Abstract
Gastric cancer is one of the most common types of human cancer, and it is additionally one of the leading causes of cancer-associated mortality worldwide. Previous studies have suggested that interleukin (IL)-10 may contribute to the pathogenesis of gastric cancer. However, the underlying mechanisms remain unclear. In the present study, it was observed that the expression of IL-10 was significantly upregulated in gastric tumor tissues and serum samples of patients with gastric cancer. Furthermore, IL-10 was increased in the cell culture supernatant of cancer-associated macrophages (CAMs). Treatment with cell culture supernatant from CAMs induced a significant increase in proliferation and migration, while it suppressed apoptosis, in MGC-803 and BGC-823 gastric cancer cells. Notably, application of an inhibitory IL-10 antibody partially blocked the cell culture supernatant of CAM-induced oncogenic effects. RNA-sequencing analysis was then performed to identify the differentially expressed genes in MGC-803 cells treated with IL-10. Based on the sequencing results and in vitro analysis, it was demonstrated that IL-10-induced carcinogenic behaviors in MGC-803 cells were potentially mediated by activation of the c-Met/STAT3 signaling pathway. In conclusion, the present results demonstrated that IL-10 secreted by CAMs may be involved in the pathogenesis of gastric cancer, suggesting that IL-10 may serve as a potential therapeutic target for the treatment of gastric cancer.
Keywords: interleukin-10, cancer-associated macrophages, gastric cancer, sequencing
Introduction
Gastric cancer is a common human gastrointestinal cancer, and it is additionally one of the leading causes of cancer-associated mortality (1). Similar to numerous other cancer types, the majority of patients with gastric cancer are diagnosed at an advanced or metastatic stage when they came to the hospital (2), leading to the poor prognosis of the disease (3–5). As a result, identifying novel biomarkers and therapeutic targets for the early diagnosis and prognosis of gastric cancer is urgently required.
Interleukin (IL)-10 is a cytokine that is encoded by the IL10 gene. The receptor consists of two different chains: IL-10 receptor 1 and IL-10 receptor 2 (6). In the human body, IL-10 is primarily produced by immune cells, including monocytes, type 2 T helper cells and regulatory T cells. IL-10 may exert its functions by regulating important signaling pathways, including the extracellular signal-regulated kinase 1/2, signal transducer and activator of transcription 3 (STAT3) and nuclear factor-κB signaling pathways, and affecting the expression of downstream genes (7,8). The roles of IL-10 in carcinogenesis have been discussed previously; however, the underlying mechanism requires further investigation.
In recent years, research on the tumor microenvironment has attracted increasing attention. Previous studies have demonstrated that the tumor microenvironment serves a key role in the progression of cancer (9,10). In the majority of solid tumors, cancer-associated macrophages (CAMs) are typically identified as M2 phenotype macrophages and an increased number of CAMs is correlated with poor prognosis in numerous types of cancer (11,12), including gastric cancer (13,14). The results of previous studies have demonstrated that the underlying interactions among CAMs, cancer cells and cytokines, serve important roles in the pathogenesis of various types of cancer, and targeting CAMs has emerged as a novel method for the treatment of cancer.
The present study aimed to examine the roles and associated mechanisms of IL-10 secreted by CAMs in the pathogenesis of gastric cancer. The expression levels of IL-10 were examined in tumor tissues and serum samples of patients with gastric cancer. The expression of IL-10 in CAMs and normal macrophages was compared. Furthermore, the roles of IL-10 in proliferation, apoptosis and migration of gastric cancer cells were investigated. RNA-sequencing analysis was performed to identify critical genes that were differentially expressed in gastric cancer cells with and without IL-10, and the effect of IL-10 on the activation of the c-Met/STAT3 signaling pathway was examined. The present results may provide novel insight for IL-10 as a potential therapeutic target for gastric cancer.
Materials and methods
Patients and clinical tissue samples
In total, 20 pairs of gastric tumor tissues and adjacent normal tissues were collected from patients (11 males and 9 females, 58–72 years old, median age 63) with gastric cancer that enrolled at the Institute of Digestive Endoscopy and Medical Center for Digestive Disease between May 2017 to March 2018 at the Second Affiliated Hospital, Nanjing Medical University (Nanjing, China). The tissue samples were immediately frozen in liquid nitrogen following surgery and stored at −80°C until required. The serum of every patient was additionally collected and stored at −80°C, and the serum samples of 20 healthy volunteers served as the control group. All patients were pathologically diagnosed with gastric cancer, and patients subjected to pre-operative radiotherapy and/or chemotherapy were excluded from the present study. All patients signed an informed consent form, and the present study was approved by the Ethical Committee of Nanjing Medical University.
Cell culture and treatment
For the in vitro differentiation of human monocytes (THP-1 cells; American Type Culture Collection) into CAMs, the cells were incubated for 48 h with 20 ng/ml IL-4 and 20 ng/ml IL-13 (PeproTech, Inc.) to obtain M2 polarized macrophages. Human gastric cell lines MGC-803 and BGC-823 (American Type Culture Collection) were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator at 5% CO2. Cells were either treated with the supernatant from CAMs or the supernatant from CAMs+IL-10 antibody (cat. no. ab133575; 1:2,000; Abcam) for 72 h for further analysis. For cell profiling, MGC-803 cells were treated with IL-10 overexpression vector (Origene) and empty vector (Origene).
ELISA
The serum expression level of IL-10 in patients and the expression of IL-10 in the cell culture supernatant of CAMs were measured using an ELISA kit (cat. no. H009; Nanjing Jiancheng Bio-Engineering Institute Co., Ltd.) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the clinical tissue samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. cDNA was synthesized using the PrimeScript RT Master Mix kit (Takara Biotechnology Co., Ltd.). Subsequently, qPCR was performed to examine the expression of IL-10 using SYBR premix Ex Taq (Takara Biotechnology Co., Ltd.) on an ABI 7500 Real-Time PCR System. Relative mRNA expression levels were calculated using the 2−∆∆Cq method (15), with GAPDH as the internal control. The thermocycling conditions were 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The primers were as follows: IL-10, 5′-GCCAGAGCCACATGCTCCTA-3′ (forward) and 5′-GATAAGGCTTGGCAACCCAAGTAA-3′ (reverse); and GAPDH, 5′-AAATGGTGAAGGTCGGTGTGAAC-3′ (forward) and 5′-CAACAATCTCCACTTTGCCACTG-3′ (reverse).
