Abstract
Tissue sampling of biliary tract carcinomas (BTCs) for molecular characterization is challenging. The aim of this study was to investigate the possibility of identifying individual actionable mutations derived from bile cell-free DNA (cfDNA) using targeted deep sequencing. Ten BTC patients, four with gallbladder carcinomas and six with cholangiocarcinomas, were enrolled in the present study. Using targeted deep sequencing with a panel of 150 tumor-related genes, paired bile cfDNA and tumor DNA were analyzed for mutational variants individually and then compared. The present study, to the best of our knowledge, is the first to reveal that bile cfDNA is predominantly comprised of long DNA fragments, which is not the case for plasma cfDNA. Herein, paired bile cfDNA and tumors from ten BTC patients were examined using targeted deep sequencing. When comparing bile cfDNA and tumor DNA for single nucleotide variation (SNV)/insertion and deletion (Indel), the results using targeted deep sequencing revealed high sensitivity (94.7%) and specificity (99.9%). Additionally, the sensitivity of detecting a copy number variation (CNV) was 75.0%, with a specificity of 98.9%. When comparing two bile extraction methods, including percutaneous transhepatic cholangial drainage and operation, no significant difference in SNV/Indel or CNV detection sensitivity was noted. Moreover, when examining the tumor stage and incidence site, AJCC stage II and the distal bile duct both had significantly decreased CNV detection sensitivities. The present study revealed that targeted deep sequencing can reliably detect mutational variants within bile cfDNA obtained from BTC patients. These preliminary results may shed light on bile cfDNA as a promising liquid biopsy for BTC patients.
Keywords: bile cell-free DNA, targeted deep sequencing, biliary tract carcinoma, individual mutation, liquid biopsy
Introduction
Biliary tract carcinomas (BTCs), which include gallbladder carcinomas (GBCs) and cholangiocarcinomas (bile duct cancer), are rapidly growing lethal tumors associated with the digestive system (1). The 5-year survival rate is <10% in patients with advanced or metastatic BTCs (2). Since the current treatments are minimally effective, a multi-omic approach combining genomic, transcriptomic, and metabonomic landscapes would be beneficial and could lead to improved treatments and possibly earlier diagnosis.
At present, the concept of precision or personalized medicine as a treatment strategy for many types of cancer has been widely recognized (3). When utilizing this type of treatment strategy, one or several actionable mutations typically provide drug targets. Thus, appropriate biopsies and technologies are required to identify relevant biomarkers, such as single nucleotide variations (SNVs), insertion and deletions (Indels), gene fusions, copy number variations (CNVs), or aberrant expression (4). Previous studies examining biliary tract tumors have utilized genetic mutational profiling (5). Moreover, in BTCs, gene mutations in ERBB2, PIK3CA, FGFR, and IDH1 have been utilized as drug targets (6–9). However, obtaining BTC tissue samples is challenging due to the anatomical complexity of the biliary tract system. These tumors are typically characterized by a small lesion size, potentially severe biliary hemorrhaging, and a high perforation risk, which limits the use of endoscopic retrograde cholangiopancreatography (ERCP) with intraductal ultrasonography (IDUS)-guided fine needle aspiration. A promising solution is the use of cell-free DNA (cfDNA), DNA released by tumor and normal cells that could potentially be the basis for liquid biopsies. This could provide tumor genomic information in a less invasive way than the current methods (10). Previous studies have revealed that cfDNA can be utilized to identify actionable mutations and predict therapeutic responses in non-small cell lung cancer (NSCLC) (11–14). In the present study, bile cfDNA that was obtained from BTC patients as a minimally invasive ‘liquid biopsy’ method was subsequently examined using targeted deep sequencing to identify somatic variants.
Materials and methods
Clinical specimens and data collection
The present study was approved by the Ethics Committee of the Shanghai Eastern Hepatobiliary Surgery Hospital, Navy Military Medical University (Second Military Medical University) (EHBHKY2018-K-003). Ten BTC patients, four with gallbladder carcinomas and six with cholangiocarcinomas (Table I), were evaluated in the present study. Each of the patients provided a bile sample and paired formalin-fixed paraffin-embedded (FFPE) tumor tissue samples were obtained from the clinical sample bank at the Department of Biliary II, Shanghai Eastern Hepatobiliary Surgery Hospital, Navy Military Medical University. The samples were collected from August 2017 to March 2018. Patient status was confirmed by surgical pathology, and FFPE specimens were reviewed by a pathologist to ensure a cancer cell content ≥20%. All of the patients provided written informed consent for the examination of their samples and the use of their clinical data.
Table I.
Characteristics of ten patients with biliary tract cancer that received surgery.
| Characteristics | N |
|---|---|
| Age, median (range) in years | 68 (44–74) |
| Sex | |
| Male | 4 |
| Female | 6 |
| Cancer type | |
| Gallbladder carcinoma | 4 |
| Cholangiocarcinomas | 6 |
| Incidence site | |
| Gallbladder | 4 |
| Intrahepatic bile duct | 1 |
| Hilar bile duct | 4 |
| Distal bile duct | 1 |
| AJCC stage | |
| I | 0 |
| II | 4 |
| III | 3 |
| IV | 3 |
| Operation type | |
| R0 | 8 |
| R1 | 1 |
| R2 | 1 |
| Gallbladder stones | |
| Yes | 4 |
| No | 6 |
| Cholecystitis | |
| Yes | 4 |
| No | 6 |
| Method of obtaining bile | |
| PTCD | 5 |
| Operation | 5 |
| Location of obtaining bile | |
| Gallbladder | 4 |
| Intrahepatic bile duct | 1 |
| Hilar bile duct | 4 |
| Distal bile duct | 1 |
| AFP, median (range) ng/ml | 3.4 (2.4–8.0) |
| CEA, median (range) ng/ml | 4.5 (1.3–83.9) |
| CA19-9, median (range) U/ml | 154.6 (8.8–1,000.0) |
| Total bilirubin, median (range) µmol/l | 42.2 (8.8–305.6) |
AFP, α-fetoprotein; CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9; PTCD, percutaneous transhepatic cholangial drainage.
