Abstract
IncRNAs play an important role in the regulation of gene expression. The present study profiled differentially expressed lncRNAs (DELs) and mRNAs (DEMs) in myelodysplastic syndrome (MDS) to construct a 4-aminobutyrate aminotransferase (ABAT)-DEL-DEM co-expression network in MDS development using the Agilent human BeadChips and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and network analyses. Compared with controls, there were 543 DELs and 2,705 DEMs in MDS patients, among which 285 (52.5%) DELs were downregulated and 258 (47.5%) DELs were upregulated, whereas 1,521 (56.2%) DEMs were downregulated and 1,184 (43.70%) DEMs were upregulated in MDS patients. The ABAT-DEL-DEM co-expression network contained six DELs that were co-expressed with ABAT in MDS. The GO analysis revealed that the co-expression network mainly participated in response to organic cyclic compound, cell proliferation, cell part morphogenesis, regulation of cell proliferation and enzyme-linked receptor protein signaling pathways, while the KEGG database showed that the co-expression network was involved in various pathways, such as phagosome and metabolic pathways. Furthermore, the expression of a selected DEL (lncENST00000444102) and ABAT was shown to be significantly downregulated in MDS patients, and in SKM-1 and THP-1 cells. The selected lncENST00000444102 was then overexpressed and ABAT expression was knocked down in the MDS cell lines using lentiviral transfection. In addition, lncENST00000444102 overexpression reduced the viability and increased the apoptosis of MDS cells, ABAT expression was upregulated by lncENST00000444102.
Keywords: myelodysplastic syndrome, 4-aminobutyrate aminotransferase, ABAT, long non-coding RNAs, microarray analysis, co-expression network
Introduction
Myelodysplastic syndrome (MDS) refers to a group of neoplastic bone marrow disorders characterized by abnormal blood cell morphology and functions due to defects in hematopoietic precursor differentiation into mature blood cells. Importantly, MDS can progress to acute myeloid leukemia (AML). Clinically, patients manifest symptoms related to anemia, neutropenia, and/or thrombocytopenia, such as chronic fatigue, shortness of breath, chilled sensations, and increased susceptibility to infection and bleeding, while other patients may lack symptoms and are diagnosed following blood analyses (1). To date, the pathogenesis of MDS is poorly understood, and risk factors include exposure to pesticides, benzene, or previous chemotherapy and/or radiotherapy, all of which cause damage to genomic DNA (1), Studies have aimed to identify the key genetic (2) and epigenetic alterations (3) involved in MDS. Moreover, overexpression of immune-related genes (4) and abnormal activation of innate immune signals (5,6) have been widely reported in MDS. However, the definite pathogenetic mechanisms of MDS are still not fully understood. Thus, research into the molecular mechanisms of MDS development and progression is urgently needed, and the findings could provide novel strategies for effective control and prevention of MDS.
To this end, a previous study revealed that aberrant gene expression through genomic DNA methylation could be a dominant mechanism by which MDS progresses to AML (7). Our previous study identified a set of methylated genes (8). One of these genes, 4-aminobutyrate aminotransferase (ABAT), was highly methylated and its expression was reduced in MDS patients compared with that noted in healthy controls (8,9). The ABAT gene is localized on chromosome 16p13.2 and encodes a protein responsible for the catabolism of γ-aminobutyric acid (GABA, an important neurotransmitter in the central nervous system) into succinic semialdehyde (10). In humans, gene mutations leading to ABAT deficiency are extremely rare, while a clinical study biochemically confirmed that ABAT deficiency contributes to symptoms related to psychomotor retardation, hypotonia, hyperreflexia, lethargy, and intractable seizures (11). Moreover, ABAT single-nucleotide polymorphisms have been associated with depression (12), sleep homeostasis (13), autism (14) and gastroesophageal reflux disease (15). In regards to human cancer, reduced ABAT expression has been associated with resistance to endocrine therapy of breast cancer, and with poor recurrence-free survival of breast cancer patients (16,17).