Cell proliferation analysis
Following treatment for 72 h, the proliferation of MGC-803 and BGC-823 cells was determined by Cell Counting Kit-8 (CCK-8), according to the manufacturer's protocol. Cells were collected and seeded onto 96-well plates, and 10 µl CCK-8 reagent was added to each well. Subsequently, the plate was incubated for 4 h. The optical density value was measured at 450 nm with a microplate reader.
Cell apoptosis analysis
The apoptosis of MGC-803 and BGC-823 cells following different treatments was analyzed using an Annexin V-fluorescein isothiocyanate (FITC) apoptosis kit (BD Biosciences) by flow cytometry. Cells were collected and stained with Annexin V-FITC and propidium iodide solutions. Subsequently, the apoptosis of the cells was examined using a FACSCalibur flow cytometer (BD Biosciences) with CellQuest 6.0 software (BD Biosciences, Franklin Lakes, NJ, USA).
Scratch wound healing assay
MGC-803 and BGC-823 cells (~2×105) were cultured until they reached 100% confluence in 6-well plates and the monolayer was then scratched using a 10 µl pipette tip. The plate was subsequently incubated at 37°C with 5% CO2 for 24 h. The images of the migration area from five fields of the control group and treatment group were captured with an inverted light microscope (magnification, ×200) and analyzed using ImageJ (National Institutes of Health).
Matrigel invasion assay
A cell invasion assay was conducted using 24-well plates with Transwell chambers (Corning, Inc.). Cells (4×104) were seeded onto the upper Matrigel-coated chambers containing DMEM culture medium (300 µl) without any bovine serum and the lower chambers were filled with DMEM medium containing 10% FBS. After incubation for 24 h, cells that had invaded the membrane were fixed with methanol and stained with crystal violet. The images of the stained membrane were captured from five randomly fields using an inverted fluorescence microscope (magnification, ×200).
Western blot analysis
Total protein was isolated from MGC-803 and BGC-823 cells using a lysis buffer (Beyotime Institute of Biotechnology). Protein concentrations were determined using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). The proteins were subsequently separated by 10% SDS-PAGE, and transferred onto a polyvinylidene fluoride membrane, blocked with 5% non-fat milk for 1 h at room temperature and incubated with primary antibodies against c-Met (cat. no. ab51067; 1:2,000), STAT3 (cat. no. ab76315; 1:5,000), phosphorylated (p)-STAT3 (cat. no. ab32143; 1:2,000), matrix metalloproteinase (MMP) 9 (cat. no. ab219372; 1:2,000) and caspase-3 (cat. no. ab13847; 1:500; all purchased from Abcam) at 4°C overnight. GAPDH was used for normalization. On day 2, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (cat. no. ab6721; 1:5,000) and detected using a ChemiDoc™ XRS+ imaging system (Bio-Rad Laboratories, Inc.). Relative protein expression was quantified by ImageJ software (version 1.47; National Institutes of Health).
RNA isolation, construction of cDNA library and RNA-sequencing
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract the total RNA from the cell samples of different treatments, according to the manufacturer's instructions. The quality of the total RNA was evaluated by Nanodrop 2000 spectrophotometry (Thermo Fisher Scientific, Inc.). RNA samples with 260/280 nm optical density ratio 1.8–2.0 were used for cDNA synthesis using the 1st Strand Enzyme Mix (Vazyme). The second strand cDNA was synthesized with 2nd Strand Marking Buffer and 2nd Strand/End Repair Enzyme Mix (Vazyme). Then, the cDNA library was constructed by PCR as previously published (16), and RNA-sequencing was performed with Illumina HiSeq XTen (Vazyme).
The quantitative analysis was based on reads per kb per million reads (RPKM). Differentially expressed genes (DEGs) were selected by adjusted P<0.05 for multiple tests using the Benjamini-Hochberg Method (17). Functional annotation of the genes was performed by Gene Ontology (GO) analysis (http://geneontology.org). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were annotated using ggplot2 program (https://ggplot2.tidyverse.org/reference/) and mapped onto KEGG pathways (18).
Statistical analysis
Data are presented as the mean ± standard deviation. Comparisons between two groups were analyzed by a t-test, and comparisons among multiple groups were analyzed by analysis of variance followed by Turkey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Increased expression of IL-10 in tissue and serum samples of patients with gastric cancer
The expression levels of IL-10 in tumor tissue sand adjacent normal tissues from patients with gastric cancer were detected by RT-qPCR. As presented in Fig. 1A, the expression of IL-10 was significantly increased in gastric tumor tissues compared with adjacent tissues (P<0.01; Fig. 1A). Furthermore, IL-10 levels in serum samples from patients with gastric cancer and healthy volunteers were compared; the results revealed that the serum levels of IL-10 were significantly increased in patients with gastric cancer compared with healthy volunteers (P<0.01; Fig. 1B).
Figure 1.
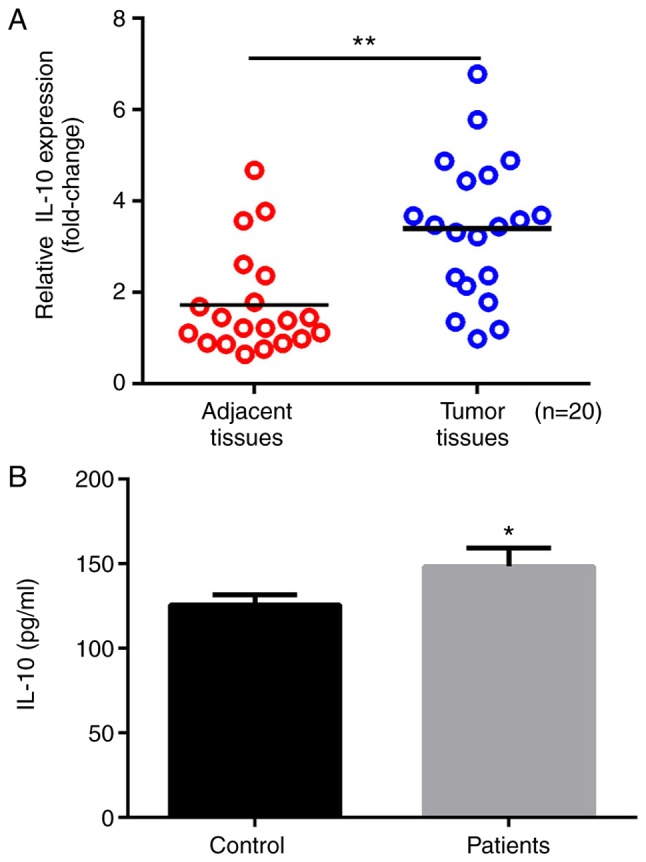
Increased expression of IL-10 in tissue samples and serum of patients with gastric cancer. (A) mRNA expression levels of IL-10 in tumor tissue samples and adjacent normal tissues from patients with gastric cancer. (B) IL-10 levels in serum of patients with gastric cancer and healthy volunteers measured by ELISA. *P<0.05 and **P<0.01. IL-10, interleukin-10.