DNA extraction from FFPE and bile samples
To extract DNA from FFPE samples, 4-µm thick sections were processed using a QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer's instructions. The DNA concentrations were determined using a Qubit™ 3.0 Fluorometer (Thermo Fisher Scientific, Inc.), and size distributions were analyzed using LabChip GX Touch 24 nucleic acid analyzer (PerkinElmer, Inc.). To obtain bile cfDNA, bile (3 ml) was collected in 4 ml BD Vacutainer blood collection tubes (BD Biosciences) and processed within 1 h after collection. First, the bile samples were centrifuged at 1,600 × g for 15 min at room temperature, and the supernatants were carefully transferred to new 2-ml tubes (Eppendorf). The samples were then centrifuged at 16,000 × g for 10 min at room temperature, and the supernatants were collected and examined under a light microscope to ensure that there were no residual cells or debris. Samples were then aliquoted and stored at −80°C. Bile cfDNA was extracted using a QIAamp Circulating Nucleic Acid kit (Qiagen) according to the manufacturer's instructions. Bile cfDNA concentrations were determined using a Qubit dsDNA HS Assay kit (Life Technologies; Thermo Fisher Scientific, Inc.), and cfDNA size distributions were analyzed by Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
Tumor tissue analysis by targeted deep sequencing and variant calling
Tumor tissues from the 10 BTC patients were analyzed utilizing a previously established targeted deep sequencing method (15). Experimental procedures and variant calling were performed as previously described, with 150 tumor-associated genes examined (Table SI). This 150-gene list of targeted deep sequencing was comprised of highly recurrent and known important regulatory genes of biliary tract carcinoma, such as TP53, KRAS, NRAS, ERBB2, ERBB3, CDKN2A, PIK3CA, NF1, among others. The cut-off of mutant allele frequencies (MAFs) in tumor tissue was ≥0.01.
Bile cell-free DNA analysis by targeted deep sequencing and variant calling
Bile cfDNA samples were evaluated using a previously established targeted deep sequencing platform for cfDNA with a unique identification (UID) indexed capturing-based sequencing (UC-Seq) to establish a mutational profile (11). The same gene panel that was used for the tumor samples was also utilized. Prior to sequencing, the bile cfDNAs were fragmented into sizes ranging from 200 to 400 bp using the Covaris S2 Sonolab (Covaris, Inc.), with the remaining procedures and variant calling performed as previously described (11). The cut-off of mutant allele frequencies (MAFs) in bile cfDNA was ≥0.001.
Statistical analysis
A Fisher's exact test was used to assess the significance and sensitivity differences for identifying mutations within groups, such as the sample collection, cancer stage, and tumor site. A P-value ≤0.05 was considered to indicate a statistically significant difference.
Results
Clinical information of BTC patients
In the present study, 10 BTC patients (four males and six females) with a median age of 68 years (range: 44–74) at the time of diagnosis (Table I) were examined. Four of the patients had gallbladder carcinomas and six had bile duct cancer (one intrahepatic, four hilar, and one distal cholangiocarcinoma). Based on the seventh edition of the American Joint Committee on Cancer (AJCC) staging system for BTC (16), four (40%) patients had stage II, three (30%) patients had stage III, and three (30%) patients had stage IV (Table I). Four patients carried gallbladder stones and cholecystitis, while the other six patients did not. Percutaneous transhepatic cholangial drainage (PTCD) and operation are regarded as two different methods of extracting the bile. The origin of the obtained bile was respectively the gallbladder (four patients), the intrahepatic bile duct (one patient), the hilar bile duct (four patients) and the distal bile duct (one patient). Out of the 10 patients with BTCs, eight received a radical resection (R0), while the other two patients underwent R1 and R2 resections.
Bile cfDNA fragment size distributions
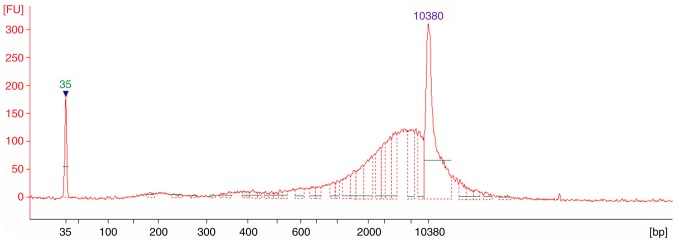
To examine the distributions of bile cfDNA fragments, samples were analyzed using an Agilent 2100 Bioanalyzer. The electropherograms revealed that all 10 of the samples had similar distributions, which sheared to ~6,000 bp (Fig. 1). This result indicated that long DNA fragments are more prevalent among bile cfDNAs, which is not consistent with findings from plasma cfDNA (11).
Figure 1.
cfDNA fragment distributions. Representative electropherogram for a BTC bile cfDNA sample as detected by an Agilent 2100 Bioanalyzer. cfDNA, bile cell-free DNA; BTC, biliary tract cancer.
