Abstract
It has been appreciated for many years that there is a strong association between metabolism and immunity in advanced metazoan organisms. Distinct immune signatures and signaling pathways have been found not only in immune but also in metabolic cells. The newly discovered DNA-sensing cGAS-cGAMP-STING pathway mediates type I interferon inflammatory responses in immune cells to defend against viral and bacterial infections. Recent studies show that this pathway is also activated by host DNA aberrantly localized in the cytosol, contributing to increased sterile inflammation, insulin resistance, and the development of nonalcoholic fatty liver disease (NAFLD). Potential interactions of the cGAS-cGAMP-STING pathway with mTORC1 signaling, autophagy, and apoptosis have been reported, suggesting an important role of the cGAS-cGAMP-STING pathway in the networking and coordination of these important biological processes. However, the regulation, mechanism of action, and tissue-specific role of the cGAS-cGAMP-STING signaling pathway in metabolic disorders remain largely elusive. It is also unclear whether targeting this signaling pathway is effective for the prevention and treatment of obesity-induced metabolic diseases. Answers to these questions would provide new insights for developing effective therapeutic interventions for metabolic diseases such as insulin resistance, NAFLD, and type 2 diabetes.
Introduction
For maintenance of normal physiological function, a living organism needs to obtain nutrients from the environment and convert it into energy through its metabolic system. Meanwhile, the organism has to protect itself from attacks of potential pathogenic invaders. Evolutionarily, it is not surprising that the functions of the immune and metabolic systems are closely linked and coordinated. It is well known that an effective immune response is highly energy dependent in order to activate innate immunity and promote adaptive immunity in response to various environmental insults. Under conditions of energy insufficiency such as famine, prolonged and intensive physical activities, and overloaded neuronal and cardiovascular functions, the immune responses in an organism may be sacrificed, leading to increased infections and other immune-related defects (1). On the other hand, overnutrition may lead to overactivated immunity, resulting in inflammation and related metabolic diseases such as insulin resistance and type 2 diabetes (2). To maintain homeostasis, an organism thus codevelops the immune and metabolic systems to adapt to environmental changes. Although metabolic cells and tissue-resident immune cells exert distinct functions, numerous studies have already demonstrated that these cells undergo intensive and dynamic cross talks to coordinate their action in preserving homeostasis. Emerging evidence accumulated over the past several years also reveals the presence of distinct immune signatures and signaling pathways in key metabolic cells and vice versa, further signifying an integral link between immune activity and metabolic function.
Activation of the cGAS-cGAMP-STING Pathway in Immunity
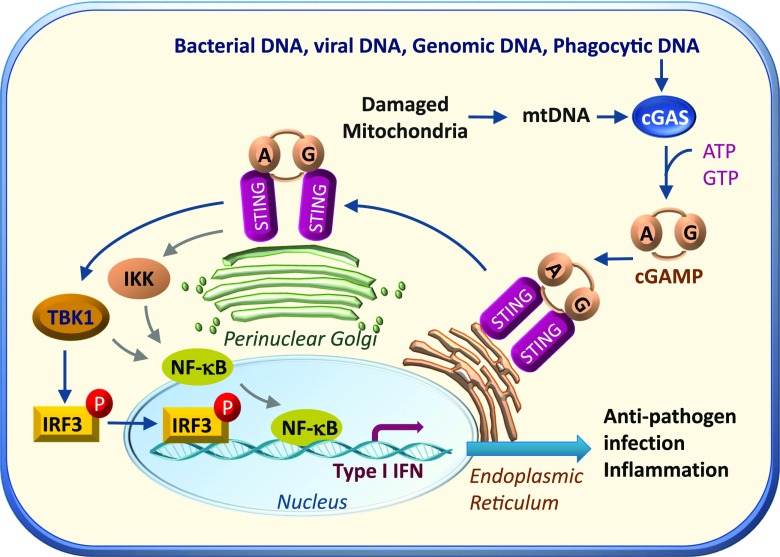
Over the past several years, a key DNA immune response pathway, the cGAMP synthase (cGAS [also known as Mb21d1])–cGAMP–stimulator of interferon genes (STING [also called TMEM173, MITA, MPYS, and ERIS]) pathway, has been discovered in immune cells. The cGAS-cGAMP-STING pathway was originally identified as a signaling cascade that is activated by double-stranded DNA (dsDNA) during pathogen infections. cGAS senses viral and bacterial dsDNA aberrantly localized in the cytosol independent of its sequence context (3–5), and binding dsDNA promotes cGAS oligomerization and activation (3,6,7). In addition to playing a critical role in antiviral immune response, cGAS has also been shown to be involved in some other important biological processes such as macular degeneration (8), cellular senescence (9–11), myocardial infarction–related inflammation (12), and macrophage transformation (13). Activated cGAS catalyzes the formation of 2′3′-cGAMP, a cyclic dinucleotide (CDN) composed of adenosine and guanosine linked via two phosphodiester linkages. Apart from cGAMP, STING is also activated by CDNs such as cyclic di-AMP or cyclic di-GMP from bacteria (14).CDNs and 2′3′-cGAMP bind to the endoplasmic reticulum (ER)-localized STING, which promotes STING dimerization and translocation from the ER to perinuclear punctuate structures (14,15). During the trafficking process, STING recruits and activates TANK binding kinase 1 (TBK1), stimulating phosphorylation and nuclear translocation of the transcription factor interferon regulatory factor 3 (IRF3), and to a lesser extent nuclear factor-κB (NF-κB), which can also be activated by IκB kinase (IKK) (16,17), leading to the production of type 1 interferons (IFNs) and many other inflammatory cytokines (18) (Fig. 1). By binding to the IFN-α/β receptor on the cell membrane of target cells, IFNs promote the expression of proteins involved in inhibiting viral replication and thus enhances the protective defenses of the immune system (7,19). In addition to initiation of IFN signaling in cells in which it is produced by cGAS, there is some evidence showing that 2′3′-cGAMP is able to promote downstream signaling in neighboring cells via distinct mechanisms such as gap junction–, membrane fusion–, or viral particle–mediated transfer (20–22), thus mediating the cross talk between immune cells and their targeting cells. Intriguingly, activation of the cGAS-cGAMP-STING pathway could also be detected in nonimmune cells such as mouse embryonic fibroblasts and adipocytes (23,24), suggesting that activation of this pathway may have broader roles in addition to immune defense functions.