Increased IL-10 levels in the cell culture supernatant of CAMs
Subsequently, THP-1 cells were treated with IL-4 and IL-13 to induce their differentiation to CAMs in vitro, cultured for 72 h, and the IL-10 levels in the cell culture supernatant were evaluated by ELISA. It was observed that, compared with the untreated THP-1 cells, the expression of IL-10 was significantly upregulated in the cell culture supernatant of CAMs (P<0.01; Fig. 2).
Figure 2.
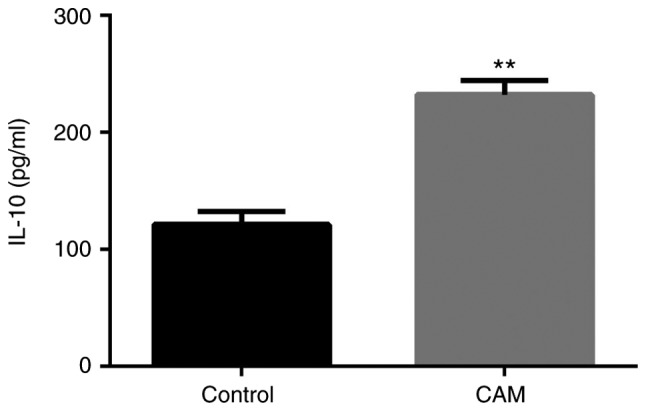
IL-10 levels in CAMs and untreated THP-1 cells. The levels of IL-10 in the cell culture supernatant of control untreated THP-1 cells and THP-1 cells differentiated to CAMs were evaluated by ELISA. **P<0.01. IL-10, interleukin-10; CAM, cancer-associated macrophage.
IL-10 affects proliferation and apoptosis in gastric cancer cells in vitro
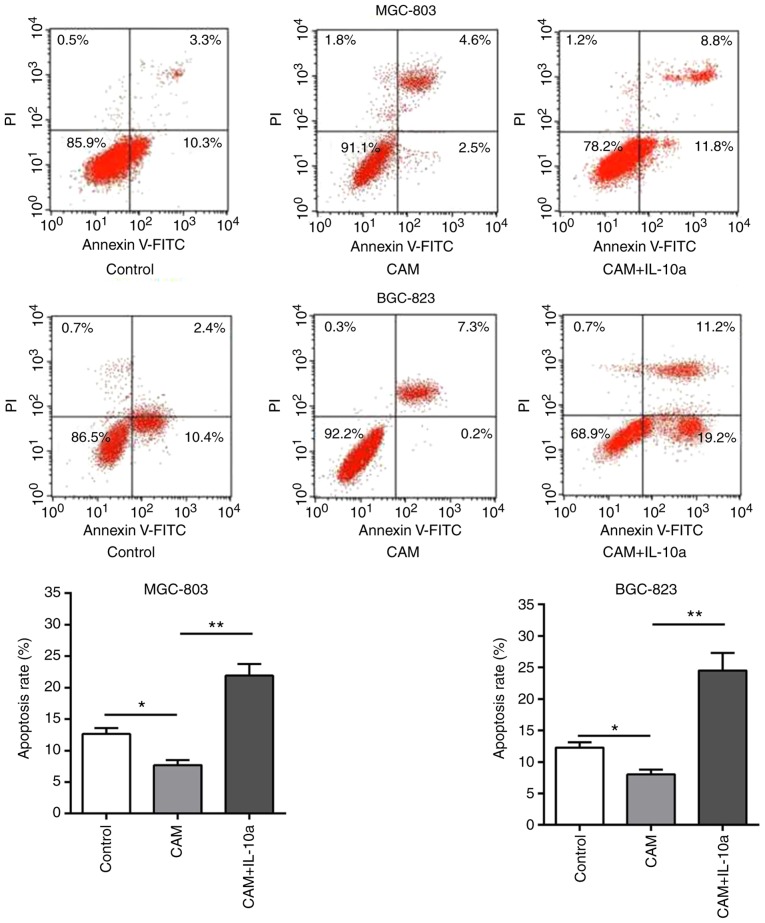
The effects of the cell culture supernatant of CAMs on the proliferation and apoptosis of MGC-803 and BGC-823 gastric cancer cells were evaluated by CCK-8 and flow cytometry, respectively. It was observed that, compared with the untreated cells, treatment with the cell culture supernatant of CAMs induced a significant increase in the proliferation (P<0.01; Fig. 3) and a decrease in the apoptosis (P<0.01; Fig. 4) of MGC-803 and BGC-823 cells. Notably, addition of an IL-10 inhibitory antibody blocked these effects (Figs. 3 and 4), suggesting that the tumor-promoting function of the CAM-conditioned media was mediated by IL-10.
Figure 3.