BTC patient bile cfDNA mutational profiles
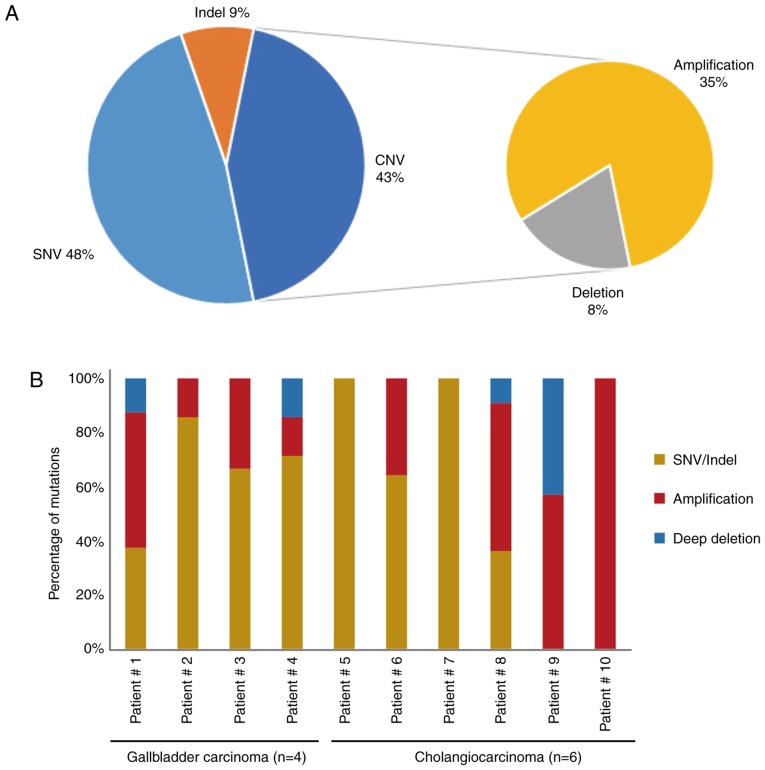
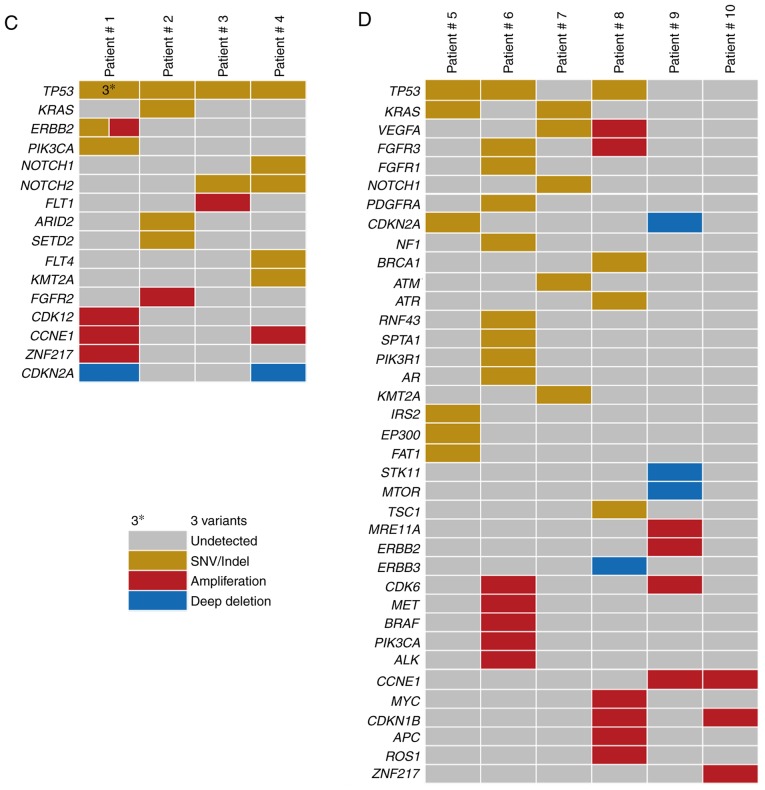
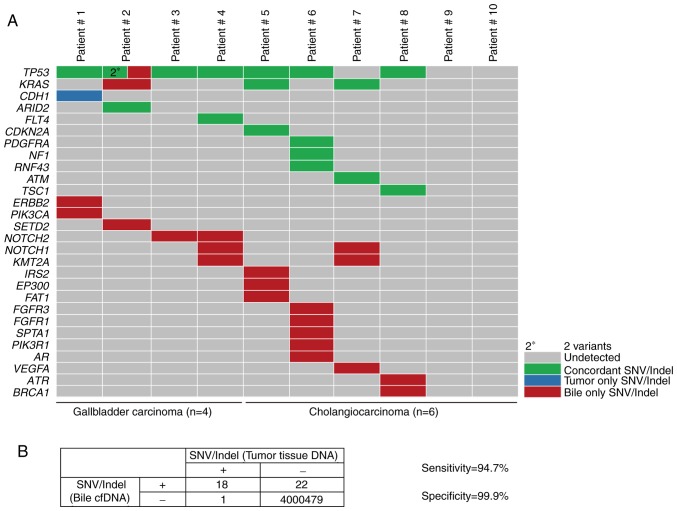
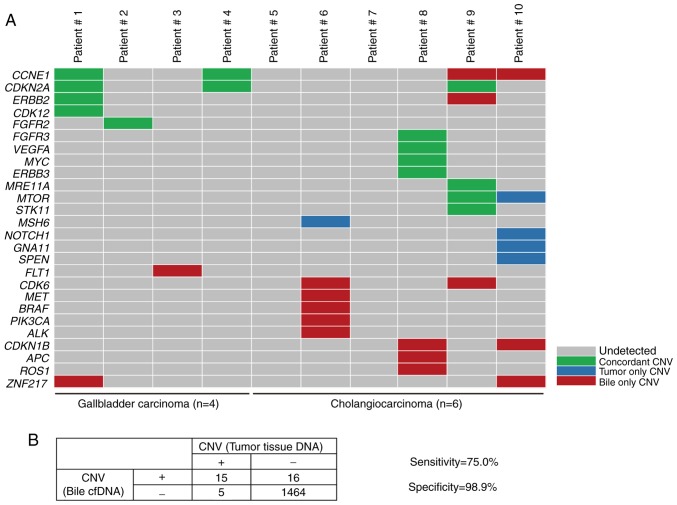
To explore the mutational landscape, the obtained bile cfDNA was sequenced using targeted deep sequencing, which is able to detect SNVs, Indels and CNVs concurrently, and a panel of 150 tumor-related genes was examined. The mean sequencing coverage for the bile cfDNA was over 3,000× with barcodes (Table SII). A total of 71 unique somatic mutations, including SNVs (48%), Indels (9%), CNVs (35%), and deep deletions (8%), were revealed (Fig. 2A and Table II). Further analysis of the gene mutational profiles revealed that in six of the patients, SNVs and Indels accounted for more than half of the mutations. In two of the bile cfDNA samples (Patients #5 and #7), only SNVs and Indels were present; while in one sample (Patient #10), only CNVs were present (Fig. 2B). Furthermore, the mutational profiles revealed that of the SNV/Indel mutations, the highest variation was observed in TP53 (70%), followed by KRAS (30%), NOTCH1 (20%), NOTCH2 (20%), and KMT2A (20%). Among the CNV mutations, low recurrent amplified genes, such as CCNE1, ERBB2, CDKN1B, ZNF217 and CDK6 were identified; with CDKN2A revealing a deep deletion (Fig. 2C). Among these genes, NOTCH1, NOTCH2, ERBB2 and CDK6 are of interest as potential drug targets.
Figure 2.