Figure 1.
Activation and regulation of the cGAS-cGAMP-STING pathway in cells. The cGAS is activated by viral and bacterial DNA as well as mtDNA and phagocytosed DNA aberrantly localized in the cytosol. Activated cGAS uses ATP and GTP as substrates to catalyze the formation of the second messenger, cGAMP, which binds to STING localized on the ER membrane. The binding of cGAMP to STING promotes STING translocation to the Golgi apparatus. During the translocation, STING recruits and activates TBK1, which in turn catalyzes the phosphorylation and nuclear translocation of IRF3, and to a lesser extent NF-κB, which can also be activated by IKK, leading to increased synthesis of IFN and other inflammatory genes.
Activation of the cGAS-cGAMP-STING Pathway by Self-DNA
In a healthy cell, host DNA normally resides in the nucleus or mitochondria. However, under certain pathophysiologic conditions such as DNA instability and/or mitochondrial stress, genomic DNA and/or mtDNA may be released into the cytoplasm, where it serves as a danger-associated molecular pattern to trigger immune responses. West et al. (23) found that, via heterozygosity of the mitochondrial transcription factor A (TFAM), disruption of mtDNA stability promoted mtDNA release into the cytosol, where it activated the cGAS-cGAMP-STING pathway and increased IFN gene expression. mtDNA-mediated activation of the cGAS-cGAMP-STING pathway is also observed by Bax/Bak-induced permeabilization of mitochondrial outer membrane (25,26). Besides mtDNA, recent studies show that the cGAS-cGAMP-STING pathway could also be activated by genomic DNA such as ruptured micronuclei and double-strand broken DNA of the primary nucleus caused by genomic instability and/or DNA damage (19,27–29). In addition to genomic DNA and mtDNA, phagocytic DNA inadequately digested in the lysosomes has been shown to activate the cGAS-cGAMP-STING pathway (7,19,30,31) (Fig. 1). These results demonstrate that self-DNA is an important source of sterile inflammatory response induction that has been widely studied in many of the autoimmune disease and cancers. These findings raise an interesting question as to whether self-DNA–induced sterile inflammation is associated with metabolic disorders.
Activation of the cGAS-cGAMP-STING Pathway in Obesity-Induced Inflammation and Metabolic Diseases
Obesity is associated with various metabolic diseases such as type 2 diabetes, nonalcoholic fatty liver disease (NAFLD), cardiovascular disease, and many types of cancer. Numerous studies have shown that chronic sterile inflammation in adipose tissue plays a key role in mediating obesity-induced insulin resistance and its associated metabolic diseases (32,33). However, the precise mechanisms by which obesity causes inflammation remain to be fully elucidated.
During the past several years, evidence has accumulated to suggest an important role of the cGAS-cGAMP-STING pathway in regulating inflammation and energy homeostasis. The expression levels and/or activities of components in this signaling cascade, including cGAS, STING, and TBK1, are significantly upregulated under obesity conditions in mice (24,34,35). Activation of the cGAS-cGAMP-STING pathway in mouse adipose tissue could be triggered by high-fat diet (HFD)–induced mtDNA release, leading to an increase in chronic sterile inflammatory response (24). HFD-induced obesity and activation of the cGAS-cGAMP-STING pathway are prevented by adipose tissue–specific overexpression of disulfide bond A oxidoreductase-like protein (DsbA-L), a chaperone-like and mitochondrial localized protein whose expression in adipose tissue is greatly suppressed by obesity (36). Alternatively, knockout of DsbA-L in adipose tissue impaired mitochondrial function, increased mtDNA release, and activated the cGAS-cGAMP-STING pathway, leading to increased inflammation and exacerbated obesity-induced insulin resistance (24) (Fig. 2). These findings reveal that activation of the cGAS-cGAMP-STING pathway may mediate obesity-induced inflammation and metabolic dysfunction, beyond its well-characterized roles in innate immune surveillance. Consistent with this view, global knockout of the cGAS-cGAMP-STING downstream target TBK1 (37) or IRF3 (38), or pharmacological inhibition of IκB kinase ε (IKKε) and TBK1 by amlexanox, reduced body weight, enhanced insulin sensitivity, and improved glucose tolerance in obese mice and in a subset of patients with type 2 diabetes (34,39). However, it should be noted that while fat-specific knockout of TBK1 increased energy expenditure and attenuated HFD-induced obesity, it also exaggerated adipose tissue inflammation and insulin resistance, suggesting that TBK1 may have a feedback role in regulating obesity-induced inflammation (35) (Fig. 2). Indeed, activation of TBK1 has been found to reduce NF-κB activity and inflammation by promoting phosphorylation-dependent degradation of NF-κB–inducing kinase (NIK), an upstream kinase of IKKs (35). TBK1 has also been shown to attenuate cGAS-cGAMP-STING–mediated response by promoting STING ubiquitination and degradation (40) (Fig. 2). These findings explain the bidirectional roles of TBK1 in regulating inflammation (35). Nevertheless, it remains to be determined what role cGAS-cGAMP-STING signaling, which activates TBK1, may play in inflammation, insulin resistance, and energy expenditure in metabolic cells.
Figure 2.
Activation of the cGAS-cGAMP-STING pathway mediates obesity-induced inflammation and metabolic disorders. Obesity reduces the expression levels of disulfide bond A oxidoreductase-like protein (DsbA-L) in adipose tissue, leading to mitochondrial stress and subsequent mtDNA release into the cytosol. Aberrant localization of mtDNA in the cytosol activates the cGAS-cGAMP-STING pathway, leading to enhanced inflammatory gene expression and insulin resistance. Phosphorylated and activated TBK1 exerts a feedback inhibitory role by promoting STING ubiquitination and degradation or stimulating phosphorylation-dependent degradation of NF-κB–inducing kinase (NIK), thus attenuating cGAS-cGAMP-STING–mediated inflammatory response.
