Abstract
Imatinib-based targeted treatment is the standard therapy for chronic myeloid leukemia (CML); however, drug resistance is an inevitable issue for imatinib-based CML treatment. Imatinib resistance can be ascribed to Bcr-Abl-dependent and independent resistance. In the present study, peripheral blood samples were collected from imatinib-sensitive (IS) and imatinib-resistant (IR) CML patients and transcriptome sequencing was carried out. From the RNA-seq data, a significantly altered IR-related gene (IRG), ribonucleotide reductase regulatory subunit M2 (RRM2) was identified. Using real-time quantitative fluorescence PCR (qF-PCR), we found that RRM2 was elevated in both IR CML patients and an IR cell line. Using reverse-transcription PCR (RT-PCR) and western blot analysis, we indicated that imatinib can increase RRM2 level in a dose-dependent manner in IR cells. We also demonstrated that RRM2 is involved in the Bcl-2/caspase cell apoptotic pathway and in the Akt cell signaling pathway, and therefore affects the cell survival following imatinib therapy. The present study, for the first time, indicates that RRM2 is responsible for drug resistance in imatinib-based therapy. Therefore, RRM2 gene can be considered as a potential therapeutic target in the clinical treatment of CML.
Keywords: CML, imatinib, RRM2, drug resistance
Introduction
Chronic myeloid leukemia (CML) is a common hematologic malignancy in China. As a myeloproliferative disease, CML results from the reciprocal translocation of chromosome 9 and chromosome 22, which leads to the Bcr-Abl gene fusion and increases constitutive tyrosine kinase activity (1).
Imatinib, a tyrosine kinase inhibitor (TKI), directly inhibits constitutive tyrosine kinase activity, which results in the modification of the functions of various genes involved in the control of the cell cycle, cell adhesion, cytoskeleton organization and ultimately in the apoptotic death of Ph(+) cells (2). Imatinib-based targeted treatment has become the standard therapy for CML, and most patients in the chronic phase (CP) of CML achieve not only a complete cytogenetic response (CCR) but also a major molecular response (MMR) (3). A 10-year follow-up of patients with CML who were treated with imatinib as initial therapy showed that imatinib can improve the prognosis of CML patients without unacceptable cumulative or late toxic effects (4).
The major reason for the therapeutic success of imatinib in CML is the well-defined molecular target toward the Bcr-Abl gene and relatively selective therapies aimed at this gene. However, drug resistance is a recurrent issue for imatinib-based CML treatment. A clinical trial indicated that for imatinib-based treatment of CML, there is a 15 to 25% rate of primary cytogenetic resistance by 18 months of therapy, and the secondary resistance rate was 7 to 15% (5). Imatinib resistance (IR) can be ascribed to two major reasons: Bcr-Abl-dependent and -independent resistance. The Bcr-Abl-dependent resistance includes Bcr-Abl duplication and mutation. In vitro cell experiments have demonstrated that continuous culture with imatinib-containing medium elevated Abl kinase activity due to a genetic duplication of the Bcr-Abl sequence (6,7). Gorre et al demonstrated that the T315I mutation creates steric hindrance to the bonding between imatinib and the Abl kinase (8). Bcr-Abl-independent resistance includes a decrease in drug influx and an increase in drug efflux (9), drug sequestration in the plasma (10), epigenetic modification (11) and alternative signaling pathway activation (12).
Ribonucleotide reductase regulatory subunit M2 (RRM2) plays a significant role in tumor progression and is frequently overexpressed in cancer. It is involved in the regulation of cell invasion, cell migration and tumor metastasis (13). Elevated RRM2 expression has been reported to be associated with a poor prognosis of several types of cancer including gastric cancer, adrenocortical cancer, and non-small cell lung cancer (13–15). RRM2 is also demonstrated to be significantly associated with drug resistance. For example, Shah et al revealed that RRM2 is a key contributor to AKT-induced tamoxifen resistance in breast cancer treatment (16), and Tu et al demonstrated that VASH2 reduced the chemosensitivity to gemcitabine in pancreatic cancer cells via the JUN-dependent transactivation of RRM2 (17).
In the present study, peripheral blood samples were collected from 22 imatinib-sensitive (IS) and 17 imatinib-resistant (IR) primary CML patients and the transcription profile of these samples was analyzed using high-throughput sequencing (RNA-seq). Numerous genes were found to be altered in IR patients. Four significantly increased genes that may correlate with IR, aryl hydrocarbon receptor nuclear translocator 2 (ARNT2), ATP binding cassette subfamily A member 13 (ABCA13), RRM2 and secreted frizzled related protein 1 (SFRP1) were screened (18–21), and three significantly decreased genes were identified that may be correlated with IR, growth differentiation factor 7 (GDF7 or BMP12), glutathione S-transferase µ1 (GSTM1) and AP-1 transcription factor subunit (c-Fos) (22–24). Among these genes, RRM2 was observed to be elevated in both IR patients and an IR cell line. It was also demonstrated that RRM2 is involved in the Bcl-2/caspase and Akt cell signaling pathways and therefore affects the cell survival in imatinib therapy.
Herein, for the first time, we report that RRM2 is responsible for drug resistance in imatinib-based CML therapy. This study evaluated RRM2 as a potential therapeutic target in the clinical treatment of CML.
Materials and methods
Patients and peripheral blood collection
Peripheral blood samples were collected from 20 IR CML patients at the First Hospital of Jilin University (Changchun, China) from April 2015 to August 2018. Among these IR patients, 11 were male and 9 were female, with a median age of 53 years (range 18–72). Two of the IR patients were in accelerated phase and 18 were in chronic phase. These patients had received imatinib as the first line therapy for 8 months to 13 years. CML diagnosis and resistance were defined on the basis of European Leukemia Net: ELN Recommendations 2013 (https://www.leukemia-net.org/content/home/index_eng.html). Peripheral blood samples were also collected from 17 IS CML patients who had achieved a major molecular response (MMR). The median age of the IS CML patients was 46 years (range 26–60). Control peripheral blood samples were obtained from 15 healthy people with a median age of 32 years (range 25–39). From these blood samples, nucleated cells including lymphocytes, monocytes and granulocytes were isolated. All of these samples were stored at −80°C until use. Permission to use the clinical samples for research purposes was obtained and approved by the Ethics Committee of the First Hospital of Jilin University. Informed consents were obtained from all patients.