Furthermore, long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides that are not usually translated into proteins, and their genes are usually located within intergenic stretches or overlapping antisense transcripts of protein coding genes (18,19). lncRNAs function to regulate chromatin remodeling, genomic imprinting, gene transcription, splicing, and translation in cells, and aberrant lncRNA expression contributes to human diseases, including the pathogenesis of hematopoietic malignancies (20). Although lncRNAs are increasingly recognized as regulators of normal and aberrant hematopoiesis (21), their role in MDS has not been investigated thoroughly. In the present study, we uncovered an ABAT-DEL-DEM co-expression network and assessed the function of network components in MDS. We first identified differentially expressed lncRNAs (DELs) and mRNAs (DEMs) in MDS samples and then performed an integrative analysis to identify the co-expressed network based on ABAT and lncRNAs, and conducted lncRNA-mRNA networks. Subsequently, we further annotated this co-expressed network using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and network terms. We aimed to provide a novel methodology with which to analyze and annotate disease-associated lncRNAs for functional validation and targeted therapies.
Materials and methods
Patients and bone marrow samples
We recruited 23 MDS patients at the Department of Hematology, Huashan Hospital, Fudan University (Shanghai, China) from January 1, 2015 to December 30, 2016. The inclusion criteria consisted of newly diagnosed patients and no previous treatment and excluded patients with myelodysplastic/myeloproliferative neoplasms (MDS/MPN) and MDS which had progressed to acute myeloid leukemia. Bone marrow samples were collected for all patients and patients were diagnosed with MDS and classified according to the 2008 revision of the World Health Organization (WHO) criteria (22). There were 15 men and 8 women with a median age of 65 years (range, 29–82 years), in the MDS group. In addition, bone marrow samples from 7 cases with non-hematological malignancies with a median age of 38 years (range 23–78 years) were obtained between January 1, 2015 and December 30, 2015, and were analyzed as controls. No statistical difference was found with regards to age and sex distributions among the subjects with MDS and non-MDS controls. For microarray analysis groups, we obtained bone marrow samples from anther 4 MDS patients (2 patients with refractory anemia with excess blasts 1 (RAEB-1) and 2 patients with RAEB-2] and 4 age-matched patients with hypersplenism from the Department of Hematology, Huashan Hospital, Fudan University (Shanghai, China) in 2014. We collected 2 ml of all bone marrow samples using heparin and stored the samples at room temperature within 8 h. Total RNA was isolated from bone marrow samples within 8 h and then stored at −80°C. The present study was approved by the Ethics Committee of Huashan Hospital, School of Medicine, Fudan University. All participants provided a written informed consent form before being enrolled into this study.
RNA isolation and reverse transcription and quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was isolated from bone marrow samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The purity of RNA was assessed by measuring the optical density (OD) 260/280 ratio using a NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). The RNA samples were reverse transcribed into cDNA using Takara PrimeScript RT Master Mix (Takara Bio Inc., Otsu, Shiga, Japan). The 10-µl reaction mixture comprised 2 µl 5X PrimeScript RT Master Mix, 500 ng RNA and RNase-Free distilled H2O, which was used to ensure that the total reached 10 µl. Subsequently, the mixture was amplified at 37°C for 15 min, 85°C for 5 sec and was stored at 4°C for further use.
For RT-qPCR, cDNA samples were amplified using an Applied Biosystems™ 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) in a 20-µl mixture containing 10 µl SYBR® Premix Ex Taq™ (Takara Bio, Inc., Otsu, Japan), 0.4 µl of each primer (10 µmol/l), 0.4 µl ROX Reference Dye II (Takara Bio, Inc.), 2 µl cDNA, and 6.8 µl ddH2O at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 34 sec. Gene expression levels were expressed relative to the expression of GADPH. The primers were designed with the Primer 3.0 online software (http://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by BioTNT (Shanghai, China). The primer sequences were as follows: lncENST00000444102, 5′-TATCGACGTAGTTAAAGCCCACT-3′ and 5′-CTTCTGCCCTTCACATCCTCT-3′; ABAT, 5′-CCGACTACAGCATCCTCTCC-3′ and 5′-GGTTCTCTTTCACAAACTCTTCC-3′; GAPDH, 5′-GACCTGACCTGCCGTCTA-3′ and 5′-AGGAGTGGGTGTCGCTGT-3′. The relative levels of gene expression were calculated using the 2−ΔΔCq method (23).