The mutational landscape of cfDNA from ten patients with BTC. (A) Gene mutational compositions in bile cfDNA from ten BTC patients (48% SNV, 43% CNV, and 9% Indel). (B) Gene mutational profiles illustrated independently. (C and D) Mutated genes harboring somatic variants and somatic copy number alterations. Mutation subtypes (SNV/Indel, CNV, and deep deletion) are each denoted with an individual color. Data presentation: (A) Pie charts presenting gene mutational compositions in bile cfDNA. (B) Histogram displaying the ratio of gene mutational types per person. (C and D) Heat maps presenting mutated genes and the types of bile cfDNA from the ten patients. cfDNA, bile cell-free DNA; BTC, biliary tract cancer; SNV/Indel, single nucleotide variation/insertion and deletion; CNV, copy number variation.
Table II.
Mutations of bile cell-free DNA from ten biliary tract cancer patients.
| Patient ID | SNV/Indel | CNV | |||
|---|---|---|---|---|---|
| Patient #1 | TP53 | p.Q38Pfs*5 | 0.498a | CDK12 | Gain |
| ERBB2 | p.D769Y | 0.004 | ERBB2 | Gain | |
| PIK3CA | c.1665-1G>T | 0.004 | CCNE1 | Gain | |
| CDKN2A | Loss | ||||
| ZNF217 | Gain | ||||
| Patient #2 | TP53 | p.C275F | 0.037 | FGFR2 | Gain |
| TP53 | p.P27Lfs*17 | 0.012 | |||
| ARID2 | p.Q794Cfs*45 | 0.014 | |||
| KRAS | p.A146V | 0.006 | |||
| TP53 | p.E180K | 0.022 | |||
| SETD2 | p.M118I | 0.015 | |||
| Patient #3 | TP53 | p.P151T | 0.022 | FLT1 | Gain |
| NOTCH2 | p.T235S | 0.011 | |||
| Patient #4 | TP53 | p.V157F | 0.062 | CCNE1 | Gain |
| FLT4 | p.A622T | 0.010 | CDKN2A | Loss | |
| NOTCH1 | p.R1824Q | 0.492 | |||
| KMT2A | p.G3131S | 0.467 | |||
| NOTCH2 | p.R29Q | 0.028 | |||
| Patient #5 | TP53 | c.783-2_821del | 0.050 | ||
| CDKN2A | p.V82E | 0.169 | |||
| KRAS | p.I36M | 0.246 | |||
| IRS2 | p.Q1206R | 0.592 | |||
| EP300 | p.P881S | 0.423 | |||
| FAT1 | p.D850E | 0.129 | |||
| Patient #6 | TP53 | p.M237I | 0.760 | CDK6 | Gain |
| PDGFRA | p.L216I | 0.516 | MET | Gain | |
| NF1 | p.E649* | 0.710 | BRAF | Gain | |
| RNF43 | p.L706Cfs*20 | 0.471 | PIK3CA | Gain | |
| FGFR3 | p.K413R | 0.320 | ALK | Gain | |
| FGFR1 | p.G35R | 0.109 | |||
| SPTA1 | p.N2076K | 0.107 | |||
| PIK3R1 | p.N564D | 0.034 | |||
| AR | p.K778M | 0.014 | |||
| Patient #7 | KRAS | p.G12D | 0.016 | ||
| ATM | p.A2626V | 0.012 | |||
| NOTCH1 | p.R592H | 0.486 | |||
| KMT2A | p.P658P | 0.483 | |||
| VEGFA | p.T8Rfs*78 | 0.385 | |||
| Patient #8 | TP53 | p.R248W | 0.533 | FGFR3 | Gain |
| TSC1 | p.R190H | 0.130 | VEGFA | Gain | |
| ATR | p.M211I | 0.441 | MYC | Gain | |
| BRCA1 | p.R762S | 0.197 | ERBB3 | Gain | |
| CDKN1B | Gain | ||||
| APC | Gain | ||||
| ROS1 | Gain | ||||
| Patient #9 | MRE11A | Gain | |||
| MTOR | Loss | ||||
| CDKN2A | Loss | ||||
| STK11 | Loss | ||||
| ERBB2 | Gain | ||||
| CCNE1 | Gain | ||||
| CDK6 | Gain | ||||
| Patient #10 | CDKN1B | Gain | |||
| CCNE1 | Gain | ||||
| ZNF217 | Gain | ||||
Mutation allele frequency in bile cell-free DNA sample. SNV/Indel, single nucleotide variation/insertion and deletion; CNV, copy number variation.
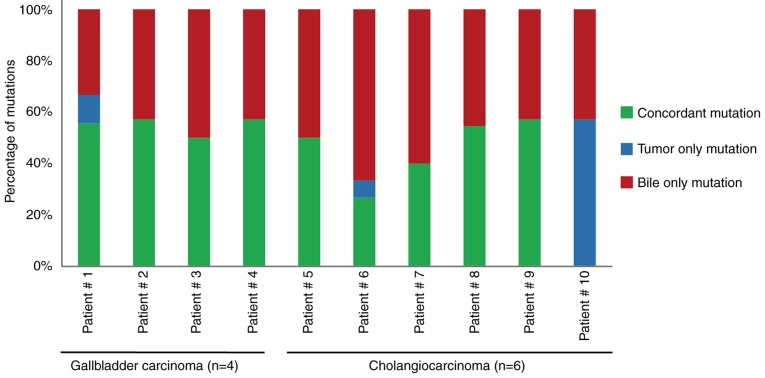
Concordance of mutations between tumor tissue and bile
To assess the performance of the targeted deep sequencing in detecting mutations in the bile cfDNA samples, the findings were compared to the paired tumor tissue DNA findings. In the tumor tissues, a total of 19 SNVs/Indels and 20 CNVs were identified as a reference (Table III). Mutational concordance percentages between paired bile cfDNA and tumor tissue DNA samples are presented in Fig. 3. With the exception of Patient #10, the other 9 BTC patients had concordant mutations. In seven patients (70%, 7/10), >50% mutational concordance between the bile cfDNA and tumor tissue DNA was revealed (Fig. 3). To further investigate the observed SNVs/Indel mutational concordance, the paired bile and tumor samples were further examined. Of the 19 variants detected in the tumor samples, 18 were also detected in bile cfDNA (Fig. 4A). The only discordant mutated gene was CDH1, whose allelic frequency was the lowest of all of the tumor tissue SNVs. Moreover, when compared with the tumor results, targeted deep sequencing of bile cfDNA revealed a high sensitivity (94.7%) and specificity (99.9%; Fig. 4B).