RNA-seq
Two IR samples and two IS samples were randomly selected and the RNA-seq was carried out. The distinct mRNAs between IS and IR patients and their related pathways were identified. Briefly, total RNA was isolated from the patient peripheral blood samples using Qiazol (Qiagen, Shanghai, China). Then the total RNA was reverse-transcribed to a cDNA library. RNA-seq was carried out on a HiSeq4000 (Illumina, Inc.) and yielded approximately 30 million reads with a length of 150 bp per sample (Shanghai Biotechnology Corp., Shanghai, China). Gene counts were normalized to the values of fragments per kilobase of transcript per million mapped reads (FPKM).
Cell culture
The human myeloid leukemia K562 cell line and basic IR type K562G cell line were purchased from the Chinese Academy of Medical Sciences (Tianjin, China). K562 cells were grown in normal RPMI-1640 media (Thermo Fisher Scientific, Inc, Beijing, China) supplemented with 10% fetal bovine serum (HyClone, Beijing, China) in 5% CO2 at 37°C. K562G cells were grown in the same culturing condition except that 8 µM imatinib (Sigma-Aldrich; Merck KGaA, Shanghai, China) was added to the media.
For the concentration-effect analysis of imatinib, K562G cells were cultured in 8 µM imatinib as the initial concentration for two weeks, then in 10 µM imatinib for another two weeks, and finally in 12 µM imatinib for two weeks.
Reverse-transcription PCR (RT-PCR) and real-time quantitative fluorescence PCR (qF-PCR)
Cells were collected and total RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, Shanghai, China). RNA (1 µg) was reverse-transcribed to cDNA using Moloney murine leukemia virus reverse transcriptase (M-MLV, Invitrogen; Thermo Fisher Scientific, Inc.). The reaction system for RT-PCR analysis contained (6 µl): 2 µl of prepared sample cDNA as the template, 3 µl 2X Taq PCR StarMix buffer (GenStar, Beijing, China) and 1 µl forward and reverse primers. The reaction system for qF-PCR analysis contained (20 µl): 2 µl of prepared sample cDNA as the template, 10 µl 2X SYBR Premixed buffer (Roche, Shanghai, China), 2 µl forward and reverse primers and 6 µl ddH2O. The primer sequences were as follows: ARNT2 forward primer, 5′-TGCATCGGAGAAGAAGATGATG-3′ and reverse primer, 5′-ATTCACTCCAGGCACATGAAC-3′ (136 bp); ABCA13 forward primer, 5′-CTGTGGAAGAATTGGCTCTGCA-3′ and reverse primer, 5′-TGTCTCTGTATCTGGGAGGTTC-3′ (127 bp); RRM2 forward primer, 5′-CTATGGCTTCCAAATTGCCATG-3′ and reverse primer, 5′-GACACAAGGCATCGTTTCAATG-3′ (127 bp); SFRP1 forward primer, 5′-TGTGCCACAACGTGGGCTAC-3′ and reverse primer, 5′-AGTTCTTGTTGAGCAGGGGCACC-3′ (114 bp); BMP12 forward primer, 5′-CACTTCATGATGTCGCTTTACC-3′ and reverse primer, 5′-CGTTAAGGCTGGACACGTCGA-3′ (181 bp); GSTM1 forward primer, 5′-TTCCCAATCTGCCCTACTTG-3′ and reverse primer, 5′-CACGAATCTTCTCCTCTTCTG-3′ (120 bp); c-FOS forward primer, 5′-GATAGCCTCTCTTACTACCAC-3′ and reverse primer, 5′-GAATGAAGTTGGCACTGGAGAC-3′; Bcl-2 forward primer, 5′-ACCTGGATCCAGGATAACGGA-3′ and reverse primer, 5′-GATAGGCACCCAGGGTGATGC-3′ (148 bp); GAPDH forward primer, 5′-TGCACCACCAACTGCTTA-3′ and reverse primer, 5′-GGATGCAGGGATGATGTTC-3′ (178 bp). The RT-PCR amplification was carried out on the Eppendorf Mastercycler Pro (Eppendorf, Shanghai, China) and the PCR process was as follows: 10 min denaturation at 95°C, 26 to 32 cycles at 95°C for 20 sec, 62°C for 15 sec and 72°C for 15 sec, and then at 72°C for 2 min. PCR productions were visualized by 3% agarose gel electrophoresis. Densitometric analysis was conducted using Quantity One software (version 4.6, Bio-Rad). The qF-PCR amplification was carried out on the ABI StepOnePlus (ABI, Beijing, China) and the PCR process was as follows: 5 min denaturation at 95°C followed by 40 cycles at 95°C for 20 sec, 60°C for 15 sec and 72°C for 15 sec. mRNA levels were normalized to GAPDH levels within the same sample. Data analysis was performed using the 2−ΔΔCq method (25).
siRNA and RNA interference
Control siRNA (siCT) and siRNA for RRM2 were synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, China). The sequence of siCT is: 5′-UAGCGACUAAACACAUCAAUU-3′ and the sequence of siRRM2 is: 5′-GCGAUUUAGCCAAGAAGUUCA-3′ (26). Amaxa® Cell Line Nucleofector® Kit V (Lonza, Basel, Switzerland) was used for the siRNA transfection. According to the manufacturer's guidelines, 1×106 K562 or K562G cells were transfected with 100 pmol siRNA. Twelve hours after the electrotransfection, the cells were centrifuged at a low speed (90 × g) to exclude the dead cells and the debris. The cells were incubated for another 12 h and the knockdown efficiency was measured by RT-PCR, qF-PCR and western blotting. All the cell experiments were performed at 24 h post-transfection.