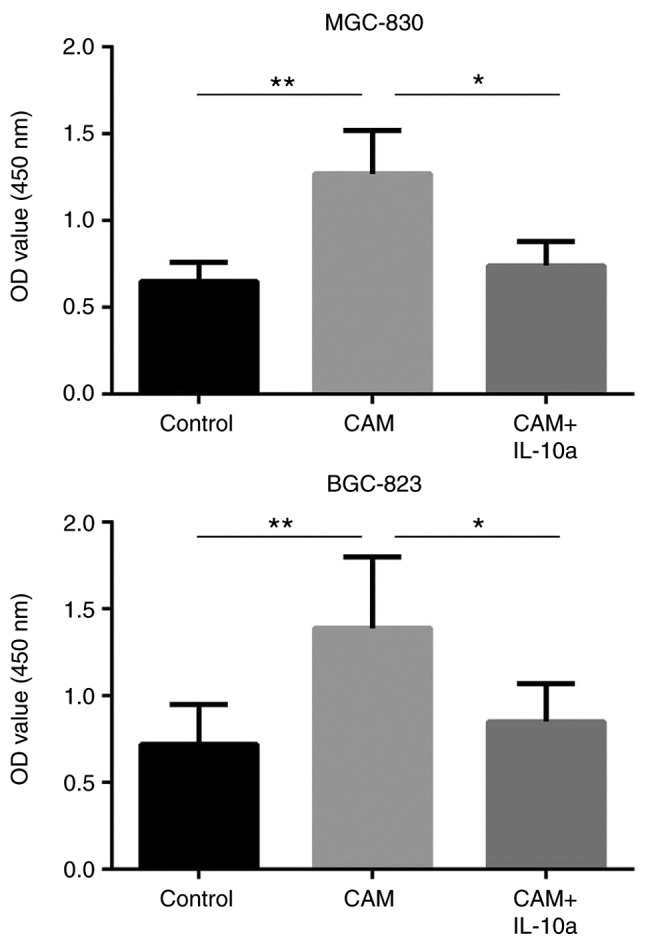
Effect of IL-10 on the proliferation of gastric cells in vitro. Proliferation of MGC-803 and BGC-823 cells following different treatments was evaluated by CCK-8 assay. *P<0.05 and **P<0.01, with comparisons indicated by lines. IL-10, interleukin-10; OD, optical density; CAM, cancer-associated macrophage; IL-10a, IL-10 inhibitory antibody.
Figure 4.
Effect of IL-10 on the apoptosis of gastric cells in vitro. The apoptosis rates of MGC-803 and BGC-823 cells following different treatments were evaluated by flow cytometry. Representative plots and quantification is shown. *P<0.05 and **P<0.01, with comparisons indicated by lines. IL-10, interleukin-10; PI, propidium iodide; FITC, fluorescein isothiocyanate; CAM, cancer-associated macrophage; IL-10a, IL-10 inhibitory antibody.
IL-10 affects migration and invasion in gastric cancer cells in vitro
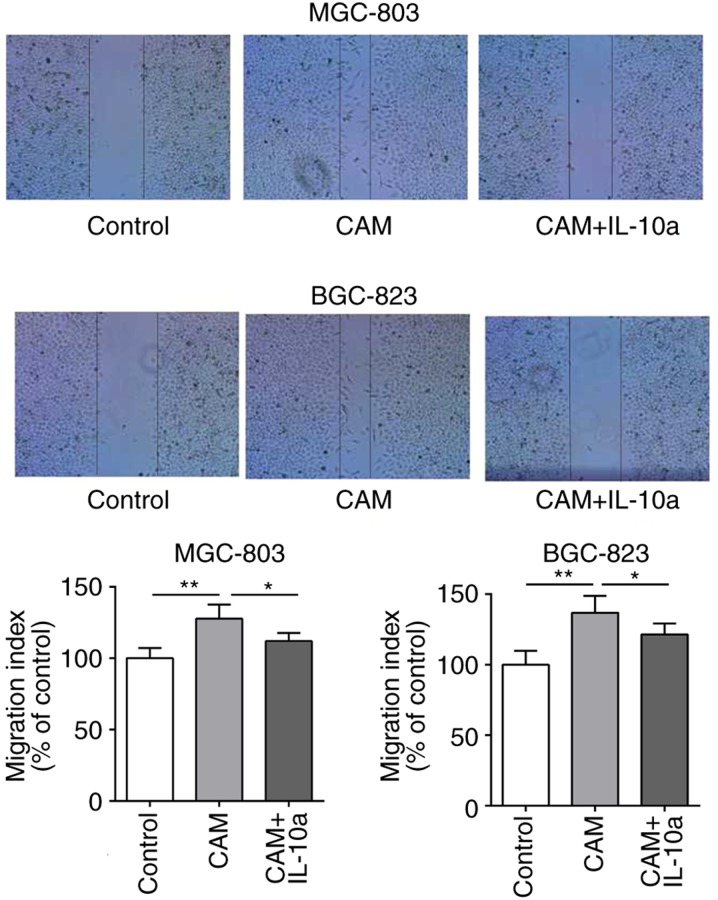
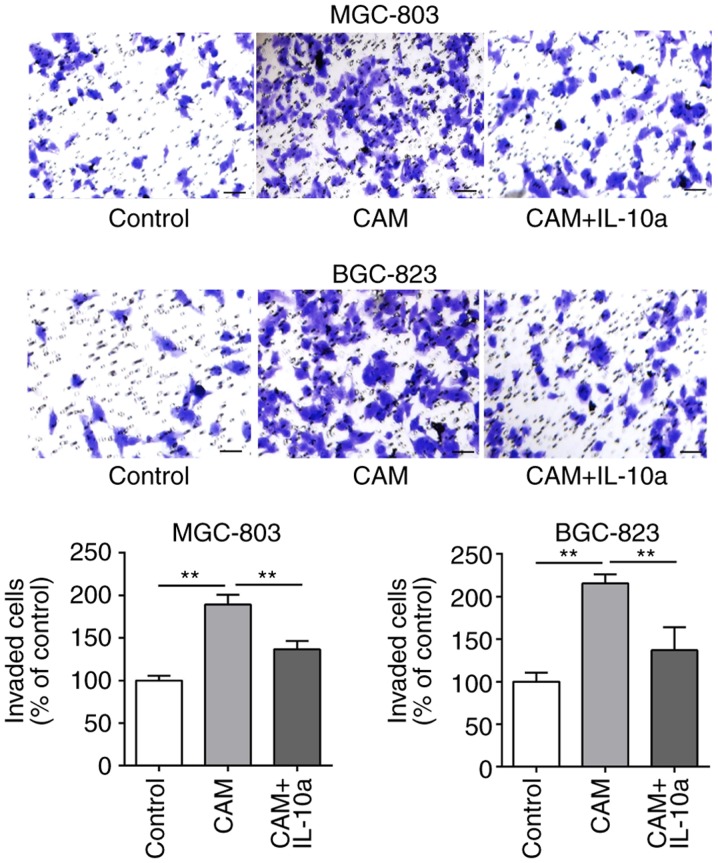
The effects of the cell culture supernatant of CAMs on the migratory and invasive abilities of MGC-803 and BGC-823 cells was examined. As presented in Figs. 5 and 6, treatment with the cell culture supernatant of CAMs induced a significant increase in the migratory and invasive abilities of MGC-803 and BGC-823 cells, whereas the addition of the IL-10 inhibitory antibody partially blocked the CAM-conditioned media-induced effects.