Table III.
Mutations of tumor tissue from ten biliary tract cancer patients.
| Patient ID | SNV/Indel | CNV | |||
|---|---|---|---|---|---|
| Patient #1 | TP53 | p.Q38Pfs*5 | 0.30a | CDK12 | Gain |
| CDH1 | p.Q610* | 0.07 | ERBB2 | Gain | |
| CCNE1 | Gain | ||||
| CDKN2A | Loss | ||||
| Patient #2 | TP53 | p.C275F | 0.22 | FGFR2 | Gain |
| TP53 | p.P27Lfs*17 | 0.10 | |||
| ARID2 | p.Q794Cfs*45 | 0.11 | |||
| Patient #3 | TP53 | p.P151T | 0.22 | ||
| Patient #4 | TP53 | p.V157F | 0.49 | CCNE1 | Gain |
| FLT4 | p.A622T | 0.21 | CDKN2A | Loss | |
| Patient #5 | TP53 | c.783-2_821del | 0.07 | ||
| CDKN2A | p.V82E | 0.19 | |||
| KRAS | p.I36M | 0.35 | |||
| Patient #6 | TP53 | p.M237I | 0.08 | MSH6 | Loss |
| PDGFRA | p.L216I | 0.12 | |||
| NF1 | p.E649* | 0.08 | |||
| RNF43 | p.L706Cfs*20 | 0.07 | |||
| Patient #7 | KRAS | p.G12D | 0.19 | ||
| ATM | p.A2626V | 0.16 | |||
| Patient #8 | TP53 | p.R248W | 0.24 | FGFR3 | Gain |
| TSC1 | p.R190H | 0.23 | VEGFA | Gain | |
| MYC | Gain | ||||
| ERBB3 | Loss | ||||
| Patient #9 | MRE11A | Gain | |||
| MTOR | Loss | ||||
| CDKN2A | Loss | ||||
| STK11 | Loss | ||||
| Patient #10 | MTOR | Gain | |||
| SPEN | Gain | ||||
| NOTCH1 | Gain | ||||
| GNA11 | Gain | ||||
Mutation allele frequency in DNA sample from tumor tissue. SNV/Indel, single nucleotide variation/insertion and deletion; CNV, copy number variation.
Figure 3.
Mutational concordance between paired bile cfDNA and tumor tissue DNA samples from ten BTC patients. Green depicts the proportion of concordant mutations in a given patient, while mutations that are exclusively tumor- or bile-associated are depicted by blue or red, respectively. cfDNA, bile cell-free DNA; BTC, biliary tract cancer.
Figure 4.
Targeted deep sequencing to identify SNVs and Indels from cfDNA. (A) The concordance of SNV/Indel mutations in paired bile cfDNA and tumor tissue DNA samples from four patients with gallbladder carcinomas and six with cholangiocarcinomas. Concordance between tumor tissue DNA and bile cfDNA samples for a given SNV/Indel is depicted in green, while mutations found to be exclusively tumor- or bile-associated are depicted as blue or red, respectively. (B) The sensitivity and specificity of utilizing targeted deep sequencing to detect SNVs/Indels in bile when compared to tumor samples, with calculations performed using the four-table method. Data presentation: (A) Heat maps presenting SNV/Indel mutated genes of bile cfDNA from ten patients. (B) The SNV/Indel results of tumor tissue is regarded as the gold standard. In the SNV/Indel detection of bile cfDNA: Sensitivity=true positive/(true positive + false negative); specificity=true negative/(true negative + false positive). SNVs, single nucleotide variations; Indels, insertions and deletions; cfDNA, bile cell-free DNA.
To further evaluate the capacity of targeted deep sequencing to detect CNVs in bile cfDNA, the paired bile and tumor samples were evaluated for concordance. Of the 20 CNVs identified in the tumor tissue, 15 were also confirmed in bile cfDNA (Fig. 5A). The genes which were identified with CNVs in bile cfDNA included CCNE1, ERBB2, FLT1, CDK6, MET, PIK3CA and ROS1, with ERBB2, CDK6, MET, PIK3CA and ROS1 being of interest as potential BTC therapeutic targets. Furthermore, when compared to the tumor sample results, targeted deep sequencing of bile cfDNA had a sensitivity of 75.0% and specificity of 98.9% (Fig. 5B). These results indicated that targeted deep sequencing of bile cfDNA can confidently detect variance.
Figure 5.
Targeted deep sequencing to detect CNVs in cfDNA. (A) The concordance of CNVs in paired bile cfDNA and tumor tissue DNA samples from ten BTC patients. Concordance between the tumor tissue DNA and bile cfDNA samples for a given CNV is depicted in green, while mutations revealed to be exclusively tumor- or bile-associated are depicted by blue or red, respectively. (B) The sensitivity and specificity of utilizing targeted deep sequencing to identify CNVs in bile cfDNA when compared to tumor samples, with calculations performed using the four-table method. Data presentation: (A) Heat maps presenting CNV mutated genes of bile cfDNA from ten patients. (B) The CNV results of tumor tissue are regarded as the gold standard. In the CNV detection of bile cfDNA: Sensitivity=true positive/(true positive + false negative); specificity=true negative/(true negative + false positive). CNVs, copy number variations; cfDNA, bile cell-free DNA.