Cell viability determination by the CCK-8 assay
K562 and K562G cells (5×103) were plated in 96-well plates and cultured for 24 h. For the cell viability assay, 10 µl CCK-8 solution (Thermo Fisher Scientific, Inc, Beijing, China) was added to each well and the plate was incubated at 37°C for 2 h. The optical density (OD) at 450 nm (OD450) was measured using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Apoptosis assay
K562G cells were maintained in RPMI-1640 medium supplemented with 8 µM imatinib. Approximately 1×106 cells were collected and washed with cold PBS. Then, the cells were incubated with Annexin V-FITC/PI (BD Biosciences, Franklin Lakes, NJ, USA) for 15 min in the dark and analyzed with fluorescence-activated cell sorting (FACScan; BD Biosciences). FlowJo software (FlowJo, LLC, Ashland, OR, USA) was used for apoptosis analysis. Cells in the different proportions represent the different cell states as follows: The dead cells are shown in the upper left portion (Q1), the late-apoptotic cells are shown in the upper right portion (Q2), the viable cells are shown in the lower left portion (Q4), and the early apoptotic cells are the cells present in the lower right portion (Q3).
Protein extraction and western blotting
Cells were lysed in RIPA buffer (KeyGen Biotech. Co., Ltd., Nanjing, China) compensated with a cocktail protease inhibitor (Roche). Lysates were centrifuged at 20,000 × g for 30 min at 4°C. Protein concentration was determined by Bradford Protein Assay Kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China). For protein expression assay, equal amount of 20 µg protein from each sample was separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The blots were then blocked with 5% skim milk in TBST at 37°C for 1 h and incubated with primary antibodies including: RRM2 (dilution 1:1,000, cat. no. ab57653; Abcam, Shanghai, China), Bcl-2 (dilution 1:1,000; cat. no. ab59348; Abcam), Bax (dilution 1:1,000; cat. no. ab32503; Abcam), cleaved caspase-3 (dilution 1:1,000; cat. no. 9664; Cell Signaling Technology, Shanghai, China), cleaved caspase-9 (dilution 1:1,000; cat. no. 9505; Cell Signaling Technology), Akt (dilution 1:1,000. cat. no. 2920; Cell Signaling Technology), phospho-Akt (Ser473, dilution 1:1,000; cat. no. 4060; Cell Signaling Technology) and β-actin (dilution 1:1,000; cat. no. sc47778; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Then the membranes were incubated with horseradish peroxidase-coupled goat anti-rabbit or goat anti-mouse secondary antibody (dilution 1:3,000; cat. nos. sc-2004 and sc-2005; Santa Cruz Biotechnology) at room temperature for 1 h. The chemiluminescence signals were detected with a chemiluminescence system (ECL, Thermo Fisher Scientific, Inc.). Densitometric analysis was conducted using Quantity One software, version 4.6 (Bio-Rad Laboratories).
Statistical analysis
All experimental results represent the average of at least three independent experiments. Data were analyzed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) and expressed as mean ± SD. Statistical analysis was performed using t-test (between two groups) or one-way analysis of variance followed by an LSD post hoc test (more than two groups). *P<0.05, **P<0.01, #P<0.05 or ##P<0.01 was considered to indicate a statistically significant difference (relevant symbols are shown in the figures and legends).
Results
Identification of IR-related genes (IRGs) using RNA-seq and qF-PCR
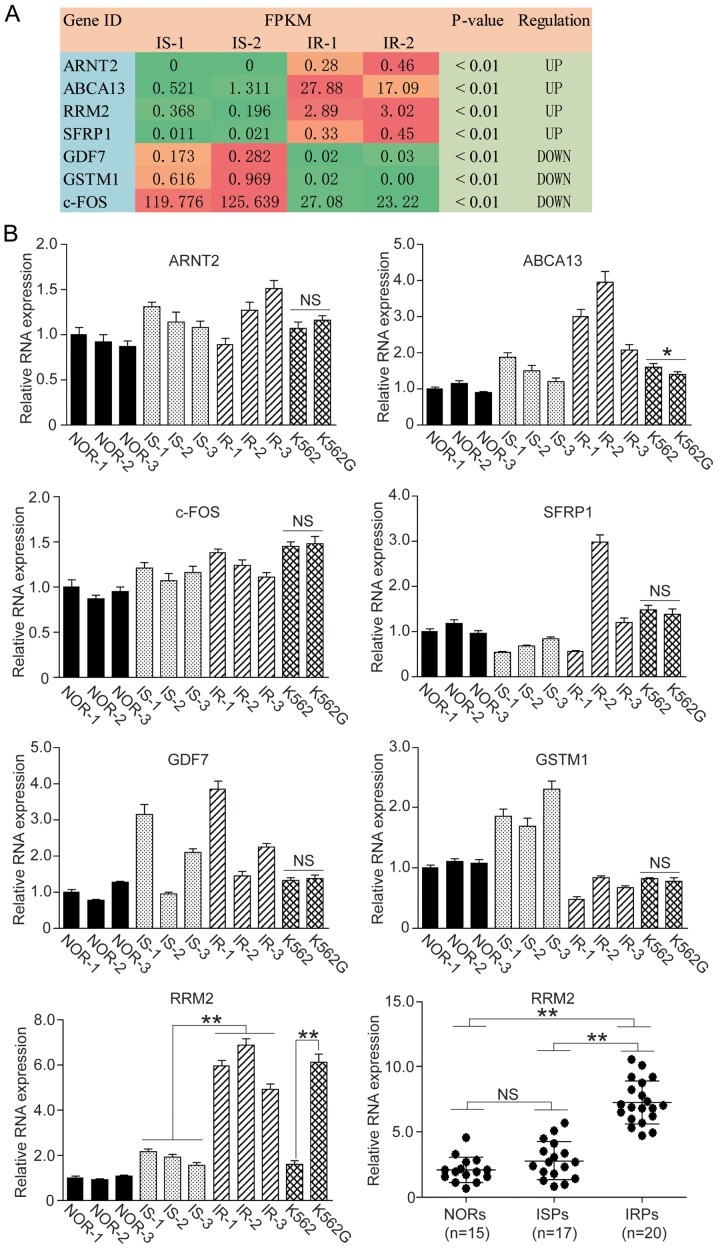
The relative mRNA levels from the RNA-seq are expressed as FPKM value and are listed in Fig. 1A. Referring to the KEGG Pathway Database (https://www.kegg.jp/kegg/pathway.html), 7 genes were screened that may be involved in the IR process. The expression levels of ARNT2, ABCA13, RRM2 and SFRP1 were significantly increased and the expression levels of GDF7, GSTM1 and c-FOS were significantly decreased.