Figure 5.
Effect of IL-10 on the migration of gastric cancer cells in vitro. The migration ability of MGC-803 and BGC-823 cells following different treatments was evaluated by scratch wound healing assay. *P<0.05 and **P<0.01, with comparisons indicated by lines. IL-10, interleukin-10; CAM, cancer-associated macrophage; IL-10a, IL-10 inhibitory antibody.
Figure 6.
Effect of IL-10 on the invasion of gastric cancer cells in vitro. The invasion ability of MGC-803 and BGC-823 cells following different treatments was evaluated by Transwell assay. Scale bar. 50 µm. **P<0.01, with comparisons indicated by lines. IL-10, interleukin-10; CAM, cancer-associated macrophage; IL-10a, IL-10 inhibitory antibody.
Differential gene expression profiling of MGC-803 cells treated with or without IL-10
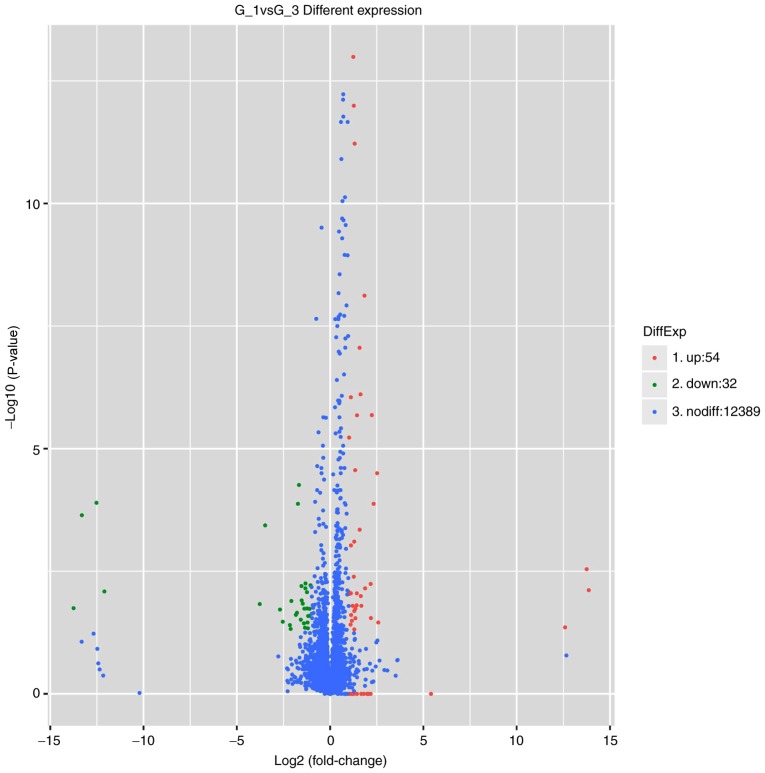
To profile the differentially expressed genes of MGC-803 cells treated with IL-10 overexpression vector (Origene) and empty vector (Origene), genome-wide RNA-sequencing was performed. As presented in Fig. 7, it was identified that 86 genes were significantly altered in MGC-803 cells treated with IL-10 compared with untreated MGC-803 cells. Of those, 54 genes were upregulated and 32 genes were downregulated. The top upregulated and downregulated genes are listed in Table I. The significantly upregulated genes included oncogenes IL-6, IL-32, insulin like growth factor binding protein 3, C-X-C motif chemokine ligand 2, Pim-2 proto-oncogene and STAT3; the significantly downregulated genes included tumor suppressors, such as tensin 2 and LIM domain and actin binding 1 (Table I).
Figure 7.
Differentially expressed genes between untreated and IL-10-treated MGC-803 cells analyzed by RNA-sequencing. The volcano plot was generated using the ggplot2 program (https://ggplot2.tidyverse.org/reference/). IL-10, interleukin-10.
Table I.
Differentially expressed genes by RNA-sequencing.
| A, Upregulated | |
|---|---|
| Gene name | Log2 fold change |
| CXCL2 | 5.4087 |
| CXCL8 | 2.58106 |
| STAT3 | 2.52124 |
| IL-6 | 2.33332 |
| IL-32 | 2.23426 |
| IGFBP3 | 2.184 |
| PIM2 | 2.17285 |
| ANGPTL4 | 2.17171 |
| RND1 | 2.0646 |
| CXCL3 | 1.98878 |
| B, Downregulated | |
| Gene name | Log2 fold change |
| SNORD3A | −3.47926 |
| HIST1H2BF | −2.68504 |
| TNS2 | −2.53616 |
| TRIM17 | −2.15445 |
| OGFRP1 | −2.11346 |
| LIMA1 | −2.07622 |
| MARCO | −1.83237 |
| CLYBL | −1.77506 |
| RDH5 | −1.72344 |
| FUT4 | −1.67064 |
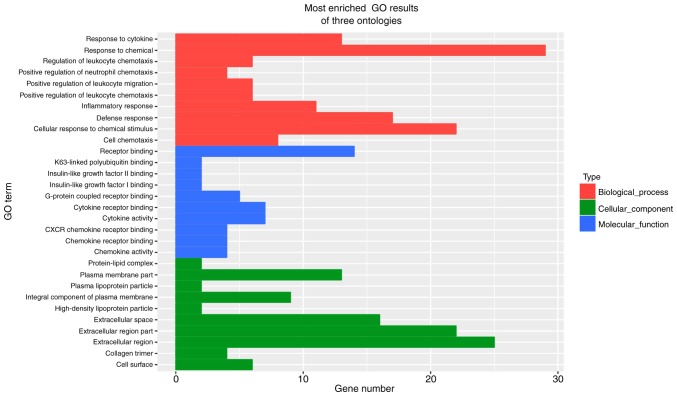
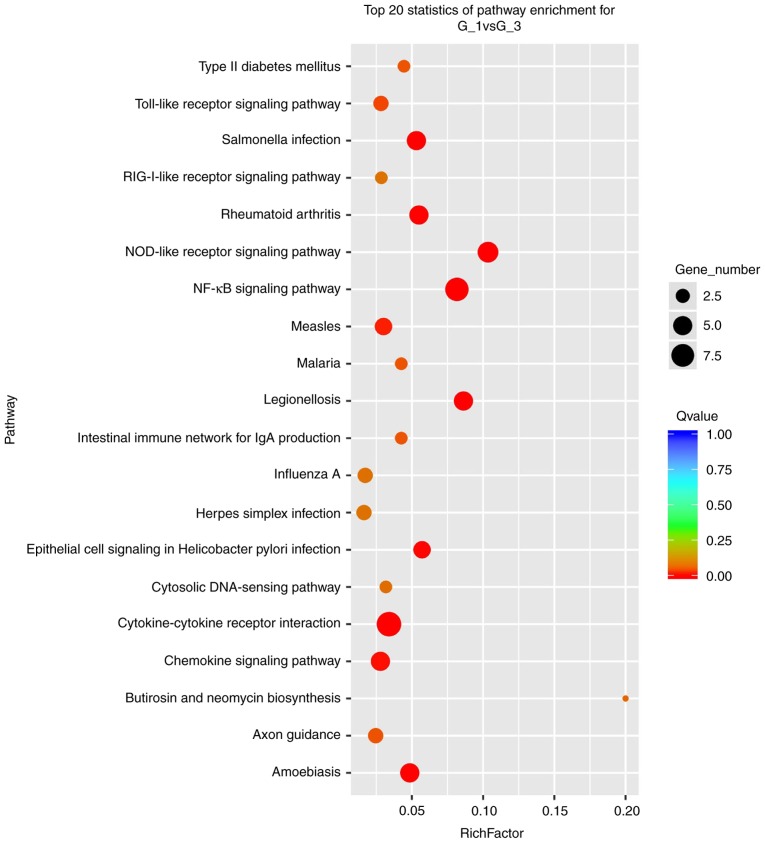
To understand the important biological processes affected by the IL-10 treatment on MGC-803 cells, a GO analysis was performed. The results revealed that IL-10-affected genes were enriched in the categories response to chemical, extracellular region, extracellular region part, cellular response to chemical stimulus, defense response, extracellular space, receptor binding, plasma membrane part, response to cytokine, inflammatory response, integral component of plasma membrane, cell chemotaxis, cytokine activity, cytokine receptor