The Potential Role of the cGAS-cGAMP-STING Pathway in NAFLD
In addition to mediating obesity-induced insulin resistance in adipose tissue, activation of the cGAS-cGAMP-STING pathway has also been implicated in other metabolic diseases including NAFLD. NAFLD is characterized by hepatic steatosis, which contributes to the development of nonalcoholic steatohepatitis (NASH), a potentially progressive liver disease that may lead to cirrhosis and hepatocellular carcinoma. There is some evidence suggesting that the innate immune response contributes to NAFLD and NASH (41–44). However, the underlying mechanisms by which the innate immune response promotes NALFD remain elusive. Luo et al. (42) and Yu et al. (45) recently independently found that activation of the liver cGAS-cGAMP-STING signaling pathway may mediate overnutrition-induced NAFLD and/or NASH. Indeed, STING levels were higher in liver tissues from NAFLD human patients compared with those without NAFLD. In addition, the mRNA levels of cGAS and STING are elevated in NASH mouse livers (42,46). Furthermore, the phosphorylation states of TBK1 and IRF3, two downstream targets of the cGAS-cGAMP-STING pathway, were significantly higher in livers of mice fed an HFD, which is coupled with NAFLD (42). Consistent with these findings, cGAS-cGAMP-STING–dependent activation of TBK1 in hepatocytes promotes the formation of insoluble p62/sequestosome 1 (SQSTM1) aggregates, a critical marker of NASH (47). On the other hand, STING deficiency attenuates steatosis, fibrosis, and inflammation in livers of mice fed with either methionine- and choline-deficient diet or HFD (42,45). Interestingly, both systemic or myeloid cell–specific knockout of STING increased resistance to HFD-induced or methionine- and choline-deficient diet–induced hepatic steatosis, inflammation, and/or fibrosis in mice (42,45). In addition, transplantation of bone marrow cells from control mice to STING knockout mice restored HFD-induced severity of steatosis and inflammation (42), suggesting that the improved metabolic phenotypes in the STING knockout mice were due to STING deficiency in liver-resident macrophages rather than in hepatocytes. These results are consistent with the finding that STING is not present in human and murine hepatocytes but is expressed at high abundance in hepatic nonparenchymal cells (48). Indeed, there is some evidence showing that hepatocytes do not express STING (45,49) and that by facilitating hypoxia-induced autophagy in hepatocytes, cGAS protects the liver from ischemia-reperfusion injury via a STING-independent mechanism. The lack of STING in human hepatocytes also explains why hepatitis virus has adapted to specifically replicate in hepatocytes (48). It is well known that selective pressures in evolution promote the development of an effective immune surveillance system to ensure survival in the face of pathogen invasion. While at early stages infection initiates various biochemical processes such as glucose release from stored glycogen, glycogenolysis, and gluconeogenesis, the glucose synthesis ability of the infected body may be greatly impaired at later stages of overwhelming infection, leading to hypoglycemia (50). Because hypoglycemia is detrimental to an organism, in the frame of evolution there is no host survival advantage for chronic pathogen infection. Therefore, a successful immune response is often short-lived, resulting in the termination of the pathogen-induced response quickly to ensure an organism’s survival. However, the lack of STING in hepatocytes results in type 1 IFN deficiency in response to hepatitis virus infection, which facilitates hepatitis viruses to escape from immune detection and causes not only acute but also chronic inflammation in the liver, leading to consequent hepatitis, cirrhosis, and hepatocellular carcinoma (48), which is often accompanied with metabolic dysfunction such as insulin resistance (51,52). Activation of the cGAS-STING pathway by overexpression of STING specifically in hepatocytes significantly suppressed the replication of hepatitis virus in vivo (48,53). Nevertheless, there are some reports showing that STING is present in hepatocytes and that knocking down either STING or IRF3 in hepatocytes alleviated lipid accumulation, hepatic inflammation, and apoptosis (54–56). These findings raise a possibility that some of the NAFLD phenotypes observed in the whole-body STING knockout mice may result from STING deficiency in the hepatocytes of the mice. Further studies are needed to clarify this discrepancy.