Figure 1.
Screening of IRGs in CML patients. RNA-seq was employed to screen specific IRGs in imatinib-sensitive (IS) and imatinib-resistant (IR) CML patients and 7 genes were identified. Among these genes, 4 genes were upregulated and 3 genes were downregulated in the IR patients. We carried out qF-PCR to confirm the levels of these IRGs in peripheral blood samples and cell lines. (A) FPKM values are presented in RNA-seq. Green box, downregulation; orange and red box, upregulation. (B) qF-PCR assay of 7 IRGs in blood samples and cell lines. IRGs, IR-related genes; CML, chronic myeloid leukemia; NORs, normal patients; ISPs, imatinib-sensitive patients; IRPs, imatinib-resistant patients. One-way ANOVA and t-test, *P<0.05; **P<0.01; NS, not significant.
We carried out qF-PCR to confirm whether the variation trends of these genes were consistent with the RNA-seq result, and we found that the variation trends of ABCA13, GSTM1 and RRM2 were consistent with the RNA-seq result, whereas ARNT2 and c-FOS were not consistent with the RNA-seq result. As for SFRP1 and GDF7, the individual variation in the gene transcription was significant (Fig. 1B).
We then detected the mRNA level of ABCA13, GSTM1 and RRM2 in normal K562 cells and the IR type K562G cells. Among these three genes, RRM2 showed the most significant difference between K562 and K562G cells (Fig. 1B, P<0.01). We went on to detect the RRM2 level in 15 normal individuals (NORs), 17 IS patients (ISPs) and 20 IR patients (IRPs), and we observed that the RRM2 level in the IR group was significantly higher than that of the IS and NOR group (Fig. 1B, P<0.01). Therefore, we selected RRM2 as our target gene and performed the following cell experiments.
Imatinib increases the RRM2 level in a dose-dependent manner in K562G cells
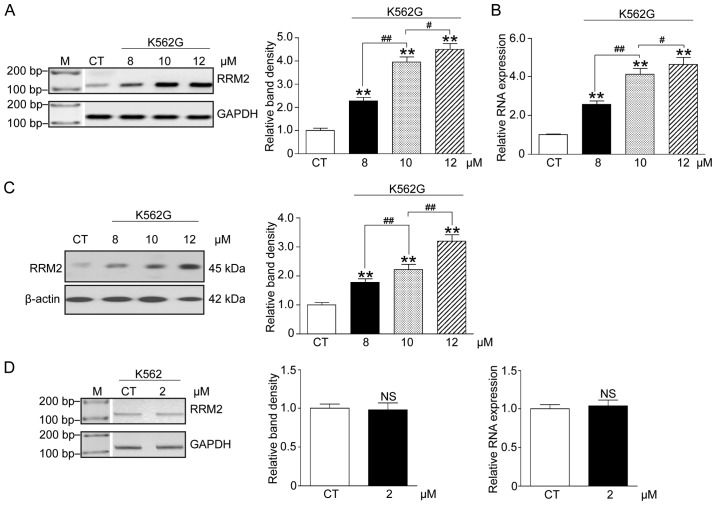
In order to confirm the correlation between imatinib therapy and RRM2 expression, K562G cells were treated with three doses of imatinib (8, 10 and 12 µM), and K562 cells were treated with 2 µM imatinib. We analyzed the RRM2 mRNA and protein levels by RT-PCR, qF-PCR and western blot analysis. The results indicated that following increasing concentrations of imatinib, both the mRNA and protein levels of RRM2 in the K562G cells were significantly elevated when compared with the CT group (Fig. 2A-C, P<0.01). However, the mRNA level in K562 cells was not significantly altered (Fig. 2D, P>0.05). These data showed an apparent concentration-effect relationship of imatinib and RRM2 in IR cells.
Figure 2.
High dose imatinib increases the RRM2 level in imatinib-resistant (IR) cells. We treated K562G cells with 8, 10 and 12 µM imatinib, and K562 cells with 2 µM imatinib. The RRM2 mRNA and protein levels were analyzed using RT-PCR, qF-PCR and western blot analysis. (A) RT-PCR results. (B) qF-PCR results. (C) Western blot analysis results and the relative band density. (D) RT-PCR results showed that 2 µM imatinib did not significantly alter the mRNA level of RRM2 in imatinib-sensitive (IS) cells. CT, K562 cells used as the control. One-way ANOVA, **P<0.01, compared with the CT group. #P<0.05, ##P<0.01. NS, not significant; RRM2, ribonucleotide reductase regulatory subunit M2.
Knockdown of RRM2 enhances the apoptosis of K562G cells
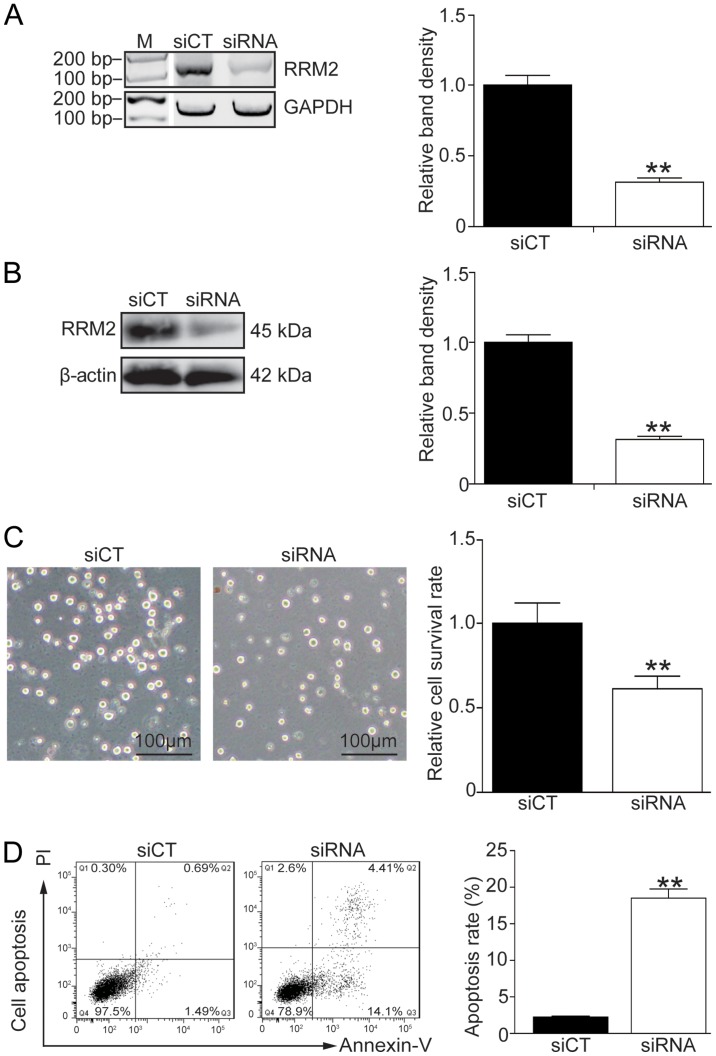
In order to address the correlation between the RRM2 level and imatinib-induced cell apoptosis, endogenous RRM2 was effectively knocked down using siRNA (Fig. 3A and B, P<0.01). Cell morphology showed that K562G cells became sensitive to 8 µM imatinib treatment after RRM2 was knocked down (Fig. 3C, left panel). Cell survival experiment and flow cytometry also demonstrated that knockdown of RRM2 inhibited the cell growth and induced a high rate of cell death following imatinib-based treatment (Fig. 3C, right panel, P<0.01; Fig. 3D, P<0.01).