binding, cell surface, positive regulation of leukocyte chemotaxis, regulation of leukocyte chemotaxis, positive regulation of leukocyte migration and G-protein coupled receptor binding (Fig. 8). Furthermore, in terms of cellular component, it was identified that the most abundant genes were focused on cytoplasm, organelle, membrane, membrane-bounded organelle, intracellular organelle, intracellular membrane-bounded organelle, extracellular region, cytoplasmic part, extracellular region part, cell periphery (Fig. 8). In addition, as far as molecular function is concerned, the most enriched GO terms were binding, protein binding, ion binding, receptor binding, catalytic activity, heterocyclic compound binding, organic cyclic compound binding, metal ion binding and cation binding (Fig. 8). Finally, the results from KEGG pathway enrichment analysis are presented in Fig. 9. The Nod-like receptor, the NF-kB and the chemokine signaling pathways were amongst the top 20 most significantly enriched.
Figure 8.
GO analysis of the differentially expressed genes between untreated and IL-10-treated MGC-803 cells. GO, gene ontology; IL-10, interleukin-10.
Figure 9.
Results of KEGG pathway analysis of the differentially expressed genes between untreated and IL-10-treated MGC-803 cells. IL-10, interleukin-10.
IL-10 regulates the expression of c-Met/STAT3 in MGC-803 cells in vitro
Based on the sequencing results, the expression of STAT3 was significantly upregulated in IL-10-treated cells, thus, RT-qPCR and western blot analysis were performed to verify the sequencing results and examine the roles of STAT3 in IL-10 induced tumor-promoting effects. As presented in Fig. 10, treating MGC-803 with the cell culture supernatant of CAMs induced a significant increase in the protein expression levels of STAT3, and additionally increased the phosphorylation of STAT3. c-Met has been confirmed as an upstream regulator of STAT3 in several previous studies (19,20); therefore, the effects of IL-10 on the expression of c-Met, as well as on the downstream effectors of STAT3 caspase-3 and MMP9, were examined. It was observed that the cell culture supernatant of CAMs induced a significant increase in the expression of c-Met at the protein level (Fig. 10). Furthermore, treatment with the cell culture supernatant of CAMs resulted in a decreased protein expression of caspase-3 and increased expression of MMP9 (Fig. 10). Notably, addition of the IL-10 inhibitory antibody significantly blocked the effects of the CAM-conditioned media on the c-Met/STAT3 signaling pathway (Fig. 10).
Figure 10.
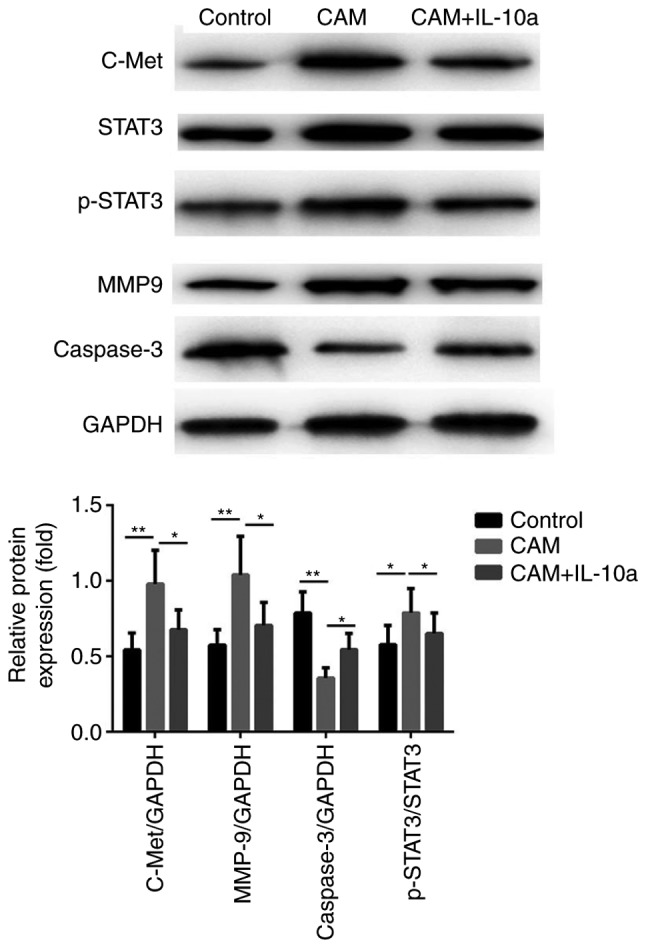
Protein expression levels of c-Met/STAT3 signaling proteins in different treatment groups. Western blot analysis was used to confirm the effects of IL-10 on proteins associated with the c-Met/STAT3 pathway. *P<0.05 and **P<0.01, with comparisons indicated by lines. c-Met, MET proto-oncogene; IL-10, interleukin-10; CAM, cancer-associated macrophage; IL-10a, IL-10 inhibitory antibody; p-, phosphorylated; MMP9, matrix metalloproteinase 9.