Microarray analysis
Subsequently, cDNA samples from microarray analysis groups were used to profile differentially expressed gene transcripts and lncRNAs using Agilent human genome-wide gene expression BeadChips (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol at 65°C. The expression values were normalized with Robust Multi-array Average (RMA) of background-adjusted, normalized and log2-transformed using the statistical software package R (24).
Construction of the ABAT-DEL-DEM co-expression network
After profiling each DEL and DEM in the MDS samples, we specifically identified the altered ABAT expression in MDS patients for construction of the ABAT-DEL-DEM network. In brief, the co-expression of ABAT and particular DELs and lncRNA-mRNA correlation were evaluated using the Pearson correlation coefficient (PCC). The ABAT-DEL-DEM co-expression network was further filtered by the overlapping DELs using software of Cytoscape 3.4.0 (The Cytoscape Consortium, San Diego, CA, USA. Weblink: http://cytoscape.org/download.html) according to a previous study (25).
GO and KEGG pathway and network analyses
Subsequently, we investigated the potential role of the identified ABAT-DEL-DEM co-expression network using the GO and KEGG pathway and network analyses using the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8 tool) (26,27). The KEGG pathway enrichment analysis was performed using the clusterProfiler package in R/bioconductor. Furthermore, the key KEGG signaling pathway was analyzed using the R package pathview (28).
Bioinformatic analysis of lncRNAs
For the bioinformatic analysis of lncRNAs, we first accessed the UCSC database (http://genome.ucsc.edu) to prioritize ABAT-associated lncRNAs and searched the NCBI human genomes database to identify the chromosomal localization of each lncRNA. We also utilized PhyloCSF, a novel comparative genomics method to analyze multispecies nucleotide sequence alignment to determine whether DNA sequences where each lncRNA resides are likely to represent a conserved protein-coding or non-coding region (29). PhyloCSF scores for selected phylogenies may be viewed in the UCSC Genome Browser by copying the URL ‘http://www.broadinstitute.org/compbio1/PhyloCSFtracks/trackHub/hub.txt’ into the ‘My Hubs’ tab under ‘track hubs’. PhyloCSF outputs a score, positive if the alignment is likely to represent a conserved coding region, and negative otherwise. Moreover, we also used computational and mathematical methods to predict advanced lncRNA structure (http://rna.tbi.univie.ac.at/RNAfold/UyEBF8akiV), and used the TRANSFAC database to predict the transcription factor binding sites (TFBS) of each lncRNA (http://www.gene-regulation.com) (30).
Cell lines and culture
A human AML cell line (THP-1) was purchased from Chinese Academy of Sciences (Shanghai, China) and a human AML cell line transformed from MDS cells (SKM-1) was obtained from the Japanese Collection of Research Bioresources (LCRB; Tokyo, Japan). 293T cells were used as a control (Chinese Academy of Sciences, Shanghai, China). All cell lines were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640; Hyclone; GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (Gibco-BRL; Thermo Fisher Scientific, Inc.) in a humidified incubator with 5% CO2 at 37°C.
Vector construction, lentivirus preparation, and stable cell infection
To confirm our ABAT-DEL-DEM co-expression network, we constructed lentiviral vectors and prepared lentivirus to stably overexpress lncRNA or knock down ABAT expression. All plasmid vectors were produced using standard cloning techniques (31). The lncRNA was overexpressed and cloned into GV470 lentiviral vectors (GeneChen, Shanghai, China). The shRNA hairpins that targeted the 3′-untranslated region (3′-UTR) of ABAT were designed and cloned into pGMLV-SC5 lentivirus vectors (Genomeditech, Shanghai, China). The sequences of oligonucleotides were 5′-GCTGGAGACGTGCATGATTAA-3′. The SKM-1 and THP-1 cells were respectively cultured in 6-well plates at a density of 1×106 cells/ml overnight and transduced with lncRNA overexpression lentivirus and shRNA lentivirus plus a scramble lentivirus (negative controls), and cells without any lentivirus transduction were considered as controls. Since the lentivirus had green fluorescence, we evaluated transduction efficiency by flow cytometry at 72 h. The percentage of positive cells was >80% by flow cytometry. The growth medium was then replaced with 1 or 2 µg/ml puromycin, respectively, to select stable cells for two weeks. These cells were then subjected to quantitative reverse transcription polymerase chain reaction (RT-qPCR) to verify expression levels of lncRNA and ABAT gene.