The Cross Talk Between the cGAS-cGAMP-STING and the mTORC1 Signaling Pathways
The mechanistic target of rapamycin complex 1 (mTORC1) is a nutrient sensor that integrates energy, hormonal, metabolic, and nutritional inputs to regulate cellular metabolism, growth, and survival. Activation of the mTORC1 signaling pathway promotes anabolic processes such as protein, nucleotide, fatty acid, and lipid biosynthesis while inhibiting catabolic processes such as lipolysis and autophagy (57). Hasan et al. (58) recently found that chronic activation of the cGAS-cGAMP-STING signaling pathway is associated with reduced mTORC1 signaling in metabolically relevant tissues such as liver, fat, and skeletal muscle of the three-prime repair exonuclease 1 knockout (Trex1−/−) mice, concurrently with increased inflammation and altered metabolic phenotypes such as reduced adiposity and increased energy expenditure. Interestingly, STING deficiency in the Trex1−/− mice rescued both inflammatory and metabolic phenotypes, but IRF3 deficiency only rescued inflammation, suggesting that a component downstream of STING but upstream of IRF3 in the cGAS-cGAMP-STING pathway may play a role in regulating metabolism. Consistent with this view, they found that TBK1, the downstream target of STING, directly inhibited mTORC1 signaling by interacting with the mTORC1 complex (58) (Fig. 3). This result is in agreement with the findings of others that TBK1 inhibits mTORC1 activity in prostate cancer cells (59,60) and in an experimental autoimmune encephalomyelitis model (60). Nevertheless, one study reported that TBK1 may activate, rather than inhibit, mTORC1 through site-specific phosphorylation of mTOR at Ser2159 in response to epidermal growth factor but not insulin treatment (61). On the other hand, there is some evidence showing that mTOR may inhibit the cGAS-cGAMP-STING antiviral pathway. Meade et al. (62) recently identified a cytoplasmically replicating poxviruses–encoded protein, F17, that binds and sequesters Raptor and Rictor. The binding of F17 to Raptor promotes mTORC1-mediated suppression of STING activity and IRF3 translocation to the nucleus. The binding of F17 to Rictor, on the other hand, facilitates mTORC2-mediated cGAS degradation. By disrupting the mTORC1-mTORC2 cross talk, F17 inhibits cGAS-cGAMP-STING signaling and thus retains the benefits of mTOR-mediated stimulation of viral protein synthesis. Likewise, the mTOR downstream effector ribosomal protein S6 kinase 1 (S6K1) has been found to interact with STING in a cGAS-cGAMP–dependent manner, and the interaction promotes TBK1-mediated phosphorylation of STING and recruitment of IRF3 for antiviral immune responses (63). Of note, the interaction of S6K1 with STING is mediated by the kinase domain but not the kinase function of S6K1, suggesting a mTORC1-independent regulation of S6K1 on STING signaling (Fig. 3). Taken together, all these findings indicate a complex cross talk between the cGAS-cGAMP-STING and the mTORC1 signaling pathways. Further investigations will be needed to clarify these controversies and to elucidate the molecular details as well as the metabolic consequences of the cross talk between the cGAS-cGAMP-STING and the mTORC1 signaling pathways.
Figure 3.
The interplay between the cGAS-cGAMP-STING pathway with mTORC1 signaling. Knockout of three-prime repair exonuclease 1 (Trex1), which degrades DNA in the cytosol, leads to the activation of the cGAS-cGAMP-STING pathway and suppression of the mTORC1 activity in mice. The cGAS-cGAMP-STING pathway may inhibit mTORC1 activity through a TBK1-dependent mechanism. Conversely, the cGAS-cGAMP-STING pathway may be inhibited by mTORC1-dependent suppression of STING activity and IRF3 translocation or by mTORC2-mediated cGAS degradation. Of note, the kinase domain but not the kinase function of ribosomal protein S6 kinase 1 (S6K1) is essential for S6K to interact with STING, which facilitates TBK1-mediated phosphorylation of STING and recruitment of IRF3 for antiviral immune responses.
The cGAS-cGAMP-STING Pathway and Autophagy/Mitophagy
In addition to cross talking to mTORC1, the cGAS-cGAMP-STING pathway has also been found to interact with the autophagy machinery in innate immune responses (Fig. 4). Autophagy exerts its quality control function by sequestering damaged organelles and protein aggregates and invading intracellular pathogens in the cytoplasm for lysosomal-mediated degradation (64). This programmed survival pathway also acts as a recycling system to maintain essential protein biosynthesis during stress conditions, such as nutrient insufficiency, growth factor depletion, and pathogen invasion (65). Thus, autophagy is important for the maintenance of the metabolic homeostasis of a cell, and its dysregulation might contribute to the development of metabolic disorders (66–68).
Figure 4.
The cross talk between the cGAS-cGAMP-STING pathway and autophagy/mitophagy. Autophagy is initiated with the activation of ULK1 complex. ULK1-induced activation of beclin-1 complex favors the nucleation of autophagosome precursors and promotes autophagy. Mitophagy is a selective form of autophagy. Activation of the cGAS-cGAMP-STING pathway may stimulate autophagy via a cGAS/beclin-1 interaction–dependent mechanism. The cGAS-cGAMP-STING pathway also promotes autophagy/mitophagy by TBK1-dependent phosphorylation and activation of receptors OPTN and p62 (SQSTM). Alternatively, activation of the cGAS-cGAMP-STING pathway promotes an autophagy-dependent negative feedback regulation by 1) the interaction of cGAS with beclin-1, which in turn inhibits cGAS activity; 2) cGAMP-induced activation of ULK1, which promotes STING degradation by phosphorylation at Ser366; and 3) TBK1-p62/SQSTM1–dependent ubiquitination and degradation of STING. PINK/Parkin-induced mitophagy also restrains cGAS-cGAMP-STING signaling and innate immunity by mitophagy-mediated mtDNA clearance.
Autophagy is initiated with the activation of the ULK1 complex, which is inhibited by mTORC1 and promoted by nutrient deprivation and AMPK activation (67–69) (Fig. 4). Autophagy is also regulated by a multiprotein complex comprising beclin-1, a mammalian ortholog of the yeast autophagy-related gene 6 (Atg6), vacuolar protein sorting 34 (VPS34), and autophagy/beclin-1 regulator 1 (AMBRA1), which favors the nucleation of autophagosome precursors (67,70). Activation of the cGAS-cGAMP-STING pathway has been found to prompt ubiquitin-mediated autophagy that delivers bacteria to autophagosomes for degradation (71). Similarly, cGAS protects hepatocytes from ischemia-reperfusion injury–induced apoptosis in vivo and in vitro through an induction of autophagy in mouse hepatocytes (49). How cGAS stimulates autophagy in hepatocytes is currently unknown but appears to be mediated by a STING-independent mechanism (49). Interestingly, cGAS has been found to competitively bind beclin-1 to dissociate the negative autophagy factor rubicon from the beclin-1–phosphatidylinositol 3-kinase class III (PI3KC3) autophagy complex, leading to PI3KC3 activation and subsequent autophagy induction (72,73). This finding provides a possible explanation for the finding that cGAS promotes autophagy via a STING-independent mechanism. It is interesting to note that in addition to stimulating autophagy, the interaction between cGAS and beclin-1 negatively regulates cGAS enzyme activity in immune cells such as RAW264.7 and L929 cells, thus promoting cytosolic DNA degradation and preventing overactivation of the cGAS-cGAMP-STING pathway–mediated IFN responses and persistent immune stimulation (72). Activation of the cGAS-cGAMP-STING pathway also promotes an autophagy-dependent negative feedback regulation of STING, which is mediated by cGAMP-induced dephosphorylation of AMPK and activation of ULK1 that phosphorylates and promotes autophagosome-dependent degradation of STING (74) or by TBK1-p62/SQSTM1–dependent ubiquitination and degradation of STING (40), providing a mechanism to prevent the persistent transcription of innate immune genes (Fig. 4). However, a very recent study showed that upon binding cGAMP, STING translocated from the endoplasmic reticulum to the endoplasmic reticulum-Golgi intermediate compartment, which served as a membrane source for LC3 lipidation, leading to autophagosome formation and autophagy (75). This study reveals that the STING-induced activation of autophagy is mediated by a mechanism that is dependent on the Trp-Asp (W-D) repeat domain phosphoinositide-interacting protein (WIPI2) and autophagy-related gene 5 (ATG5), but independent of TBK1, ULK, or VPS34-beclin kinase complexes. Interestingly, p62/SQSTM1 has been shown to interact with mTOR and Raptor, which is critical for mTOR recruitment to lysosomes and for amino acid signaling–induced activation of S6K1 and 4EBP1 (76). However, it remains unknown whether the cGAS-cGAMP-STING signaling–induced and TBK1-mediated phosphorylation of p62/SQSTM1 plays a role in regulating mTORC1 signaling and function.