Effects of various clinical conditions on the detection of mutations in bile cfDNA
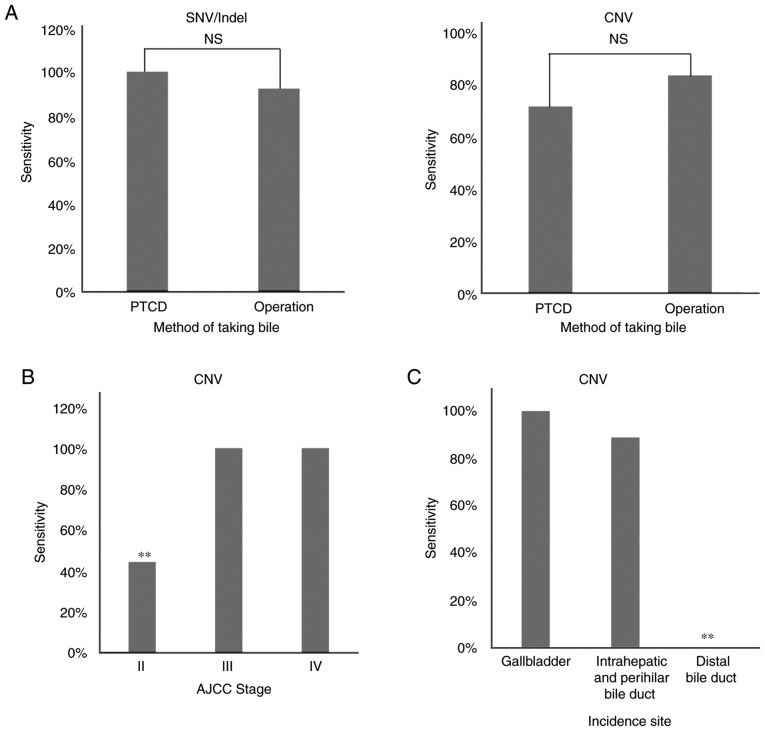
To further evaluate the use of bile cfDNA, the methods of obtaining bile (namely percutaneous transhepatic cholangial drainage (PTCD) or operation) and the tumor stage or incidence site were examined to determine if variant detection sensitivity is impacted. In five of the patients, bile was obtained using PTCD, while in the other five it was obtained by operation (Table I). When examining the detected SNV/Indels and CNVs, no significant difference in sensitivity was noted between the PTCD and operation methods (P>0.00 and P>0.00; Fig. 6A). Next, any potential effects on sensitivity due to tumor stage or incidence site were evaluated. The results revealed that AJCC stage II had a significantly decreased sensitivity in CNV detection when compared with other tumor stages (P<0.05), as did having an incidence site of the distal bile duct (P<0.05; Fig. 6B). These findings indicated that the sensitivity of tumor-matched mutation detection in bile cfDNA can be affected by tumor stage and incidence site.
Figure 6.
Potential effects of the bile extraction method, tumor stage and incidence site on the sensitivity of targeted deep sequencing in identifying mutations in cfDNA. (A) Bile cfDNA was obtained by PTCD or operation and the sensitivity in detecting SNVs/Indels and CNVs using targeted deep sequencing was analyzed. Sensitivities were examined based on (B) tumor stage and (C) incidence sites, with findings displayed as histograms. Statistical analysis: Fisher's exact test; n.s., P>0.05 and **P<0.05 as indicated. cfDNA, bile cell-free DNA; PTCD, percutaneous transhepatic cholangial drainage; SNVs/Indels, single nucleotide variations/insertions and deletions; n.s., not significant.
Discussion
Despite a real unmet need for improved BTC therapies (17), clinical efforts to develop genotype-specific targeted therapies have largely failed due to a lack of reliable, safe, and reproducible genomic analyses. Current approaches rely on obtaining a tumor mutational profile that can then be utilized to predict sensitivity or resistance to particular therapies (18). However, tissue sampling in BTC patients is challenging due to the anatomical complexity of the biliary tract system (19), with many patients at an advanced stage losing the opportunity to receive personalized targeted therapy. Herein, for the first time, the integral mutational landscape of bile cfDNA was examined in 10 BTC patients. The examination of these bile cfDNA samples with targeted deep sequencing revealed a high sensitivity and specificity in detecting SNVs/Indels (94.7 and 99.9%, respectively) and CNVs (75.0 and 98.9%, respectively). Overall, this study provides a promising liquid biopsy method for evaluating clinical BTC patient samples.
Previous studies have reported that plasma cfDNA has a higher proportion of small fragments, with sizes between 150 to 200 bp, in many types of tumors or cancer cell lines (11,20). However, DNA fragment sizes in bile cfDNAs from BTC patients had not been elucidated until now. Herein, bile cfDNA was revealed to be composed of predominantly long DNA fragments, which was not consistent with the findings in plasma cfDNA. These differences may be attributed to DNA restriction endonuclease variety or activity differences between the bile and plasma.
The landscape of BTC has been reported in several previous studies using whole exome sequencing and whole genome sequencing (21–24). The most recurrently mutated genes were TP53, CDKN2A/B, KRAS, ARID1A, and IDH1 in intrahepatic cholangiocarcinoma (ICC); KRAS, TP53, CDKN2A/B, and SMAD4 in extrahepatic cholangiocarcinoma; and TP53, CDKN2A/B, ARID1A, and ERBB2 in gallbladder cancer (25). Ras/phosphatidylinositol-4,5-bisphosphate 3-kinase signaling, p53/cell cycle signaling, and transforming growth factor-β/Smad signaling are substantially affected in ICC (26), while ErbB signaling is the most extensively mutated pathway in gallbladder cancer samples (22). In the present study, similar to previous research of tumor tissue, mutated TP53, KRAS, CDKN2A, PIK3CA, and ERBB2 were also detected in bile cfDNA. NOTCH1, and NOTCH2 mutations were recurrent in bile cfDNA from BTC patients, which was not observed in previous studies.
In a previous study examining SNVs in GBCs, bile containing cfDNA was revealed to have a high concordance rate (87.5%) with tumor tissues (19). Considering the anatomical complexity of the biliary tract system (27), patients not only with GBC, but also cholangiocarcinomas with different incidence sites, were evaluated in this cohort. In addition to SNVs, gene mutations such as Indels and CNVs were also profiled. This study found a high concordance between bile cfDNA and tumor tissues when examining SNVs/Indels, but a high concordance was also found when identifying CNVs, which has not been previously reported. Of the identified CNVs, ERBB2, CDK6, MET, PIK3CA, and ROS1 are of interest as potential BTC therapeutic targets. These results support that targeted deep sequencing with barcodes can effectively isolate somatic mutations in bile cfDNA from BTC patients.