Cell viability CCK-8 assay
Cell viability was assessed using the Cell Counting Kit-8 (CCK-8) assay kit from Dojindo Laboratories (Kumamoto, Japan). In brief, stably transfected and control cells were seeded into a 96-well culture plate at a density of 1×103 cells/well and cultured for up to five days, with media replaced every three days. At the end of each experiment, 5 µl of the CCK-8 reagent was added into each well and the cells were further cultured for 4 h. Then, the optical density of cells was measured at 450 nm using a spectrophotometer (Tecan, Männedorf, Switzerland). The experiments were performed in triplicate and repeated at least three times.
Flow cytometric Annexin V apoptosis assay
Cells cultured for 48 h (6-well plate, 1×106 cells/ml) were collected and washed twice with phosphate-buffered saline (PBS), and then re-suspended in 200 µl of the binding buffer. Annexin V-APC (5 µl) and 7-AAD (5 µl; Thermo Fisher Scientific, Inc.) were added and the mixture was incubated in the dark at room temperature for 15 min. The rate of cellular apoptosis was then measured using a flow cytometer (BD Accuri C6; BD Biosciences, San Jose, CA, USA). Data were statistically analyzed using the software Flowjo 7.6 (Flow Jo, LLC, Ashland, OR, USA). The experiments were performed in duplicate and repeated at least once.
Statistical analysis
Student's t-test was used to identify DELs and DEMs between patients with MDS and hypersplenism by calculation of P<0.05 and fold change (FC)>2 for each DEM and DEL. The correlation of ABAT-DEL-DEM co-expression was evaluated using Pearson correlation coefficient (PCC). Pairs with a PCC threshold >0.95 and P-value <0.05 were used to construct the ABAT and DEL co-expression association matrix. Pairs with a PCC threshold >0.99 and P-value <0.01 were selected as the meaningful value to construct the DEL and DEM co-expression network. A P-value <0.05 using Fisher's exact test and a kappa (κ) coefficient of 0.4 were used as threshold values in GO and KEGG pathway and network analyses. The in vitro experimental data were analyzed using Graphpad Prism 6 software (GraphPad Software, La Jolla, CA, USA). Comparisons between two groups were analyzed by Student's t-test when data conformed to normal distribution, if not, a non-parametric Kruskal-Wallis test was performed. Following a Kruskal-Wallis test, Dunn's Multiple Comparisons test was used as the post test to compare the difference in the sum of ranks between two columns with the expected average difference. A P<0.05 was considered statistically significant.