Mitophagy is a selective form of autophagy that mitigates inflammation by removing damaged mitochondria from cells (77). Mitochondrial damage induces mitophagy by promoting the accumulation of the ubiquitin kinase PINK1 on the outer membrane of the damaged mitochondria, which in turn phosphorylates Parkin at Ser65, leading to the activation of this E3 ubiquitin ligase and subsequent degradation of ubiquitinated substrates (78). A recent study showed that Parkin and PINK deficiency promoted mtDNA release and activation of the cGAS-cGAMP-STING pathway in mouse heart tissue, leading to a strong inflammatory phenotype (77). These results support a role of PINK/Parkin-mediated mitophagy in restraining cGAS-cGAMP-STING signaling and innate immunity. By quantitative proteomics analysis, Richter et al. (79) recently found that the cGAS-cGAMP-STING downstream target TBK1 phosphorylated several mitophagy receptors such as optineurin (OPTN) and p62 (SQSTM) at their autophagy-relevant sites, which creates a signal loop amplifying mitophagy (Fig. 4). However, while these findings reveal a link between the cGAS-cGAMP-STING pathway and autophagy/mitophagy, the detailed biochemical mechanism underlying the link remains largely elusive, especially in metabolic tissues. Seeking answers to these questions would be an attractive subject for further investigation.
The cGAS-cGAMP-STING Signaling and Apoptosis
Apoptosis is a programmed cell death process that provides a mechanism to maintain organismal homeostasis (80). Apoptosis is regulated by prodeath proteins such as Bak and Bax and prosurvival proteins such as BCL-2 and BCL-XL. Bak/Bax activation promotes mitochondrial outer-membrane permeabilization and the release of apoptotic proteins such as cytochrome c to the cytosol, leading to further activation of the downstream pathway of intrinsic apoptosis through initiator and executioner caspases cascade (81) (Fig. 5). Activation of caspases, which is a hallmark of apoptosis, has been found to not only promote cell death but also prevent dying cells from triggering a host immune response (82). However, while apoptosis has long been known as an immunologically silent form of cell death, the molecular basis underlying the suppression of immune responses remains unknown. Several recent studies suggest that caspase-mediated inhibition of the cGAS-cGAMP-STING signaling pathway may contribute to the silencing of immune process in apoptotic cells. In agreement with this, TBK1 phosphorylation and IFNα-stimulated gene expression are increased in caspase-9 knockout or caspase-3/-7 double knockout mice and cells (25). Constitutive activation of the type I IFN response was also observed in caspase-9–deficient mouse embryonic fibroblasts (26). In addition, inhibition of caspases led to increased phosphorylation of TBK1 and IRF3 in Bax/Bak-sufficient cells but not in Bax/Bak knockout cells. Furthermore, the caspase deficiency–induced IFNβ response was prevented by knocking out cGAS or STING (25). These findings suggest that apoptosis may suppress immune responses by inhibiting the cGAS-cGAMP-STING pathway. However, the precise biochemical mechanism(s) by which caspases negatively regulate the cGAS-cGAMP-STING pathway remains unclear but could result from multiple redundant processes such as attenuated gene expression, cleavage and inactivation of a component or components of the type I IFN production pathway, and caspase-mediated degradation of mtDNA, thereby disrupting its interaction with cGAS, thus preventing the activation of the cGAS-cGAMP-STING signaling pathway and its downstream IFN action (26). Consistent with this, Wang et al. (83) found that cGAS could be cleaved by several inflammatory caspases including caspase-1, which can be activated by mtDNA release–induced formation of inflammasome, or by caspase-4, -5, and -11. Alternatively, activation of the cGAS-cGAMP-STING pathway may promote apoptosis via both transcriptional and nontranscriptional mechanisms (84,85). It has been shown that STING activation in T cells induces apoptosis through an IRF3- and p53-mediated transcriptional proapoptotic program (86). A proapoptotic but transcription-independent role of STING is observed in hepatocytes that is mediated by the association of IRF3 with the proapoptotic molecule Bax/Bak, which contributes to alcoholic liver disease (55). The proapoptotic role of STING was also found in other cells such as B cells and endothelial cells (87,88). Collectively, these findings demonstrate a complicated cross talk between cGAS-cGAMP-STING signaling and apoptosis. Given the importance of apoptosis in regulating metabolic homeostasis, it would be of great interest to determine the functional roles of the cross talk between cGAS-cGAMP-STING signaling and apoptosis in metabolic tissues.
Figure 5.