Discussion
IL-10 has been demonstrated to serve regulatory roles in inflammatory diseases; however, the roles of IL-10 in carcinogenesis remain unclear. In the present study, the expression levels of IL-10 in clinical gastric tumor tissues and serum samples from patients with gastric cancer were examined, and it was demonstrated that IL-10 was significantly upregulated in gastric cancer. Furthermore, IL-10 levels were significantly increased in the cell culture supernatant of CAMs compared with untreated THP-1 cells. In addition, it was identified that IL-10 was able to regulate the proliferation, apoptosis and migration of gastric cancer cells in vitro via activation of the c-Met/STAT3 signaling pathway.
Accumulating evidence has revealed that the tumor microenvironment serves important roles in the pathogenesis of gastric cancer. Numerous studies have demonstrated that M2 macrophages contribute to cancer progression, including tumor growth, invasion and metastasis directly or indirectly, through paracrine activation by various growth factors and cytokines (21,22). Notably, the activation of these procarcinogenic macrophages, termed CAMs, is not reversible. In the present study, it was demonstrated for the first time, to the best of our knowledge, that IL-10 secreted by CAMs enhanced cell proliferation, migration and invasion in gastric cancer cells.
In the present study, genome-wide RNA-sequencing was performed to further elucidate the global genetic alterations resulting by IL-10 stimulation in gastric cancer cells. Notably, 86 genes were identified to be significantly altered between untreated cells and cells treated with IL-10. GO analysis demonstrated that IL-10 treatment regulated genes involved in a number of principal biological events, including in response to chemical, response to cytokine, inflammatory response, cell chemotaxis, cytokine activity, positive regulation of leukocyte chemotaxis, regulation of leukocyte chemotaxis, positive regulation of leukocyte migration and G-protein coupled receptor binding. Therefore, CAMs may contribute to the reprogramming of the cancer cells via cytokine secretion, such as IL-10.
STAT3 was identified as one of the significantly upregulated genes upon IL-10 stimulation, and the present results confirmed that treatment of gastric cancer cells with the cell culture supernatant of CAMs induced a significant increase in the protein expression levels and the phosphorylation of STAT3. c-Met, also termed hepatocyte growth factor receptor, is an upstream regulator of STAT3, which has been identified as an oncogene and potential therapeutic target (23,24). Upregulation of c-MET may increase the expression and phosphorylation of STAT3, wherein the activated STAT3 directly regulates genes involved in proliferation (including cyclin D1, c-Myc, p53 and p21), survival (including Bcl-2, Bcl-xL, MCL1 apoptosis regulator and survivin), metastasis (including MMP2 and MMP9) and angiogenesis (including vascular endothelial growth factor) in numerous types of cancer. The role of c-MET/STAT3 signaling in gastric cancer has been previously discussed in multiple studies (25,26), including a previous study from our group (27). To further examine whether c-Met/STAT3 signaling may serve a key role in IL-10-induced tumor-promoting behaviors of gastric cancer cells, the expression levels of c-Met, and of downstream effectors STAT3, MMP9 and caspase-3, were examined following treatment with CAM-conditioned media. It was observed that treatment with the cell culture supernatant of CAMs induced a significant increase in the expression of c-Met, a decrease in the expression of caspase-3, and an increase in the expression of MMP9; however, addition of an inhibitory IL-10 antibody significantly blocked these effects, suggesting that IL-10 secreted by CAMs may regulate the proliferation, apoptosis and migration of the gastric cancer cells by activating the c-Met/STAT3 signaling pathway. Notably, it was observed that the cell culture supernatant of CAMs had no effect on the expression of c-Met at the mRNA level (data not shown), suggesting that the effect of IL-10 on the expression of c-Met may be post-transcriptional.
Consistent with the present results, previous studies have suggested that CAMs may be novel prospective therapeutic targets in cancer immunotherapy. As a crucial component within the cancer microenvironment, CAMs are activated at the early stages and widely contribute to tumor initiation, growth, invasion, metastasis and therapy resistance, via growth factors, cytokines and other biological molecules. Current CAM-targeted cellular therapies have undergone preclinical evaluation and clinical trials (28), including inhibition of CAM activation, blockage of CAM-cancer cell interaction, and destruction of metabolism uncoupling between CAMs and cancer cells. For example, inhibition of IL-10 secretion from CAMs in gastric cancer may be a promising therapeutic strategy.