Results
Profile of differentially expressed lncRNAs and mRNAs in MDS bone marrow samples
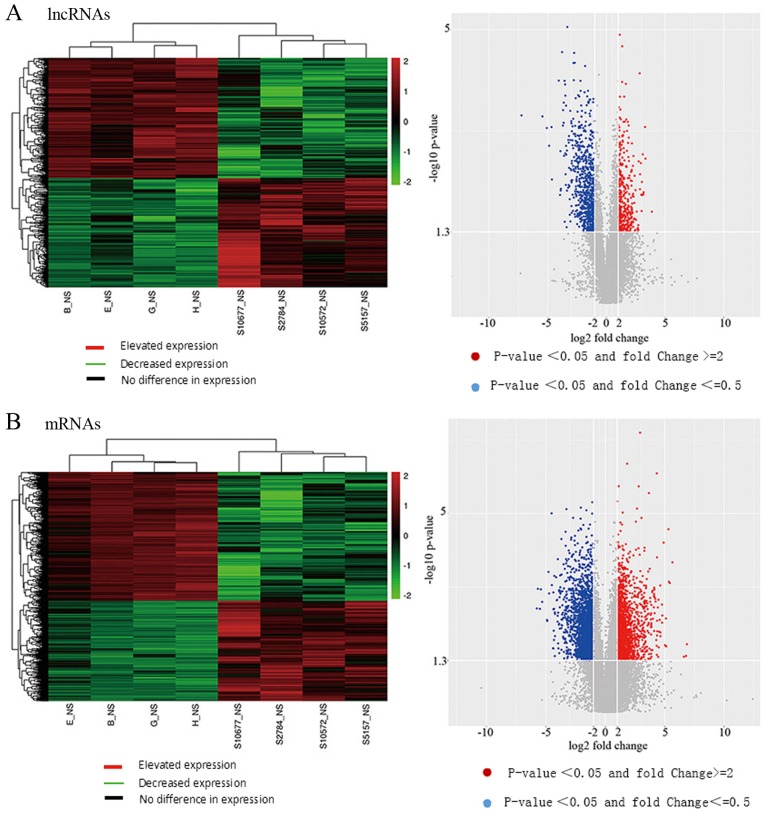
The microarray data were then scanned using SureScan Scanner (Agilent Technologies) and analyzed as previously reported (8). Bone marrow cells from 4 MDS patients were compared with cells from 4 age-matched hypersplenism controls, and the microarray analysis revealed a total of 543 DELs and 2,705 DEMs. Among them, 285 (52.5%) DELs were downregulated and 258 (47.5%) DELs were upregulated in MDS patients, whereas 1,521 (56.2%) DEMs were downregulated and 1,184 (43.70%) DEMs were upregulated in MDS. The volcano plots and heatmaps provide an overview of the DEL and DEM microarray data, respectively (Fig. 1).
Figure 1.
Differential expression of lncRNAs and mRNAs in the bone marrow of MDS patients vs. controls. (A) Heatmap and volcano diagram of differentially expressed lncRNAs (DELs) in myelodysplastic syndromes (MDS). (B) Heatmap and volcano diagram of differentially expressed mRNAs (DEMs) in MDS. Hyperthyroidism groups: B_NS, E_NS, G_NS and H-NS; MDS groups: S10677_NS, S2784_NS, S10572_NS and S5157_NS.
Identification of the ABAT-DEL-DEM co-expression network
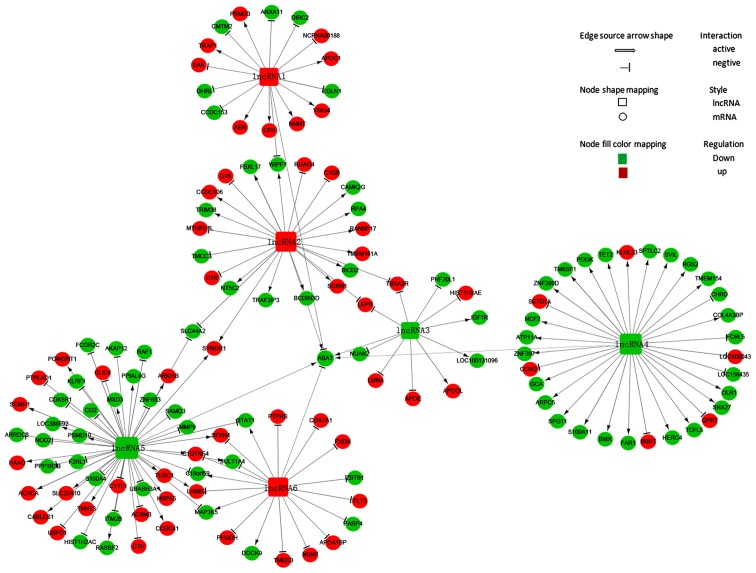
The ABAT-DEL-DEM co-expression network was constructed using the Cytoscape program. To generate the ABAT-DEL-DEM co-expression network, we related ABAT expression in MDS samples to each of the DELs using the PCC test. We were able to select six DELs (Table I) that were co-expressed with ABAT with a PCC threshold >0.95 and P<0.05 and 135 co-expressed mRNAs with a PCC threshold >0.99 and P<0.01 in MDS to construct the lncRNA and mRNA co-expression network (Table SI). Next, their potential interaction was determined using the Cytoscape program. The data showed that the co-expression network was composed of six DELs related to ABAT and 135 co-expressed mRNAs (Fig. 2). In this network, three lncRNAs were up-regulated (lncRNA1, lncRNA2 and lncRNA6), whereas the other three were downregulated (lncRNA3, lncRNA4 and lncRNA5). The network showed that a particular mRNA could correlate with numerous lncRNAs, while a single lncRNA was also able to correlate to various mRNAs, implying that an inter-regulation of lncRNAs and mRNAs occurs in MDS.