The association of cGAS-cGAMP-STING signaling with apoptosis. The activation of prodeath proteins such as Bax/Bak promotes mitochondrial outer-membrane permeabilization and the release of cytochrome c to the cytosol, leading to activation of intrinsic apoptosis through initiator and executioner caspases cascade. Defects in apoptosis by knocking out caspase-9 or caspase-3/-7 lead to constitutive activation of the cGAS-cGAMP-STING pathway; however, the precise mechanisms by which caspase-induced apoptosis inhibits innate immune remain unknown. Inflammatory caspases including caspase-1, which can be activated by mtDNA release–induced formation of inflammasome, or caspase-4, -5, and -11 cleave cGAS, thus preventing the activation of immune response. Alternatively, activation of the cGAS-cGAMP-STING pathway promotes apoptosis via both transcriptional and nontranscriptional programs in a TBK1-IRF3–dependent manner.
Concluding Remarks
Inflammation is now clearly recognized as a major risk factor for obesity-induced metabolic disorders. Suppressing inflammatory pathways, therefore, holds promise for developing effective therapeutic treatment of obesity-related diseases. However, identification of pharmacological targets to suppress inflammation is usually a challenge, requiring better understanding of the mechanisms underlying obesity-induced inflammation. The identification of the cGAS-cGAMP-STING pathway as a key player in mediating obesity-induced chronic low-grade inflammation has pointed out an exciting new direction to elucidate the mechanism underlying obesity-induced metabolic diseases and to develop potential therapeutic strategies to improve metabolic homeostasis. However, a number of important questions remain to be answered.
First, while the pivotal roles of the cGAS-cGAMP-STING pathway in immune defense against various microbial pathogens have been extensively studied (3–5), its function in nonimmune cells remains largely unexplored. The findings that the cGAS-cGAMP-STING pathway components such as cGAS, STING, and TBK1 are highly expressed in adipocytes and that the expression levels of these molecules are stimulated under obesity conditions (24) raise a possibility that activation of this pathway may play an important role in metabolic diseases. However, it should be acknowledged that HFD feeding activates the cGAS-cGAMP-STING pathway not only in adipocytes but also in macrophages (24,42), the predominant proinflammatory immune cell type in obese adipose tissue (89), suggesting that activation of the cGAS-cGAMP-STING pathway in adipose tissue–resident macrophages may make a significant contribution to the deteriorated metabolic phenotypes of the obese mice. Further studies will be warranted to dissect the relative contribution of the cGAS-cGAMP-STING pathway in metabolic relevant cells, such as such hepatocytes and adipocytes, and tissue-resident immune cells and the potential cross talk between these cells triggered by cGAS-cGAMP-STING pathway activation.
It is interesting to note that the function of cGAS and STING is regulated by mTOR complexes and vice versa (63,74) (Fig. 3), suggesting that activation of the cGAS-cGAMP-STING pathway may be modulated by environmental inputs such as cellular nutrient status and/or growth factors’ stimulation. However, the underlying mechanism by which DNA-induced activation of the cGAS-cGAMP-STING pathway is moderated by environmental changes remains unexplored. In addition, given that mTORC1 plays a pivotal role in regulating cell metabolism and energy homeostasis, it is possible that activation of the cGAS-cGAMP-STING pathway may mediate some of the multifaceted roles of mTORC1 signaling pathway. Interestingly, activation of TBK1 has been shown to inhibit mTORC1 activity (58), suggesting a potential negative regulation of mTORC1 signaling by activation of the cGAS-cGAMP-STING pathway. Further studies will be needed to elucidate the precise mechanism underlying the cross talk between these two signaling pathways and the physiological roles of the interaction. In addition to the mTORC1 signaling pathway, available evidence suggests that the cGAS-cGAMP-STING pathway also cross talks to both autophagy and apoptosis (Figs. 4 and 5). Autophagy and apoptosis regulate the turnover of cellular organelles and cells within organisms, and their interaction is highly context dependent and in most of cases mutually inhibitory (90). The identification of the cGAS-cGAMP-STING pathway linking to both autophagy and apoptosis suggests an important new layer of regulation of these distinct mechanisms for the control of cell homeostasis. However, an integrated understanding of the interplay between the cGAS-cGAMP-STING and these cellular pathways remains largely unclear. It is also unknown when and which inputs to the network are dominant and how this depends on the physiological or pathophysiological context. Answers to these questions will likely require both deeper biochemical and physiological studies of the interaction both in vitro and with tissue-specific transgenic and/or knockout mouse models that enable more specific perturbation and monitoring of these pathways in vivo.
Lastly, it remains to be established whether and how targeting the cGAS-cGAMP-STING pathway is a suitable strategy to treat metabolic diseases. Given that the cGAS-cGAMP-STING pathway is activated by mitochondrial stress under obesity conditions, it is tempting to speculate that at least a part of the effects of increased inflammation may be due to activation of the cGAS-cGAMP-STING pathway. In fact, much progress has been made on the development of small-molecule inhibitors targeting components in the cGAS-cGAMP-STING pathway over the past several years. As of now, several cGAS inhibitors have been reported. By in silico screening of drug libraries using mouse cGAS/DNA target (PDB 4LEZ), An et al. (91) recently identified hydroxychloroquine, quinacrine, and 9-amino-6-chloro-2-methoxyacridine as potential cGAS inhibitors. These molecules do not bind to the active site of cGAS but are instead found to localize to the minor groove of DNA between the cGAS/DNA interface (91). RU.521 and RU.365, however, are found to inhibit cGAS by binding at the cGAS active site (92). Wang et al. (93) showed that suramin, a drug used clinically for the treatment of African sleeping sickness (94), binds to the DNA binding site of cGAS and inhibits cGAS activity by disrupting dsDNA/cGAS binding. In addition to targeting cGAS, several small-molecule inhibitors of STING have also been reported. By a cell-based chemical screen, Haag et al. (95) recently identified two nitrofuran derivatives—C-178 and C-176—that strongly reduced STING-mediated, but not RIG-I– or TBK1-mediated, IFNβ reporter activity. Excitingly, they found that these derivatives reduce STING-mediated inflammatory cytokine production in both human and mouse cells and attenuate pathological features of autoinflammatory disease in mice (95). Very recently, Dai et al. (96) reported that aspirin, a nonsteroidal anti-inflammatory drug, can directly bind and enforce the acetylation of cGAS, leading to a robust inhibition of cGAS enzymatic activity and self-DNA–induced immune response in both Aicardi-Goutières syndrome patient cells and a mouse model of Aicardi-Goutières syndrome. These results provide proof of concept that targeting the cGAS-cGAMP-STING pathway may be efficacious in the treatment of inflammatory diseases, which opens new avenues for developing novel therapies for inflammation-related metabolic diseases. Future work on the molecular details of the regulation and network of the cGAS-cGAMP-STING pathway and its tissue-specific function should enable the rational targeting of this signaling to unravel the full therapeutic potential of this extraordinary pathway in metabolism and relevant biological functions.