Percutaneous transhepatic cholangial drainage (PTCD) has been revealed to be one of the most important treatments for benign and malignant obstructive jaundice (28–30). Therefore, PTCD and operation were used as two different methods for obtaining bile in this study. Variant detection sensitivities were compared between the two methods and no significant difference was noted when detecting SNV/Indel or CNV mutations. These results suggest that bile cfDNA could serve as a clinically applicable liquid biopsy by means of PTCD. Retrograde endoscopy is one of the main means of minimally invasive treatment of cholangiopancreatography (31). Inhepatobiliary pancreatic surgery, retrograde endoscopy is often recommended for cholangiocarcinoma. The effect of endoscopic bile extraction is a future study we plan to carry out.
Notably, several clinical features have been revealed to have a marked influence on variant detection sensitivity in bile cfDNA when compared with tumor tissues. In our previous study, lung cancer patients with different tumor stages exhibited a significant difference in mutational concordance between plasma cfDNA and tumor tissues (11). Additionally, tumor stage has been revealed to affect gene mutation detection in cfDNA (32,33). Herein, AJCC stage II was revealed to significantly decrease the sensitivity when detecting CNVs in bile cfDNA compared with other tumor stages (P<0.05). While these findings indicate that tumor stage can potentially affect CNV detection, the sample size examined herein was small and this issue should be further examined with a larger sample size in the future. Next, any potential effects on CNV detection associated with tumor incidence site were examined. The results revealed that the distal bile duct significantly decreases the CNV detection sensitivity when compared with other incidence sites. Thus, these findings demonstrated that tumor stage and incidence site can impact variant detection sensitivity in bile cfDNA samples.
In conclusion, the present study revealed that long DNA fragments are prevalent in bile cfDNA samples and that targeted deep sequencing can reproducibly identify somatic variants in BTC bile cfDNA samples. Obtaining bile cfDNA, which is considered a minimally invasive liquid biopsy, and utilizing targeted deep sequencing has the following unique advantages: i) In BTC patients, bile is commonly drained using PTCD, thus bile samples can be obtained without additional trauma; ii) due to tumor heterogeneity, the information obtained from bile cfDNA is more comprehensive in comparison to a needle biopsy; iii) targeted deep sequencing of bile cfDNA samples is feasible, accurate, and sensitive in mutational variants, thus enabling a more personalized targeted therapeutic approach; and iv) bile cfDNA may provide a means to identify benign and malignant diseases or judge tumor recurrence in the future.
Supplementary Material
Acknowledgments
The authors would like to thank Jing Ma, Zuo Zhang, Dalei Wang, Chang Jiang, Yun Wang and Chao Sun for their assistance in data collection and analysis for this study. Additionally, the authors would like to thank all of the patients and their families for their support of this study.
Glossary
Abbreviations
- BTC
biliary tract cancer
- GBC
gallbladder carcinoma
- cfDNA
cell-free DNA
- SNV
single nucleotide variations
- Indel
insertion and deletion
- CNV
copy number variation
- FFPE
formalin-fixed paraffin-embedded
- PTCD
percutaneous transhepatic cholangial drainage
Funding
The present study was funded by the Youth Fund of Shanghai Municipal Health Planning Commission (20144Y0164).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
NS designed the bile extraction study, contributed to data interpretation and wrote the manuscript. DZ designed the extraction of bile cell-free DNA (cfDNA) and wrote the manuscript. LYi performed the bile extraction from gallbladder carcinoma patients using operation and interpreted the data. YQ collected the bile samples by means of percutaneous transhepatic cholangial drainage (PTCD) and contributed to data interpretation. JL collected the tumor tissue from gallbladder carcinoma patients and contributed to data interpretation. WY performed the separation and storage of bile samples and collected the clinical information. XF performed the collection and isolation of patients' blood and interpreted the data. BZ performed the extraction of genomic DNA and contributed to data interpretation. XX established the methodology of extracting the bile cfDNA and interpreted the data. AD performed bile cfDNA extraction and contributed to data interpretation. ZC performed the quality control analyses on bile cfDNA extraction and interpreted the data. XW performed the construction of sequencing library and contributed to data interpretation. XC performed the analyses on the SNV/Indel of bile cfDNA and tumor tissue samples, contributed to data interpretation and wrote the manuscript. TZ analyzed the clinical information of patients and contributed to data interpretation. ZZ analyzed the copy number variation of bile cfDNA and tumor tissue samples and contributed to data interpretation. LYu performed the targeted deep sequencing of bile cfDNA and interpreted the data. HQ performed the the analyses on sensitivity and specificity of bile cfDNA in detecting the mutations and data interpretation. ZF performed the targeted deep sequencing of DNA from tumor tissues and contributed to data interpretation. JYL discussed the hypothesis and contributed to data interpretation. YL collected the tumor tissue samples from cholangiocarcinoma patients and interpreted the data. LX led the consistency analyses on mutations bewteen bile cfDNA and tumor tissue DNA and contributed to data interpretation. BY collected the bile samples from cholangiocarcinoma patients and interpreted the data. FL conceived the study, led the project and wrote the manuscript. YZ conceived the study and participated in its design and coordination. All of the authors have read and approved the final version of the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Bile and tumor tissue samples were collected with ethics approval obtained from the Eastern Hepatobiliary Surgery Hospital (EHBHKY2018-K-003). Informed consent was obtained from the patients for the examination of their samples and the use of their clinical data.
Patient consent for publication
Not applicable.
Competing interests
All of the authors affiliated to 3D Medicines, Inc. are current or former employees. No potential competing interests were disclosed by the other authors.