Table I.
Six DELs that are co-expressed with the ABAT gene.
| DELs | Name | Location | Strand | Regulation type | PCC | P-value |
|---|---|---|---|---|---|---|
| lncRNA1 | None | chr4:13067152-13347902 | Forward | Up | −0.968 | 0.018 |
| lncRNA2 | Jh591181.2/kb663606.1 | chr10:46972944-46982894 | Forward | Up | 0.963 | 0.036 |
| lncRNA3 | Loc100131564/RP4-717I23.3 | chr1:93796837-93806487 | Reverse | Down | 0.979 | 0.021 |
| lncRNA4 | lncENST00000444102 | chr6:167382710-167411729 | Reverse | Down | 0.988 | 0.011 |
| lncRNA5 | Jh806582.2/g1383563.2 | chr17:49425-59050 | Reverse | Down | 0.959 | 0.041 |
| lncRNA6 | None | chr5:61931044-61948469 | Reverse | Up | −0.954 | 0.046 |
Student's t-test was used to identify DELs and DEMs between MDS and non-MDS by calculation of P<0.05 and fold change (FC) >2 for each DEL. Pairs with a PCC threshold >0.95 and P<0.05 were used to construct the ABAT and DEL co-expression association matrix. ABAT, 4-aminobutyrate aminotransferase; MDS, myelodysplastic syndrome; DELs, differentially expressed lncRNAs; DEMs, differentially expressed mRNAs; PCC, Pearson correlation coefficient.
Figure 2.
ABAT-DEL-DEM co-expression network. This network was constructed using differentially expressed lncRNAs (DELs) and mRNAs (DEMs) stratified by ABAT expression and is composed of six DELs related to the ABAT gene and 135 co-expressed DEMs by Cytoscape software. lncRNA1, lncRNA-chr4:13067152-13347902 forward; lncRNA2, lncRNA-chr10:46972944-46982894 forward; lncRNA3, lncRNA-chr1:93796837-93806487 reverse; lncRNA4, lncRNA-chr6:167382710-167411729 reverse; lncRNA5, lncRNA-chr17:49425-59050 reverse; lncRNA 6, lncRNA-chr5:61931044-61948469; ABAT, reverse. 4-aminobutyrate aminotransferase.
Gene enrichment and pathway of the co-expression network
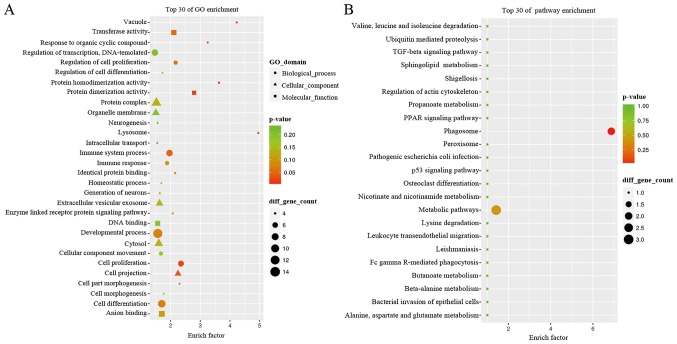
We next performed analyses using GO terms and the KEGG pathways and networks, and found that the co-expression network was mostly enriched in several biological processes, such as cellular components and molecular functions. In particular, the GO analysis revealed that the co-expression network mainly participated in response to organic cyclic compounds, cell proliferation, cell part morphogenesis, regulation of cell proliferation, and enzyme-linked receptor protein signaling pathway (Fig. 3A) for molecular functions of protein homodimerization activity. The GO term analysis also revealed that the co-expression network is localized in different areas within cells, such as the lysosome, vacuole, cell projection, and extracellular vesicular exosome. The KEGG data showed that the co-expression network was involved in different pathways, such as the phagosome and metabolic pathways (Fig. 3B). The differentially expressed gene of neutrophil cytosolic factor 2 (NCF2) plays role in neutrophil phagosome. In addition, the genes of serine palmitoyltransferase long chain base subunit 2 (SPTLC2), 4-aminobutyrate aminotransferase (ABAT) and quinolinate phosphoribosyltransferase (QPRT) were key metabolic enzymes which play a role in metabolic pathways (https://www.ncbi.nlm.nih.gov/gene) (Table II).