Article Information
Funding. This work was supported in part by grants from the Foundation for the National Institutes of Health (DK114479 to F.L.), the National Natural Science Foundation of China (81730022 to F.L. and 81870601 to J.B.), and Innovative Basic Science Awards of the American Diabetes Association (1-18-IBS-344 to F.L. and 1-19-IBS-147 to J.B.).
Duality of Interest. No potential conflicts of interest relevant to this article was reported.
Data Availability. No applicable resources were generated or analyzed during the current study.
References
- 1.Bourke CD, Berkley JA, Prendergast AJ. Immune dysfunction as a cause and consequence of malnutrition. Trends Immunol 2016;37:386–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rathmell JC. Metabolism and autophagy in the immune system: immunometabolism comes of age. Immunol Rev 2012;249:5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013;339:786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Shu C, Yi G, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 2013;39:1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Wu J, Du F, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Reports 2014;6:421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013;341:1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016;17:1142–1149 [DOI] [PubMed] [Google Scholar]
- 8.Kerur N, Fukuda S, Banerjee D, et al. cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat Med 2018;24:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou Z, Ghosh K, Vizioli MG, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017;550:402–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glück S, Guey B, Gulen MF, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 2017;19:1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci USA 2017;114:E4612–E4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King KR, Aguirre AD, Ye YX, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med 2017;23:1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao DJ, Schiattarella GG, Villalobos E, et al. Cytosolic DNA sensing promotes macrophage transformation and governs myocardial ischemic injury. Circulation 2018;137:2613–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marinho FV, Benmerzoug S, Oliveira SC, Ryffel B, Quesniaux VFJ. The emerging roles of STING in bacterial infections. Trends Microbiol 2017;25:906–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 2015;18:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu SQ, Cai X, Wu JX, et al. Chen ZJJ: Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015;347:aaa2630 [DOI] [PubMed] [Google Scholar]
- 17.Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol 2014;88:5328–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu HB, Wang YY. Adding to the STING. Immunity 2014;41:871–873 [DOI] [PubMed] [Google Scholar]
- 19.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 2018;215:1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ablasser A, Schmid-Burgk JL, Hemmerling I, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 2013;503:530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q, Boire A, Jin X, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016;533:493–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S, Ducroux A, Ponnurangam A, et al. cGAS-mediated innate immunity spreads intercellularly through HIV-1 Env-induced membrane fusion sites. Cell Host Microbe 2016;20:443–457 [DOI] [PubMed] [Google Scholar]
- 23.West AP, Khoury-Hanold W, Staron M, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015;520:553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai J, Cervantes C, Liu J, et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc Natl Acad Sci U S A 2017;114:12196–12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rongvaux A, Jackson R, Harman CC, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 2014;159:1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White MJ, McArthur K, Metcalf D, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 2014;159:1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Zhang H, Wu X, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018;563:131–136 [DOI] [PubMed] [Google Scholar]
- 29.Mackenzie KJ, Carroll P, Martin CA, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017;548:461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci USA 2012;109:19386–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn J, Xia T, Rabasa Capote A, Betancourt D, Barber GN. Extrinsic phagocyte-dependent STING signaling dictates the immunogenicity of dying cells. Cancer Cell 2018;33:862–873.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest 2011;121:2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saltiel AR. New therapeutic approaches for the treatment of obesity. Sci Transl Med 2016;8:323rv2. [DOI] [PubMed] [Google Scholar]
- 34.Reilly SM, Chiang SH, Decker SJ, et al. An inhibitor of the protein kinases TBK1 and IKK-ɛ improves obesity-related metabolic dysfunctions in mice. Nat Med 2013;19:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao P, Wong KI, Sun X, et al. TBK1 at the crossroads of inflammation and energy homeostasis in adipose tissue. Cell 2018;172:731–743.e12 [DOI] [PMC free article] [PubMed]
- 36.Liu M, Zhou L, Xu A, et al. A disulfide-bond A oxidoreductase-like protein (DsbA-L) regulates adiponectin multimerization. Proc Natl Acad Sci U S A 2008;105:18302–18307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz VH, Arner EN, Wynne KW, Scherer PE, Brekken RA. Loss of Tbk1 kinase activity protects mice from diet-induced metabolic dysfunction. Mol Metab 2018;16:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari M, Wang X, Lantier L, et al. IRF3 promotes adipose inflammation and insulin resistance and represses browning. J Clin Invest 2016;126:2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oral EA, Reilly SM, Gomez AV, et al. Inhibition of IKKɛ and TBK1 improves glucose control in a subset of patients with type 2 diabetes. Cell Metab 2017;26:157–170.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prabakaran T, Bodda C, Krapp C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J 2018;37:e97858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W, Metlakunta A, Dedousis N, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010;59:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo X, Li H, Ma L, et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology 2018;155:1971–1984.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H, Li H, Woo SL, et al. Myeloid cell-specific disruption of Period1 and Period2 exacerbates diet-induced inflammation and insulin resistance. J Biol Chem 2014;289:16374–16388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng ZB, Liu Y, Liu C, et al. Immature myeloid cells induced by a high-fat diet contribute to liver inflammation. Hepatology 2009;50:1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Liu Y, An W, Song J, Zhang Y, Zhao X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J Clin Invest 2019;129:546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong X, Wang Q, Wang S, et al. Mapping the molecular signatures of diet-induced NASH and its regulation by the hepatokine Tsukushi. Mol Metab 2019;20:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho CS, Park HW, Ho A, et al. Lipotoxicity induces hepatic protein inclusions through TANK binding kinase 1-mediated p62/sequestosome 1 phosphorylation. Hepatology 2018;68:1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomsen MK, Nandakumar R, Stadler D, et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 2016;64:746–759 [DOI] [PubMed] [Google Scholar]