Figure 3.
RRM2 knockdown enhances the imatinib sensitivity in K562G cells. RRM2 was knocked down in K562G cells using siRNA and cell viability and apoptosis were analyzed in the presence of 8 µM imatinib. (A) Effective knockdown of RRM2 by siRNA. The RRM2 mRNA level was determined by RT-PCR. (B) The RRM2 protein level was determined by western blot analysis. (C) Analysis of the cell survival by CCK-8 assay. (D) Analysis of the cell apoptosis by flow cytometry. t-test, **P<0.01, as compared with the siCT group. RRM2, ribonucleotide reductase regulatory subunit M2.
Bcl-2 is upregulated in IR patients and in K562G cells
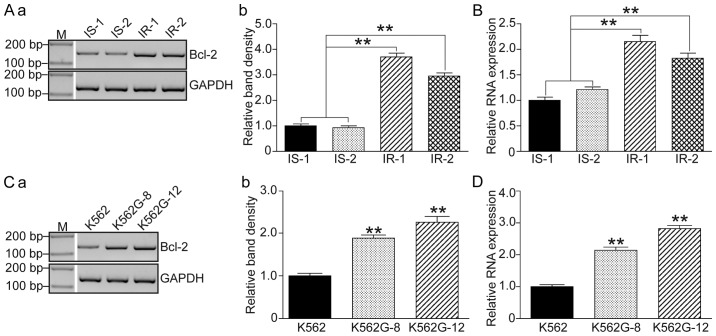
Since it was demonstrated that a high level of RRM2 induced imatinib resistance and that knockdown of RRM2 increased drug sensitivity of K562G cells to imatinib, we then focused on the variations in apoptosis-related genes. Bcl-2 levels were detected in 2 IS patients (IS-1 and IS-2), 2 IR patients (IR-1 and IR-2) and the imatinib-treated K562G cells. As shown in Fig. 4, both the IR patients and the K562G cells showed significant higher levels of Bcl-2 compared to the IS patients and the normal K562 cells (P<0.01).
Figure 4.
RRM2 is consistent with Bcl-2 in IR CML patients and IR CML cells. (A) Analysis of Bcl-2 mRNA level in IR (IR-1 and IR-2) and IS (IS-1 and IS-2) CML patients by RT-PCR. (a) PCR band and (b) relative band density. (B) Analysis of the Bcl-2 mRNA level in IR (IR-1 and IR-2) and IS (IS-1 and IS-2) CML patients by qF-PCR. (C) Analysis of the Bcl-2 mRNA level in CML cell lines by RT-PCR. (a) PCR band and (b) the relative band density. (D) Analysis of the Bcl-2 mRNA level in IS (K562) and IR (K562G-8 and K562G-12) CML cell lines by qFPCR. One-way ANOVA and t-test, **P<0.01. RRM2, ribonucleotide reductase regulatory subunit M2; CML, chronic myeloid leukemia; IR, imatinib-resistant; IS, imatinib-sensitive. K562G-8, cells were cultured in 8 µM imatinib as the initial concentration for two weeks; K562G-12, cells were cultured in 12 µM imatinib for two weeks.
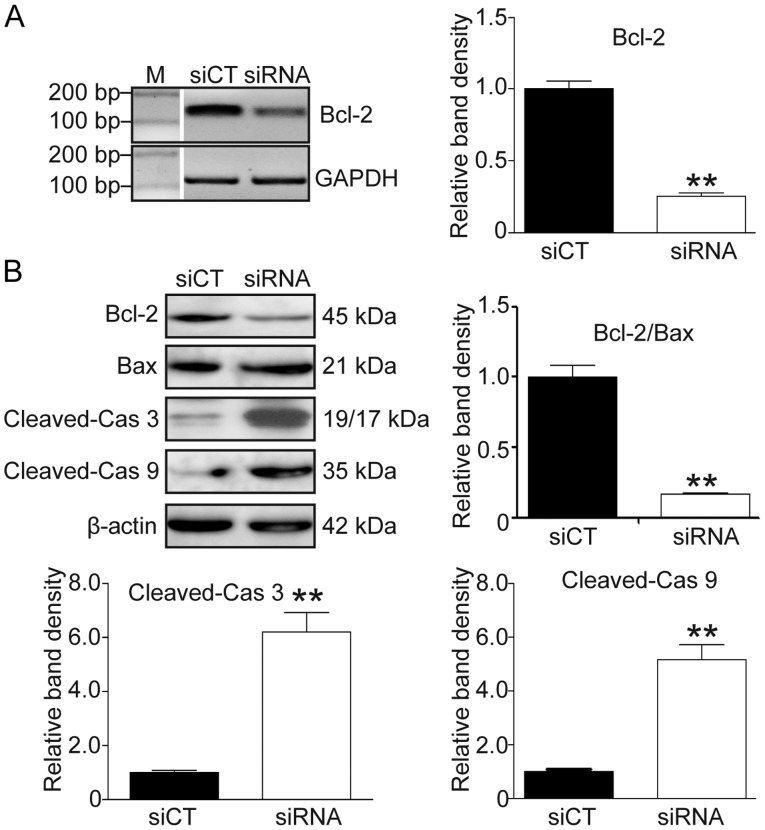
RRM2-induced IR is associated with the Bcl-2/caspase cell apoptotic pathway
To confirm the role of Bcl-2/Bax and the caspase apoptotic pathway in the RRM2-induced imatinib resistance, RRM2 siRNA was used to knock down RRM2 and RT-PCR, qF-PCR and western blot analysis were utilized to detect the expression of Bcl-2, Bax, cleaved caspase-3 and −9. As shown in Fig. 5, after RRM2 was knocked down, the Bcl-2 level was decreased (Fig. 5A and B, P<0.01) and the Bax level was increased while the Bcl-2/Bax ratio was decreased (Fig. 5B, P<0.01). Simultaneously, the levels of cleaved caspase-3 and −9 were significantly elevated (Fig. 5B, P<0.01).