Figure 3.
The GO terms and KEGG pathway analyses of the co-expressed network. (A) The Gene Ontology (GO) terms. The top 30 enrichment functions of GO terms in this ABAT-DEL-DEM co-expression network in myelodysplastic syndromes (MDS). (B) The KEGG pathway analysis. The top 30 pathways associated with the ABAT-DEL-DEM co-expression network in MDS. ABAT, reverse. 4-Aminobutyrate aminotransferase; DEL, differentially expressed lncRNA; DEM, differentially expressed mRNA.
Table II.
Data for the KEGG analysis of the co-expression network.
| Pathways | Genes | Enrich_factor | P-value |
|---|---|---|---|
| Phagosome | NCF2 | 6.87 | <0.05 |
| Metabolic pathways | SPTLC2/ABAT/QPRT | 1.42 | <0.05 |
NCF2, neutrophil cytosolic factor 2; SPTLC2, serine palmitoyltransferase long chain base subunit 2; ABAT, 4-aminobutyrate aminotransferase; QPRT, quinolinate phosphoribosyltransferase. The KEGG pathway enrichment analysis was performed using the clusterProfiler package in R/bioconductor.
Bioinformatic analysis of potential lncENST00000444102 targeting genes
In this study, we revealed six lncRNAs and ranked lncENST00000444102 as the most important lncRNA with the highest correlation index related to the ABAT gene (Table I). lncENST00000444102 (lncRNA4) was co-expressed with the genes, NCF2, SPTLC2, ABAT and QPRT which all take part in the pathways (Table II and Fig. 2), thus we focused on lncENST00000444102 for further research. We then used tools in the UCSC database (http://genome.ucsc.edu) to re-annotate them and focused on chr6: 167382710-167411729, reverse strand (Fig. 4A). This lncRNA (ENST00000444102, named lncENST00000444102) was one of the most downregulated lncRNAs in MDS, residing on chromosome 6 in the human genome as an antisense RNA, consisting of three exons and spanning nearly 29 kilobases (kb). The advanced structure of lncENST00000444102 was predicted by RNAfold WebServer. We typed in the RNA sequence and chose the options of fold algorithms, and the minimum free energy (MFE) structure and the centroid secondary structure were obtained (Fig. 4B) (32). PhyloCSF scores for lncENST00000444102 were viewed in the UCSC Genome Browser tracks. We found that the scores were negative, with which we confirmed that the lncENST00000444102 was a non-coding RNA (Fig. 5). The gene was located among chemokine receptor 6 (CCR6), FGFR1 oncogene partner (FGFR1OP), and ribonuclease T2 (RNASET2) (Fig. 5), indicating that this particular lncRNA may be able to regulate the transcription of these three genes in cells. To confirm this hypothesis, we predicted the transcription factor binding sites (TFBS) of lncENST00000444102 using tools listed in the TRANSFAC database and found that lncENST00000444102 was able to target chicken ovalbumin upstream promoter-transcription factor (COUP-TF), which plays an important role in the regulation of organogenesis, neurogenesis, metabolic homeostasis, and cellular differentiation during embryonic development, and hepatocyte nuclear factor 4 (HNF-4), mainly playing a role in hepatic diseases (Table III).
Figure 4.
Bioinformatic analysis of lncENST00000444102. (A) The localization of lncENST00000444102 in the human genome. It was located on chromosome 6 in the human genome as an antisense RNA. (B) The predicted structure of lncENST00000444102 in the RNAfold webServer. We typed in the RNA sequence and chose the options of fold algorithms, and the minimum free energy (MFE) structure and the centroid secondary structure were obtained.
Figure 5.
Prediction of lncENST00000444102 and targeting genes using the PhyloCSF method. Chemokine receptor 6 (CCR6), FGFR1 oncogene partner (FGFR1OP), and ribonuclease T2 (RNASET2) are the targeted genes cis-controlled by lncENST00000444102.
