pathway (P = 2.04 × 10−7, z = −2.53). The IPA results also suggested enrichment of disease or biological function, including metabolism of triacylglycerol (17 genes, P = 4.54 × 10−13), fatty acid metabolism (51 genes, P = 1.54 × 10−10), and IR (25 genes, P = 1.63 × 10−8) (Fig. 4D and Supplementary Table 10). Regulator effects analysis considered DEGs in ECHDC3-KD cells and suggested an effect of upstream regulators on the expression of downstream target molecules and impact on biological function and diseases. The top two regulatory networks with a consistency score >20 suggested derangement in the accumulation of lipid, adipogenesis, and fatty acid metabolism in ECHDC3-KD cells (Fig. 4E and Supplementary Table 11). Thus, ECHDC3 knockdown in adipocytes may cause IR by deranging multiple key biological pathways.
Figure 4.
Knocking down ECHDC3 gene in adipocytes modulates many pathways including genes in insulin sensitivity. A: Bar graph showing results from the qRT-PCR analysis of expression of adiponectin (ADIPOQ), peroxisome proliferator–activated receptor γ (PPARG), and facilitated glucose transporter-4 (GLUT4/SLC2A4) mRNA (normalized to RPLP0) in SGBS cells stably expressing control shRNA or ECHDC3 shRNA at 14 days post–initiation of differentiation. Bar graph indicates the mean ± SD of six biological replicates for each condition from two independent experiments. *P < 0.01. B: Heat map from unsupervised hierarchical clustering showing expression of 691 DEGs in ECHDC3-KD (E) compared with control shRNA–treated (C) SGBS cells at day 14 of differentiation. The expression levels of genes from RNA-seq analysis (FPKM values) were z score transformed for clustering analysis. The intensity of purple and blue color in heat map indicates the degree of high and low expression of transcripts, respectively. C: qRT-PCR analysis validates downregulation of fatty acid desaturase 1 (FADS1) in ECHDC3-KD SGBS cells in six biological replicates for each condition from two independent experiments. D: A network diagram from IPA that includes selected DEGs in ECHDC3-KD cells, significantly enriched for genes involved in the metabolism of triacylglycerol, fatty acid metabolism, and IR. E: Top regulatory network (consistency score = 20.85 and 37 nodes) identified by regulator effects analysis in IPA connected 23 DEGs with eight putative network regulators in ECHDC3-KD cells and suggested its impact on multiple biological processes in adipocytes including accumulation of lipid and adipogenesis.
Discussion
Our results suggest that gene expression levels in adipose tissue are key trans-ethnic (trans-ancestral) biomarkers strongly correlated with modulation of insulin sensitivity. Expression levels of transcripts for thousands of protein-coding genes in adipose tissue correlated with the Matsuda index, a well-validated estimate of insulin sensitivity measured by physiologically relevant OGTT (29). Thus, IR involves activation and repression of multiple pathways. We found that expression levels of a subset of these IR-correlated transcripts in adipose tissue are regulated by DNA sequence variants. The identification of eQTL for IR-correlated transcripts in adipose tissue suggests that the expression of these transcripts is at least partially genetically driven and causal and not just the result of altered insulin sensitivity. Comparing two cohorts, one of African Americans and one of European ancestry, provides independent validation and generalizability of our findings. The causal role of the ECHDC3 gene in IR, a candidate gene identified through variation-based statistical genetic analyses, was validated through genetic silencing and functional studies in an adipocyte model.
In this study, we compared gene expression profiles from subcutaneous adipose tissues in an African American and a European ancestry cohort, primarily to identify trans-ancestral genetically regulated mechanisms of IR. We identified 3,755 genes with transcript levels significantly correlated with the Matsuda index and in the same direction of effect in both cohorts. These data support the concept of common molecular mechanisms of IR in both European and African ancestry. We further focused on these replicated insulin sensitivity–correlated genes and their regulation mediated by QTLs to define common causal mechanisms of IR. However, the genetic regulatory architecture of a subset of IR-correlated genes differs between ethnic groups, and comparison of ethnicities may be instrumental in identifying putatively ancestry-specific biological mechanisms of IR.