Figure 5.
RRM2 suppression induces cell apoptosis through the Bcl-2/caspase pathway. (A) Knockdown of Bcl-2 in K562G cells. Left panel, PCR band; right panel, the relative band density. (B) Knockdown of RRM2 activated the Bcl-2/Bax and caspase pathways. The levels of Bcl-2, Bax, cleaved-caspase (cas) 3 and 9 were determined by western blot analysis. Upper left panel, western blot band; histograms, the relative band density. t-test, **P<0.01, as compared with the siCT group. RRM2, ribonucleotide reductase regulatory subunit M2.
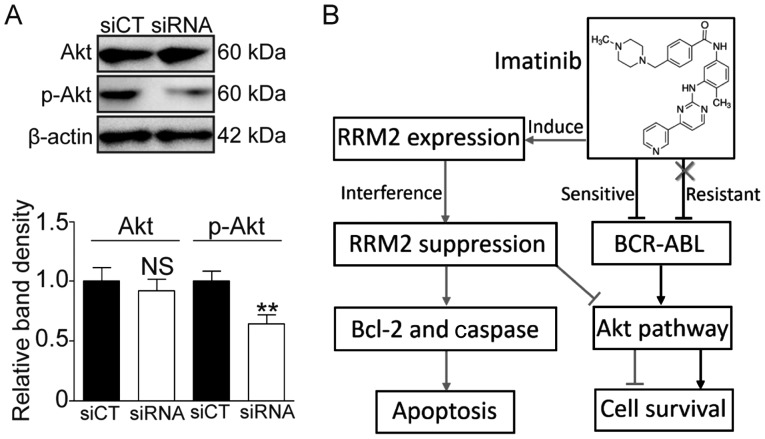
Knockdown of RRM2 suppresses activation of the Akt pathway
It was reported that RRM2 can activate Akt and promote cell invasion in gastric cancer (13). Herein, we also investigated whether or not RRM2 activates the Akt pathway. As shown in Fig. 6, the phosphorylated Akt (p-Akt) level was significantly decreased following RRM2 knockdown (Fig. 6A, P<0.01). These data suggest that RRM2 suppression may enhance the sensitivity of CML cells to imatinib via the Akt pathway.
Figure 6.
RRM2 knockdown suppresses the activation of the Akt pathway. (A) RRM2 knockdown decreased the level of phosphorylated Akt (p-Akt). Upper panel, western blot band; lower histogram, the relative band density. **P<0.01, as compared with the siCT group. NS, not significant. (B) Diagram depicts the role of RRM2 in IR, a novel type of Bcr-Abl-independent resistance in CML. RRM2, ribonucleotide reductase regulatory subunit M2; IR, imatinib-resistant; CML, chronic myeloid leukemia.
Discussion
Ribonucleotide reductase (RNR) is a multimeric enzyme that catalyzes the conversion of ribonucleoside diphosphates to deoxyribonucleoside diphosphates. It contains two subunits, a large subunit RRM1 and a small subunit RRM2. RRM1 and RRM2 form an active enzyme (27). RRM1 is a dimer of Mr 170,000 that contains nucleotide binding sites and is responsible for the complex allosteric regulation of the enzyme. RRM2 is a dimer of Mr 88,000 that contains stoichiometric amounts of non-heme iron and a unique tyrosyl free radical, which is essential for its activity (28).
RRM2 is a pivotal molecule that modulates the activity of RNR, which is essential for DNA synthesis. It was reported that RRM2 is a determinant of malignant cellular behavior in a variety of human cancers, such as breast cancer (29), gastric cancer (30), cervical cancer (31), glioblastoma (32), and head and neck cancer (33). RRM2 also plays a role in the chemoresistance process. Duxbury et al indicated that RRM2 overexpression is associated with gemcitabine chemoresistance in pancreatic adenocarcinoma cells, and that the suppression of RRM2 enhances gemcitabine-induced cytotoxicity both in vitro and in vivo (34). Huang et al used an integrative bioinformatics approach and found that RRM2 was overexpressed in tamoxifen-resistant breast tumors (35).
The present study was designed to identify various novel genes that are responsible for imatinib resistance (IR) in chronic myeloid leukemia (CML) treatment. The RNA-seq approach was employed, and we observed that the transcription profiles of many genes were altered in IR patients. KEGG pathway analysis revealed 7 genes that may be involved in the IR process. After confirming these genes by qF-PCR in both the patient samples and cell lines, we finally selected RRM2 as the optimal gene and investigated its molecular function in the IR process.
The concentration effect experiment showed that imatinib can induce a dose-dependent increase in RRM2 levels in IR cells, and knockdown of RRM2 enhanced the sensitivity of K562G cells to imatinib therapy (Figs. 2 and 3). Further investigation revealed that the overexpression of RRM2 may induce imatinib drug resistance and antiapoptosis through the Bcl-2/caspase pathway (Figs. 4 and 5). Bcl-2 family members regulate the mitochondrial pathway of apoptosis (36). The ratio between Bcl-2 and Bax is important for regulating the release of cytochrome c from mitochondria, which activates caspase-3 and induces apoptosis (37). Rahman et al demonstrated that RRM2 depletion significantly reduced Bcl-2 protein expression in head and neck squamous cell carcinoma (HNSCC) and non-small cell lung cancer (NSCLC) cells. They observed that RRM2 regulates Bcl-2 protein stability, with RRM2 suppression leading to increased Bcl-2 degradation, and demonstrated their colocalization (26). Our results are consistent with those of Rahman et al.