References
- 1.Augustine MM, Fong Y. Epidemiology and risk factors of biliary tract and primary liver tumors. Surg Oncol Clin N Am. 2014;23:171–188. doi: 10.1016/j.soc.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Marcano-Bonilla L, Mohamed EA, Mounajjed T, Roberts LR. Biliary tract cancers: Epidemiology, molecular pathogenesis and genetic risk associations. Chin Clin Oncol. 2016;5:61. doi: 10.21037/cco.2016.10.09. [DOI] [PubMed] [Google Scholar]
- 3.Streeter OE, Jr, Beron PJ, Iyer PN. Precision medicine: Genomic profiles to individualize therapy. Otolaryngol Clin North Am. 2017;50:765–773. doi: 10.1016/j.otc.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran W, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 6.Nam AR, Kim JW, Cha Y, Ha H, Park JE, Bang JH, Jin MH, Lee KH, Kim TY, Han SW. Therapeutic implication of HER2 in advanced biliary tract cancer. Oncotarget. 2016;7:58007–58021. doi: 10.18632/oncotarget.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakamoto Y, Yamagishi S, Tanizawa Y, Tajimi M, Okusaka T, Ojima H. PI3K-mTOR pathway identified as a potential therapeutic target in biliary tract cancer using a newly established patient-derived cell panel assay. Jpn J Clin Oncol. 2018;48:396–399. doi: 10.1093/jjco/hyy011. [DOI] [PubMed] [Google Scholar]
- 8.Rizvi S, Yamada D, Hirsova P, Bronk SF, Werneburg NW, Krishnan A, Salim W, Zhang L, Trushina E, Truty MJ, Gores GJ. A hippo and fibroblast growth factor receptor autocrine pathway in cholangiocarcinoma. J Biol Chem. 2016;291:8031–8047. doi: 10.1074/jbc.M115.698472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha SK, Gordan JD, Kleinstiver BP, Vu P, Najem MS, Yeo JC, Shi L, Kato Y, Levin RS, Webber JT, et al. Isocitrate dehydrogenase mutations confer dasatinib hypersensitivity and SRC dependence in intrahepatic cholangiocarcinoma. Cancer Discov. 2016;6:727–739. doi: 10.1158/2159-8290.CD-15-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 11.Yang N, Li Y, Liu Z, Qin H, Du D, Cao X, Cao X, Li J, Li D, Jiang B, et al. The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer. 2018;18:319. doi: 10.1186/s12885-018-4199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 13.Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer. 2015;90:128–134. doi: 10.1016/j.lungcan.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Jiang T, Li X, Wang J, Su C, Han W, Zhao C, Wu F, Gao G, Li W, Chen X, et al. Mutational landscape of cfDNA identifies distinct molecular features associated with therapeutic response to first-line platinum-based doublet chemotherapy in patients with advanced NSCLC. Theranostics. 2017;7:4753–4762. doi: 10.7150/thno.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su D, Zhang D, Chen K, Lu J, Wu J, Cao X, Ying L, Jin Q, Ye Y, Xie Z, et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J Exp Clin Cancer Res. 2017;36:121. doi: 10.1186/s13046-017-0591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe H, Okada M, Kaji Y, Satouchi M, Sato Y, Yamabe Y, Onaya H, Endo M, Sone M, Arai Y. New response evaluation criteria in solid tumours-revised RECIST guideline (version 1.1) Gan To Kagaku Ryoho. 2009;36:2495–2501. (In Japanese) [PubMed] [Google Scholar]
- 17.Ribiere SC, Léandri C, Guillaumot MA, Brezault C, Coriat R. Therapeutic advances in the management of biliary tract carcinoma. Presse Med. 2018;47:419–422. doi: 10.1016/j.lpm.2018.03.008. (In French) [DOI] [PubMed] [Google Scholar]
- 18.Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, Hendifar AE, Wang Z, Atreya CE, Kelley RK, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5:1040–1048. doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinugasa H, Nouso K, Ako S, Dohi C, Matsushita H, Matsumoto K, Kato H, Okada H. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol Ther. 2018;19:934–938. doi: 10.1080/15384047.2018.1456604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie D, Ren Z, Fan J, Gao Q. Genetic profiling of intrahepatic cholangiocarcinoma and its clinical implication in targeted therapy. Am J Cancer Res. 2016;6:577–586. [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z, Liu C, Shen B, Wang XA, Wu W, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872–876. doi: 10.1038/ng.3030. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira DV, Zhang S, Chen X, Calvisi DF, Andersen JB. Molecular profiling of intrahepatic cholangiocarcinoma: The search for new therapeutic targets. Expert Rev Gastroenterol Hepatol. 2017;11:349–356. doi: 10.1080/17474124.2017.1292127. [DOI] [PubMed] [Google Scholar]
- 24.Lee H, Ross JS. The potential role of comprehensive genomic profiling to guide targeted therapy for patients with biliary cancer. Therap Adv Gastroenterol. 2017;10:507–520. doi: 10.1177/1756283X17698090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, Kang HC, Catenacci D, Ali S, Krishnan S, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer. 2016;122:3838–3847. doi: 10.1002/cncr.30254. [DOI] [PubMed] [Google Scholar]
- 26.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 27.Chandra A, Gupta V, Rahul R, Kumar M, Maurya A. Intraoperative ultrasonography of the biliary tract using saline as a contrast agent: A fast and accurate technique to identify complex biliary anatomy. Can J Surg. 2017;60:316–322. doi: 10.1503/cjs.011116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao H, Liu J, Li T, Cao G, Xu G, Zhai S, Xue J, Wang Z, Shi S, Bai W. Interventional therapy for the treatment of severe hemobilia after percutaneous transhepatic cholangial drainage: A case series. Int Surg. 2013;98:223–228. doi: 10.9738/INTSURG-D-13-CC194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Pang Q, Liu H, Li Z, Wang Y, Lu Y, Zhou L, Pan H, Huang W. Prognostic value of inflammation-based markers in patients with recurrent malignant obstructive jaundice treated by reimplantation of biliary metal stents: A retrospective observational study. Medicine (Baltimore) 2017;96:e5895. doi: 10.1097/MD.0000000000005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Yu J, Liang P, Yu X, Cheng Z, Han Z, Duan S, Zheng J. Ultrasound-guided percutaneous microwave ablation assisted by three-dimensional visualization operative treatment planning system and percutaneous transhepatic cholangial drainage with intraductal chilled saline perfusion for larger hepatic hilum hepatocellular (D ≥ 3 cm): Preliminary results. Oncotarget. 2017;8:79742–79749. doi: 10.18632/oncotarget.19275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njei B, McCarty TR, Varadarajulu S, Navaneethan U. Systematic review with meta-analysis: endoscopic retrograde cholangiopancreatography-based modalities for the diagnosis of cholangiocarcinoma in primary sclerosing cholangitis. Aliment Pharmacol Ther. 2016;44:1139–1151. doi: 10.1111/apt.13817. [DOI] [PubMed] [Google Scholar]
- 32.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Dittmar RL, Xia S, Zhang H, Du M, Huang CC, Druliner BR, Boardman L, Wang L. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol Oncol. 2017;11:1099–1111. doi: 10.1002/1878-0261.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.