We identified 654 transcripts significantly correlated with the Matsuda index in our African American cohort (AAGMEx, FDR P ≤ 0.01), but probes representing corresponding transcripts in the European ancestry cohort (METSIM) showed no significant correlation (P > 0.05); whether these transcript–Matsuda index correlations represent true African-derived mechanisms or false statistical correlations (i.e., type 1 errors) remains to be determined. As examples, expression of SFFV proviral integration oncogene-1 (SPI1) in adipose tissues of AAGMEx participants was significantly inversely correlated with Matsuda index (ILMN_1696463, NM_001080547, β = −1.19, P = 2.97 × 10−13; ILMN_2392043, NM_003120, β = −1.13, P = 1.21 × 10−11), but its expression was not significantly correlated in the METSIM cohort (represented by probe 11723165_a_at). The SPI1 gene encodes for PU.1, a key transcription factor involved in monocyte-to-macrophage differentiation (30), and also plays a role in adipocyte inflammatory mechanisms causing IR (31,32). Despite the larger size of the METSIM cohort, this lack of replication may suggest that the effect of SPI1 gene predominates in individuals of African ancestry. However, we also noted an inverse correlation between SPI1 expression in adipose tissue and insulin sensitivity (Spearman semipartial ρ = −0.40, P < 0.0001 for ILMN_1696463) in another small cohort (N = 99) of European American individuals recruited from Arkansas in a previously published study (33). Gene expression data in the METSIM cohort were generated using the Affymetrix U219 array, while Illumina HumanHT-12 v4 Expression BeadChips were used for the AAGMEx cohort. Probes may have quantified different transcript isoforms or may have different efficiencies in the quantification of this gene. Thus, lack of replication, at least for a subset of these genes, may reflect technical rather than true biological difference between ethnicities and may be explained by such differences in probes. Our future studies will use alternative strategies that focus on ancestry-specific or ethnically predominant mechanisms of IR.
This study identified cis-eQTL for 587 Matsuda index–correlated transcripts in both the AAGMEx and METSIM cohorts. Thus, in line with the anticipated polygenic nature of the insulin sensitivity phenotype, genetic regulation of a large subset of genes in adipose tissue may, at least in part, be causally involved in the pathogenesis of IR in African and European ancestry populations. These IR-correlated cis-eGenes are enriched for cellular functions and biological pathways. However, the precise role of many of these genes in IR is not known. To prioritize and begin to understand the role of genes in modulating cellular and molecular mechanisms causing IR, we ranked this large list of transcripts based on their statistical significance of correlation with the Matsuda index and association with genotype. ECHDC3, macrophage migration inhibitory factor (MIF), centromere protein V (CENPV), d-dopachrome tautomerase (DDT), and apolipoprotein B (APOB) were among the top Matsuda index–correlated cis-eGenes. Some of these genes have a known role in adipose tissue biology and IR. For example, macrophage migration inhibitory factor superfamily gene members MIF and DDT bind to the CD74/CD44 receptor complex and have distinct roles in adipogenesis. MIF positively correlates with IR and contributes to adipose tissue inflammation by modulating adipose tissue macrophage functions, while DDT reverses glucose intolerance (34,35).
The ECHDC3 gene emerged as the most significant Matsuda index–correlated cis-eGene. Multitissue expression data from the GTEx study showed high ECHDC3 expression in fatty acid metabolizing tissues, including liver, muscle, and adipose. Strong cis-eQTL associations were detected for ECHDC3 expression in subcutaneous adipose tissue in GTEx V7-P2 (rs10906007, P = 1.40 × 10−10 [https://gtexportal.org/]) and STARNET (Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task) (36) (rs718641, P = 4.78 × 10−10) studies. However, its role in adipose or other tissue is unknown. Based on protein sequence homology, peroxisomal enoyl-CoA hydratase (ECH1) and short-chain acyl-CoA dehydrogenase (ECHS1/SCHE) are the closest human homologs. This suggests that ECHDC3 may act as enoyl-CoA hydratase. The enoyl-CoA hydratase is a key mitochondrial enzyme involved in fatty acid β-oxidation and catalyzes the addition of H2O across the double bond of trans-2-enoyl-CoA, resulting in formation of a β-hydroxyacyl-CoA (37). Biochemical analyses suggest that human ECHDC1, a likely homolog of ECHDC3, lacks enoyl-CoA hydratase activity (38). A recent study suggested that ECHS1, a close homolog of ECHDC3, senses nutrient signals and acts as the converging enzyme for fatty acid and branched-chain amino acid oxidation (39).