Activation of the alternative signaling pathway is an important mechanism in the Bcr-Abl-independent resistance of imatinib. The crosstalk between RRM2 and the Akt pathway may be a novel alternative signaling pathway for IR. Zhong et al found that the overexpression of RRM2 promotes gastric cancer cell invasion via the Akt/NF-κB signaling pathway (13). Shah et al found that inhibition of RRM2 significantly reversed Akt-induced tamoxifen-resistant cell growth and inhibited breast cancer cell motility. They also indicated that Akt-expression upregulated RRM2 levels, leading to increased DNA repair and protection from tamoxifen-induced apoptosis (16). The Akt signaling pathway plays an important role in the response to extracellular stimuli to regulate cell growth, apoptosis and survival (38). In our experimental model, we observed that the knockdown of RRM2 suppressed the activation of Akt (Fig. 6A). We speculate that there exists feedback regulation between RRM2 and Akt, and therefore RRM2 affects IR through the Akt signaling pathway (Fig. 6B).
To conclude, in the present study, RRM2 was found to be elevated in both the IR CML patients and an IR cell line. It was also demonstrated that RRM2 may affect the cell survival via the Bcl-2/caspase cell apoptotic pathway and the Akt cell signaling pathway in imatinib therapy. It is noteworthy that different CML phases (chronic phase or accelerated phase) probably affect the level of a certain carcinoma biomarker (39), however, this was not taken into consideration in the present study. Therefore, to gain deeper insight into the predictive value of RRM2 for IR, a larger sample size and a more detailed sample classification are still needed in subsequent investigations.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by grants from the Provincial Science Fund of Jilin Provincial Department of Science and Technology (no. 20190201041JC to CL) and the Ability Improvement Fund of Health Commission of Jilin Province (no. 2018J065).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SG contributed to the design of the study and wrote the manuscript. CL and YL performed the experiments and analyzed the data. RH was involved in conducting the experiments and analyzing of the data. WH provided constructive comments and discussions. All authors have read and approved this manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Permission to use the clinical samples for research purposes was obtained and approved by the Ethics Committee of the First Hospital of Jilin University. Informed consents were obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
- 1.Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, Geurts van Kessel A, Bootsma D, Grosveld G, Ferguson-Smith MA, Davies T, Stone M, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–280. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- 2.Deininger MW. Milestones and monitoring in patients with CML treated with imatinib. Hematology Am Soc Hematol Educ Program. 2008:419–426. doi: 10.1182/asheducation-2008.1.419. [DOI] [PubMed] [Google Scholar]
- 3.Quintás-Cardama A, Cortes JE, Kantarjian HM. Early cytogenetic and molecular response during first-line treatment of chronic myeloid leukemia in chronic phase: Long-term implications. Cancer. 2011;117:5261–5270. doi: 10.1002/cncr.26196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochhaus A, Larson RA, Guilhot F, Radich JP, Branford S, Hughes TP, Baccarani M, Deininger MW, Cervantes F, Fujihara S, et al. Long-term outcomes of imatinib treatment for chronic myeloid leukemia. N Engl J Med. 2017;376:917–927. doi: 10.1056/NEJMoa1609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bixby D, Talpaz M. Mechanisms of resistance to tyrosine kinase inhibitors in chronic myeloid leukemia and recent therapeutic strategies to overcome resistance. Hematology. Hematology. Am Soc Hematol Educ Program. 2009:461–476. doi: 10.1182/asheducation-2009.1.461. [DOI] [PubMed] [Google Scholar]
- 6.Weisberg E, Griffin JD. Mechanism of resistance to the ABL tyrosine kinase inhibitor STI571 in BCR/ABL-transformed hematopoietic cell lines. Blood. 2000;95:3498–3505. [PubMed] [Google Scholar]
- 7.le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, Marchesi E, Supino R, Gambacorti-Passerini C. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–1766. [PubMed] [Google Scholar]
- 8.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 9.Alves R, Fonseca AR, Goncalves AC, Ferreira-Teixeira M, Lima J, Abrantes AM, Alves V, Rodrigues-Santos P, Jorge L, Matoso E, et al. Drug transporters play a key role in the complex process of Imatinib resistance in vitro. Leuk Res. 2015;39:355–360. doi: 10.1016/j.leukres.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Gambacorti-Passerini C, Zucchetti M, Russo D, Frapolli R, Verga M, Bungaro S, Tornaghi L, Rossi F, Pioltelli P, Pogliani E, et al. Alpha1 acid glycoprotein binds to imatinib (STI571) and substantially alters its pharmacokinetics in chronic myeloid leukemia patients. Clin Cancer Res. 2003;9:625–632. [PubMed] [Google Scholar]
- 11.Bozkurt S, Özkan T, Özmen F, Baran Y, Sunguroğlu A, Kansu E. The roles of epigenetic modifications of proapoptotic BID and BIM genes in imatinib-resistant chronic myeloid leukemia cells. Hematology. 2013;18:217–223. doi: 10.1179/1607845412Y.0000000056. [DOI] [PubMed] [Google Scholar]
- 12.Hentschel J, Rubio I, Eberhart M, Hipler C, Schiefner J, Schubert K, Loncarevic IF, Wittig U, Baniahmad A, von Eggeling F. BCR-ABL- and Ras-independent activation of Raf as a novel mechanism of Imatinib resistance in CML. Int J Oncol. 2011;39:585–591. doi: 10.3892/ijo.2011.1062. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Z, Cao Y, Yang S, Zhang S. Overexpression of RRM2 in gastric cancer cell promotes their invasiveness via AKT/NF-κB signaling pathway. Pharmazie. 2016;71:280–284. [PubMed] [Google Scholar]