- 49.Lei Z, Deng M, Yi Z, et al. cGAS-mediated autophagy protects the liver from ischemia-reperfusion injury independently of STING. Am J Physiol Gastrointest Liver Physiol 2018;314:G655–G667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beisel WR. Metabolic response to infection. Annu Rev Med 1975;26:9–20 [DOI] [PubMed] [Google Scholar]
- 51.Desbois AC, Cacoub P. Diabetes mellitus, insulin resistance and hepatitis C virus infection: a contemporary review. World J Gastroenterol 2017;23:1697–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bose SK, Ray R. Hepatitis C virus infection and insulin resistance. World J Diabetes 2014;5:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo F, Tang L, Shu S, et al. Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother 2017;61:e00771–e00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iracheta-Vellve A, Petrasek J, Gyongyosi B, et al. Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J Biol Chem 2016;291:26794–26805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrasek J, Iracheta-Vellve A, Csak T, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci U S A 2013;110:16544–16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiao JT, Cui C, Qing L, et al. Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism 2018;81:13–24 [DOI] [PubMed] [Google Scholar]
- 57.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell 2017;168:960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasan M, Gonugunta VK, Dobbs N, et al. Chronic innate immune activation of TBK1 suppresses mTORC1 activity and dysregulates cellular metabolism. Proc Natl Acad Sci U S A 2017;114:746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JK, Jung Y, Wang J, et al. TBK1 regulates prostate cancer dormancy through mTOR inhibition. Neoplasia 2013;15:1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu J, Zhou X, Chang M, et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun 2015;6:6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bodur C, Kazyken D, Huang K, et al. The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J 2018;37:19–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meade N, Furey C, Li H, et al. Poxviruses evade cytosolic sensing through disruption of an mTORC1-mTORC2 regulatory circuit. Cell 2018;174:1143–1157.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Alain T, Szretter KJ, et al. S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3. Nat Immunol 2016;17:514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011;469:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res 2007;17:839–849 [DOI] [PubMed] [Google Scholar]
- 66.Rabinowitz JD, White E. Autophagy and metabolism. Science 2010;330:1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell 2014;159:1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim KH, Lee MS. Autophagy--a key player in cellular and body metabolism. Nat Rev Endocrinol 2014;10:322–337 [DOI] [PubMed] [Google Scholar]
- 69.Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem 2017;86:225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011;18:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012;150:803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang Q, Seo GJ, Choi YJ, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 2014;15:228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang Q, Seo GJ, Choi YJ, et al. Autophagy side of MB21D1/cGAS DNA sensor. Autophagy 2014;10:1146–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013;155:688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gui X, Yang H, Li T, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway (Letter). Nature 2019;567:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duran A, Amanchy R, Linares JF, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell 2011;44:134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sliter DA, Martinez J, Hao L, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018;561:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015;85:257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richter B, Sliter DA, Herhaus L, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 2016;113:4039–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013;5:a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suhaili SH, Karimian H, Stellato M, Lee TH, Aguilar MI. Mitochondrial outer membrane permeabilization: a focus on the role of mitochondrial membrane structural organization. Biophys Rev 2017;9:443–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol 2001;41:367–401 [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Ning X, Gao P, et al. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 2017;46:393–404 [DOI] [PubMed] [Google Scholar]
- 84.Diner BA, Lum KK, Toettcher JE, Cristea IM. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during Herpesvirus infection. MBio 2016;7:e01553–e01516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lum KK, Song B, Federspiel JD, Diner BA, Howard T, Cristea IM. Interactome and proteome dynamics uncover immune modulatory associations of the pathogen sensing factor cGAS. Cell Syst 2018;7:627–642.e6 [DOI] [PMC free article] [PubMed]
- 86.Gulen MF, Koch U, Haag SM, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun 2017;8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang CH, Zundell JA, Ranatunga S, et al. Agonist-mediated activation of STING induces apoptosis in malignant B cells. Cancer Res 2016;76:2137–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weiss JM, Guérin MV, Regnier F, et al. The STING agonist DMXAA triggers a cooperation between T lymphocytes and myeloid cells that leads to tumor regression. OncoImmunology 2017;6:e1346765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 2010;72:219–246 [DOI] [PubMed] [Google Scholar]
- 90.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014;15:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An J, Woodward JJ, Sasaki T, Minie M, Elkon KB. Cutting edge: antimalarial drugs inhibit IFN-β production through blockade of cyclic GMP-AMP synthase-DNA interaction. J Immunol 2015;194:4089–4093 [DOI] [PubMed] [Google Scholar]
- 92.Vincent J, Adura C, Gao P, et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat Commun 2017;8:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang M, Sooreshjani MA, Mikek C, Opoku-Temeng C, Sintim HO. Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-β levels. Future Med Chem 2018;10:1301–1317 [DOI] [PubMed] [Google Scholar]
- 94.Hawking F. Suramin: with special reference to onchocerciasis. Adv Pharmacol Chemother 1978;15:289–322 [DOI] [PubMed] [Google Scholar]
- 95.Haag SM, Gulen MF, Reymond L, et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018;559:269–273 [DOI] [PubMed] [Google Scholar]
- 96.Dai J, Huang YJ, He X, et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 2019;176:1447–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]