Supporting a connection between decreased ECHDC3 expression in adipose tissue and IR, in vitro knockdown of ECHDC3 expression in our study significantly downregulated insulin signaling and insulin-stimulated glucose uptake in human adipocytes. Our unbiased transcriptome-wide analysis suggested that downregulation of ECHDC3 in human adipocytes may cause IR by significantly inactivating fatty acid biosynthesis, including the γ-linolenate biosynthesis, cholesterol biosynthesis, and LXR/RXR activation pathways. Genetic polymorphisms in the ECHDC3 regulatory element (eQTL) determine a significant proportion of its variable expression in adipose tissue, likely by their differential ability to interact (as suggested by SNP2TFBS and RegulomeDb) with transcription factors. A regulatory element in the intron of ECHDC3 (near SNP rs34844369) was observed in both European and African ancestry cohorts, while an additional regulatory element upstream to ECHDC3 (−400 bp to −4 Kb, near SNP rs3814627) was observed only in European ancestry cohorts. These two regions are not in linkage disequilibrium in African ancestry cohorts (D′ = 0.21, r2 = 0.007, 1KGP Phase 3_V5 data from LDlink: https://ldlink.nci.nih.gov/), but are in strong linkage disequilibrium in European ancestry cohorts (D′ = 0.95, r2 = 0.63). Future studies will define the transcriptional regulatory mechanisms mediated by these regulatory elements and their interactions that may have ancestry-specific contributions in determining genetic regulatory networks important for the pathogenesis of IR.
In summary, genetic regulation of a large subset of genes in adipose tissue is causally involved in the pathogenesis of IR in African and European ancestry individuals. Our variation-based genomic analysis with concurrent analyses of glucose-homeostasis phenotypes, adipose tissue transcriptome, and in vitro genetic silencing studies identified several genetic regulatory mechanisms, including the novel role of the ECHDC3 gene in causing IR putatively by modulating fatty acid metabolism and other key pathways in adipocytes.
Supplementary Material
Article Information
Acknowledgments. The authors thank Drs. Jorge Calles-Escandon, Jamel Demons, Samantha Rogers, and Barry Freedman and the dedicated staff of the Clinical Research Unit at Wake Forest School of Medicine (WFSM) for support of the clinical studies and assistance with clinical data management for the AAGMEx cohort. The authors thank Lata Menon, Joyce Byers, Ethel Kouba, and Donna Davis for support in participant recruitment for the AAGMEx cohort. The authors thank the staff in the Genomics Core Laboratory at the Center for Genomics and Personalized Medicine Research for their extensive support in genotyping and gene expression analysis using the Illumina microarray platform for the AAGMEx cohort. The authors thank the METSIM study investigators for publicly sharing their data. The authors acknowledge the WFSM Center for Public Health Genomics for computational resources. The authors also acknowledge the editorial assistance of Karen Klein at the Wake Forest Clinical and Translational Science Institute.
Funding. This work was primarily supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health, research grants R01 DK090111 and R01 DK118243 to S.K.D. This work was also supported by the National Heart, Lung, and Blood Institute (NHLBI) grant R00 HL121172 and NIDDK grant R01 DK118287 to M.C. and NHLBI grant R01 HL119962 to J.S.P.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. N.K.S. performed genomic and cell biological experiments, analyzed data, and reviewed and edited the manuscript. C.-C.C.K. performed cell biological experiments, analyzed data, and reviewed and edited the manuscript. M.C. analyzed METSIM cohort data and reviewed and edited the manuscript. M.W. supplied a critical reagent (SGBS cells) for this study and reviewed and edited the manuscript. M.E.C. performed statistical genetic analysis and reviewed and edited the manuscript. C.D.L. supervised statistical genetic analysis, interpreted data, and reviewed and edited the manuscript. J.S.P. supervised cell biological studies, interpreted data, and reviewed and edited the manuscript. S.K.D. designed the study, analyzed and interpreted data, and wrote the manuscript. C.D.L. and S.K.D. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, 22–26 June 2018.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1229/-/DC1.