In summary, it was demonstrated that there was an increased expression of IL-10 in gastric cancer tumor specimens, which was correlated with the advanced clinicopathological stages of patients. Additionally, IL-10 was markedly increased in the CAMs. Furthermore, treatment with IL-10 induced an increase in the proliferation and migration of MGC-803 and BGC-823 cells; however, it decreased apoptosis via activation of the c-Met/STAT3 signaling pathway. IL-10 regulates a global altered gene expression profile in gastric cancer cells, including metabolic signaling pathways, cell proliferation and migration. Together, these results suggested that IL-10 produced by CAMs promoted the proliferation and invasion of gastric cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LC, XZ, WG, MZ and YC conducted the experiments. XZ, WG, MZ and YC analyzed the data. LT, XY, QY and ZL interpreted the data. LC drafted the manuscript. YS was involved in the conception and design of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients signed an informed consent form, and the study was approved by the Ethical Committee of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yao Q, Tu C, Lu D, Zou Y, Liu H, Zhang S. Clinicopathological significance of the microRNA-146a/WASP-family verprolin-homologous protein-2 axis in gastric cancer. Cancer Sci. 2017;108:1285–1292. doi: 10.1111/cas.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang DY, Wang J, Zhang GQ, Chu XQ, Zhang JL, Zhou Y. Correlations of MMP-2 and TIMP-2 gene polymorphisms with the risk and prognosis of gastric cancer. Int J Exp Med. 2015;8:20391–20401. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, Wang Z, Fang JY, Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto N, Tsujimoto H, Takahata R, Cao B, Zhao P, Ito N, Shimazaki H, Ichikura T, Hase K, Vande Woude GF, Shinomiya N. MET4 expression predicts poor prognosis of gastric cancers with Helicobacter pylori infection. Cancer Sci. 2017;108:322–330. doi: 10.1111/cas.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higuchi K, Inokuchi M, Takagi Y, Ishikawa T, Otsuki S, Uetake H, Kojima K, Kawano T. Cadherin 5 expression correlates with poor survival in human gastric cancer. J Clin Pathol. 2017;70:217–221. doi: 10.1136/jclinpath-2016-203640. [DOI] [PubMed] [Google Scholar]
- 6.Mosser DM, Zhang X. Interleukin-10: New perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira L, Font-Nieves M, Van den Haute C, Baekelandt V, Planas AM, Pozas E. IL-10 regulates adult neurogenesis by modulating ERK and STAT3 activity. Front Cell Neurosci. 2015;9:57. doi: 10.3389/fncel.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu BS, Cao Y, Huizinga TW, Hafler DA, Toes RE. TLR-mediated STAT3 and ERK activation controls IL-10 secretion by human B cells. Eur J Immunol. 2014;44:2121–2129. doi: 10.1002/eji.201344341. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Xie J, Fiskesund R, Dong W, Liang X, Lv J, Jin X, Liu J, Mo S, Zhang T, et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat Commun. 2018;9:873. doi: 10.1038/s41467-018-03225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno Y, Toyoshima Y, Yurino H, Monma N, Xiang H, Sumida K, Kaneumi S, Terada S, Hashimoto S, Ikeo K, et al. Lack of interleukin-6 in the tumor microenvironment augments type-1 immunity and increases the efficacy of cancer immunotherapy. Cancer Sci. 2017;108:1959–1966. doi: 10.1111/cas.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan C, Huang X, Lin S, Huang H, Cai Q, Wan T, Lu J, Liu J. Expression of M2-polarized macrophages is associated with poor prognosis for advanced epithelial ovarian cancer. Technol Cancer Res Treat. 2013;12:259–267. doi: 10.7785/tcrt.2012.500312. [DOI] [PubMed] [Google Scholar]
- 12.Herrera M, Herrera A, Domínguez G, Silva J, García V, García JM, Gómez I, Soldevilla B, Muñoz C, Provencio M, et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013;104:437–444. doi: 10.1111/cas.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JY, Peng CW, Yang GF, Hu WQ, Yang XJ, Huang CQ, Xiong B, Li Y. Distribution pattern of tumor associated macrophages predicts the prognosis of gastric cancer. Oncotarget. 2017;8:92757–92769. doi: 10.18632/oncotarget.21575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin S, Huang J, Li Z, Zhang J, Luo J, Lu C, Xu H, Xu H. The prognostic and clinicopathological significance of tumor-associated macrophages in patients with gastric cancer: A meta-analysis. PLoS One. 2017;12:e0170042. doi: 10.1371/journal.pone.0170042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Salvi A, Abeni E, Portolani N, Barlati S, De Petro G. Human hepatocellular carcinoma cell-specific miRNAs reveal the differential expression of miR-24 and miR-27a in cirrhotic/non-cirrhotic HCC. Int J Oncol. 2013;42:391–402. doi: 10.3892/ijo.2012.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 18.He F, Ai B, Tian L. Identification of genes and pathways in esophageal adenocarcinoma using bioinformatics analysis. Biomed Rep. 2018;9:305–312. doi: 10.3892/br.2018.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu JC, Wang CT, Hung HC, Wu WJ, Wu DC, Chang MC, Sung PJ, Chou YW, Wen ZH, Tai MH. Heteronemin is a novel c-Met/STAT3 inhibitor against advanced prostate cancer cells. Prostate. 2016;76:1469–1483. doi: 10.1002/pros.23230. [DOI] [PubMed] [Google Scholar]
- 20.Syed ZA, Yin W, Hughes K, Gill JN, Shi R, Clifford JL. HGF/c-met/Stat3 signaling during skin tumor cell invasion: Indications for a positive feedback loop. BMC Cancer. 2011;11:180. doi: 10.1186/1471-2407-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding H, Zhao L, Dai S, Li L, Wang F, Shan B. CCL5 secreted by tumor associated macrophages may be a new target in treatment of gastric cancer. Biomed Pharmacother. 2016;77:142–149. doi: 10.1016/j.biopha.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi T, Fushida S, Yamamoto Y, Tsukada T, Kinoshita J, Oyama K, Miyashita T, Tajima H, Ninomiya I, Munesue S, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. 2016;19:1052–1065. doi: 10.1007/s10120-015-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gholamin S, Fiuji H, Maftouh M, Mirhafez R, Shandiz FH, Avan A. Targeting c-MET/HGF signaling pathway in upper gastrointestinal cancers: Rationale and progress. Curr Drug Targets. 2014;15:1302–1311. doi: 10.2174/1389450115666141107105456. [DOI] [PubMed] [Google Scholar]
- 24.Marano L, Chiari R, Fabozzi A, De Vita F, Boccardi V, Roviello G, Petrioli R, Marrelli D, Roviello F, Patriti A. C-Met targeting in advanced gastric cancer: An open challenge. Cancer Lett. 2015;365:30–36. doi: 10.1016/j.canlet.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Ding X, Ji J, Jiang J, Cai Q, Wang C, Shi M, Yu Y, Zhu Z, Zhang J. HGF-mediated crosstalk between cancer-associated fibroblasts and MET-unamplified gastric cancer cells activates coordinated tumorigenesis and metastasis. Cell Death Dis. 2018;9:867. doi: 10.1038/s41419-018-0922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto W, Okamoto I, Arao T, Yanagihara K, Nishio K, Nakagawa K. Differential roles of STAT3 depending on the mechanism of STAT3 activation in gastric cancer cells. Br J Cancer. 2011;105:407–412. doi: 10.1038/bjc.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei X, Juan ZX, Min FX, Nan C, Hua ZX, Qing FZ, Zheng L. Recombinant immunotoxin anti-c-Met/PE38KDEL inhibits proliferation and promotes apoptosis of gastric cancer cells. J Exp Clin Cancer Res. 2011;30:67. doi: 10.1186/1756-9966-30-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.