- 14.Grolmusz VK, Karászi K, Micsik T, Tóth EA, Mészáros K, Karvaly G, Barna G, Szabó PM, Baghy K, Matkó J, et al. Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am J Cancer Res. 2016;6:2041–2053. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Meng L, Wang XW, Ma GY, Chen JH. Expression of RRM1 and RRM2 as a novel prognostic marker in advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol. 2014;35:1899–1906. doi: 10.1007/s13277-013-1255-4. [DOI] [PubMed] [Google Scholar]
- 16.Shah KN, Mehta KR, Peterson D, Evangelista M, Livesey JC, Faridi JS. AKT-induced tamoxifen resistance is overturned by RRM2 inhibition. Mol Cancer Res. 2014;12:394–407. doi: 10.1158/1541-7786.MCR-13-0219. [DOI] [PubMed] [Google Scholar]
- 17.Tu M, Li H, Lv N, Xi C, Lu Z, Wei J, Chen J, Guo F, Jiang K, Song G, et al. Vasohibin 2 reduces chemosensitivity to gemcitabine in pancreatic cancer cells via Jun proto-oncogene dependent transactivation of ribonucleotide reductase regulatory subunit M2. Mol Cancer. 2017;16:66. doi: 10.1186/s12943-017-0619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura Y, Kasamatsu A, Nakashima D, Yamatoji M, Minakawa Y, Koike K, Fushimi K, Higo M, Endo-Sakamoto Y, Shiiba M, et al. ARNT2 regulates tumoral growth in oral squamous cell carcinoma. J Cancer. 2016;7:702–710. doi: 10.7150/jca.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joha S, Dauphin V, Leprêtre F, Corm S, Nicolini FE, Roumier C, Nibourel O, Grardel N, Maguer-Satta V, Idziorek T, et al. Genomic characterization of Imatinib resistance in CD34+ cell populations from chronic myeloid leukaemia patients. Leuk Res. 2011;35:448–458. doi: 10.1016/j.leukres.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Yang B, Zhang H, Song J, Zhang Y, Xing J, Yang Z, Wei C, Xu T, Yu Z, et al. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int J Biol Sci. 2019;15:533–543. doi: 10.7150/ijbs.30114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An C, Guo H, Wen XM, Tang CY, Yang J, Zhu XW, Yin JY, Liu Q, Ma JC, Deng ZQ, et al. Clinical significance of reduced SFRP1 expression in acute myeloid leukemia. Leuk Lymphoma. 2015;56:2056–2060. doi: 10.3109/10428194.2014.977883. [DOI] [PubMed] [Google Scholar]
- 22.Ni M, Rui Y, Chen Q, Wang Y, Li G. Effect of growth differentiation factor 7 on tenogenic differentiation of bone marrow mesenchymal stem cells of rat in vitro. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2011;25:1103–1109. (In Chinese) [PubMed] [Google Scholar]
- 23.Weich N, Ferri C, Moiraghi B, Bengió R, Giere I, Pavlovsky C, Larripa IB, Fundia AF. GSTM1 and GSTP1, but not GSTT1 genetic polymorphisms are associated with chronic myeloid leukemia risk and treatment response. Cancer Epidemiol. 2016;44:16–21. doi: 10.1016/j.canep.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Čokić VP, Mojsilović S, Jauković A, Kraguljac-Kurtović N, Mojsilović S, Šefer D, Mitrović Ajtić O, Milošević V, Bogdanović A, Đikić D, et al. Gene expression profile of circulating CD34(+) cells and granulocytes in chronic myeloid leukemia. Blood Cells Mol Dis. 2015;55:373–381. doi: 10.1016/j.bcmd.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z, Wang Z, Sica G, Deng X, Chen ZG, Shin DM. RRM2 regulates Bcl-2 in head and neck and lung cancers: A potential target for cancer therapy. Clin Cancer Res. 2013;19:3416–3428. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordheim LP, Sève P, Trédan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12:693–702. doi: 10.1016/S1470-2045(10)70244-8. [DOI] [PubMed] [Google Scholar]
- 28.Engström Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 29.Liang WH, Li N, Yuan ZQ, Qian XL, Wang ZH. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog. 2019;58:461–473. doi: 10.1002/mc.22941. [DOI] [PubMed] [Google Scholar]
- 30.Kang W, Tong JH, Chan AW, Zhao J, Wang S, Dong Y, Sin FM, Yeung S, Cheng AS, Yu J, To K. Targeting ribonucleotide reductase M2 subunit by small interfering RNA exerts anti-oncogenic effects in gastric adenocarcinoma. Oncol Rep. 2014;31:2579–2586. doi: 10.3892/or.2014.3148. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Zhan T, Ke T, Huang X, Ke D, Wang Q, Li H. Increased expression of RRM2 by human papillomavirus E7 oncoprotein promotes angiogenesis in cervical cancer. Br J Cancer. 2014;110:1034–1044. doi: 10.1038/bjc.2013.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Zheng J, Chen S, Huang B, Li G, Feng Z, Wang J, Xu S. RRM2 promotes the progression of human glioblastoma. J Cell Physiol. 2018;233:6759–6767. doi: 10.1002/jcp.26529. [DOI] [PubMed] [Google Scholar]
- 33.Rahman MA, Amin AR, Wang X, Zuckerman JE, Choi CH, Zhou B, Wang D, Nannapaneni S, Koenig L, Chen Z, et al. Systemic delivery of siRNA nanoparticles targeting RRM2 suppresses head and neck tumor growth. J Control Release. 2012;159:384–392. doi: 10.1016/j.jconrel.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 35.Huang L, Zhao S, Frasor JM, Dai Y. An integrated bioinformatics approach identifies elevated cyclin E2 expression and E2F activity as distinct features of tamoxifen resistant breast tumors. PLoS One. 2011;6:e22274. doi: 10.1371/journal.pone.0022274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 38.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh N, Tripathi AK, Sahu DK, Mishra A, Linan M, Argente B, Varkey J, Parida N, Chowdhry R, Shyam H, et al. Differential genomics and transcriptomics between tyrosine kinase inhibitor-sensitive and -resistant BCR-ABL-dependent chronic myeloid leukemia. Oncotarget. 2018;9:30385–30418. doi: 10.18632/oncotarget.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.