References
- 1.Czech MP. Insulin action and resistance in obesity and type 2 diabetes. Nat Med 2017;23:804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 2009;373:2215–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercado MM, McLenithan JC, Silver KD, Shuldiner AR. Genetics of insulin resistance. Curr Diab Rep 2002;2:83–95 [DOI] [PubMed] [Google Scholar]
- 4.Liu CT, Raghavan S, Maruthur N, et al.; AAAG Consortium; CARe Consortium; COGENT-BP Consortium; eMERGE Consortium; MEDIA Consortium; MAGIC Consortium . Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet 2016;99:56–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning AK, Hivert MF, Scott RA, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012;44:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R, Hivert MF, Langenberg C, et al.; GIANT consortium; MAGIC investigators . Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 2010;42:142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowles JW, Xie W, Zhang Z, et al.; RISC (Relationship between Insulin Sensitivity and Cardiovascular Disease) Consortium; EUGENE2 (European Network on Functional Genomics of Type 2 Diabetes) Study; GUARDIAN (Genetics UndeRlying DIAbetes in HispaNics) Consortium; SAPPHIRe (Stanford Asian and Pacific Program for Hypertension and Insulin Resistance) Study . Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J Clin Invest 2015;125:1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer ND, Goodarzi MO, Langefeld CD, et al. Genetic variants associated with quantitative glucose homeostasis traits translate to type 2 diabetes in Mexican Americans: the GUARDIAN (Genetics Underlying Diabetes in Hispanics) Consortium. Diabetes 2015;64:1853–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walford GA, Gustafsson S, Rybin D, et al. Genome-wide association study of the modified Stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 2016;65:3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamazon ER, Segrè AV, van de Bunt M, et al.; GTEx Consortium . Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 2018;50:956–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannon ME, Mohlke KL. Deciphering the emerging complexities of molecular mechanisms at GWAS loci. Am J Hum Genet 2018;103:637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006;444:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badoud F, Perreault M, Zulyniak MA, Mutch DM. Molecular insights into the role of white adipose tissue in metabolically unhealthy normal weight and metabolically healthy obese individuals. FASEB J 2015;29:748–758 [DOI] [PubMed] [Google Scholar]
- 15.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest 2016;126:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes 2011;60:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature 2008;452:423–428 [DOI] [PubMed] [Google Scholar]
- 18.Sharma NK, Sajuthi SP, Chou JW, et al. Tissue-specific and genetic regulation of insulin sensitivity-associated transcripts in African Americans. J Clin Endocrinol Metab 2016;101:1455–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sajuthi SP, Sharma NK, Chou JW, et al. Mapping adipose and muscle tissue expression quantitative trait loci in African Americans to identify genes for type 2 diabetes and obesity. Hum Genet 2016;135:869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Civelek M, Wu Y, Pan C, et al. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am J Hum Genet 2017;100:428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wabitsch M, Brenner RE, Melzner I, et al. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int J Obes Relat Metab Disord 2001;25:8–15 [DOI] [PubMed] [Google Scholar]
- 22.Vaittinen M, Kaminska D, Käkelä P, et al. Downregulation of CPPED1 expression improves glucose metabolism in vitro in adipocytes. Diabetes 2013;62:3747–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGillicuddy FC, Chiquoine EH, Hinkle CC, et al. Interferon gamma attenuates insulin signaling, lipid storage, and differentiation in human adipocytes via activation of the JAK/STAT pathway. J Biol Chem 2009;284:31936–31944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25–R10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarazona S, Furió-Tarí P, Turrà D, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res 2015;43:e140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodbell M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J Biol Chem 1964;239:375–380 [PubMed]
- 29.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22:1462–1470 [DOI] [PubMed] [Google Scholar]
- 30.Gosselin D, Link VM, Romanoski CE, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014;159:1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin L, Pang W, Chen K, et al. Adipocyte expression of PU.1 transcription factor causes insulin resistance through upregulation of inflammatory cytokine gene expression and ROS production. Am J Physiol Endocrinol Metab 2012;302:E1550–E1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefterova MI, Steger DJ, Zhuo D, et al. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol 2010;30:2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das SK, Sharma NK, Hasstedt SJ, et al. An integrative genomics approach identifies activation of thioredoxin/thioredoxin reductase-1-mediated oxidative stress defense pathway and inhibition of angiogenesis in obese nondiabetic human subjects. J Clin Endocrinol Metab 2011;96:E1308–E1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleemann R, Bucala R. Macrophage migration inhibitory factor: critical role in obesity, insulin resistance, and associated comorbidities. Mediators Inflamm 2010;2010:610479 [DOI] [PMC free article] [PubMed]
- 35.Kim BS, Pallua N, Bernhagen J, Bucala R. The macrophage migration inhibitory factor protein superfamily in obesity and wound repair. Exp Mol Med 2015;47:e161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franzén O, Ermel R, Cohain A, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 2016;353:827–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agnihotri G, Liu HW. Enoyl-CoA hydratase. Reaction, mechanism, and inhibition. Bioorg Med Chem 2003;11:9–20 [DOI] [PubMed] [Google Scholar]
- 38.Linster CL, Noël G, Stroobant V, et al. Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading. J Biol Chem 2011;286:42992–43003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YK, Qu YY, Lin Y, et al. Enoyl-CoA hydratase-1 regulates mTOR signaling and apoptosis by sensing nutrients. Nat Commun 2017;8:464–00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.