Table III.
Transcription factor binding sites (TFBS) of lncRNA-ENST00000444102.
| Model | Factor | Strand | Start (bp) | End (bp) | Score |
|---|---|---|---|---|---|
| M00158 | COUP-TF, HNF-4 | − | 614 | 627 | 10.4 |
| M00923 | Adf-1 | + | 1,135 | 1155 | 9.4 |
| M00411 | HNF4α1 | + | 615 | 628 | 7.7 |
| M00034 | p53 | + | 909 | 928 | 7.5 |
| M00638 | HNF4α | − | 616 | 628 | 7.3 |
| M00401 | ABF1 | − | 942 | 968 | 7.2 |
| M00932 | Sp1 | − | 1,249 | 1,259 | 7.1 |
| M00932 | Sp1 | + | 1,388 | 1,398 | 7.1 |
| M00002 | E47 | − | 753 | 767 | 6.9 |
| M00034 | p53 | − | 909 | 928 | 6.9 |
| M00665 | Sp3 | + | 1,337 | 1,350 | 6.6 |
| M00761 | p53 decamer | + | 909 | 928 | 6.6 |
Τranscription factor binding sites (TFBS) of lncRNAs were predicted using the TRANSFAC database (http://www.gene-regulation.com). The higher the score, the more possibility of binding. This indicated that the factors of COUP-TF and HNF-4 may be the transcription factor binding sites (TFBS) of lncRNA-ENST00000444102.
Expression of lncENST00000444102 and ABAT in MDS patients and cell lines
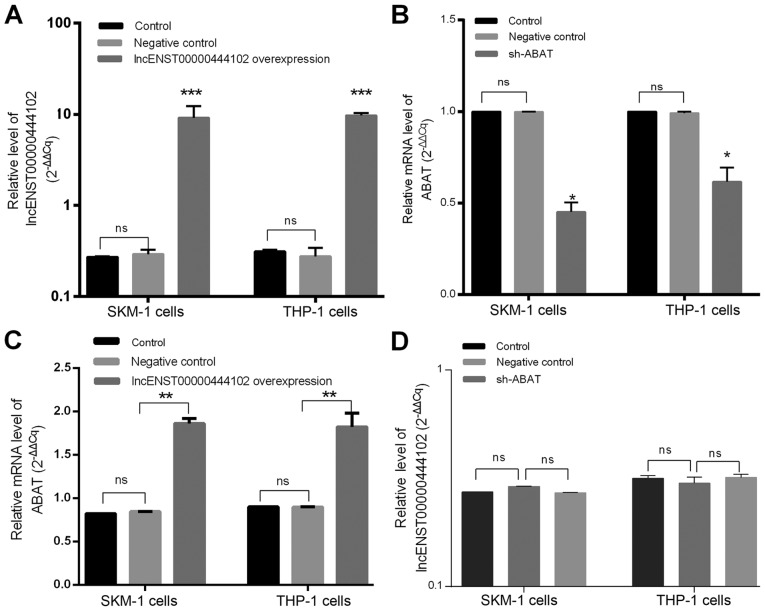
The alterations of these two molecules were next confirmed in MDS bone marrow samples from 23 MDS patients, 7 non-hematological malignancies, and two cell lines (Fig. 6). lncENST00000444102 expression was found to be significantly downregulated in MDS patients (P<0.0001, Fig. 6A) when compared to non-MDS cases as well as in the SKM-1 and THP-1 cell lines (P<0.0001, Fig. 6B) when compared to HEK-293 cells. ABAT expression was also downregulated in the bone marrow from MDS cases (P<0.001, Fig. 6C). We then overexpressed lncENST00000444102 and knocked down ABAT expression in SKM-1 and THP-1 cells (Fig. 7A and B). We found that stable overexpression of lncENST00000444102 induced ABAT expression by 2-fold in the SKM-1 and THP-1 cell lines (P<0.01, Fig. 7C). However, when ABAT expression was stably knocked down in the SKM-1 and THP-1 cell lines, the level of lncENST00000444102 was not significantly (ns) altered (P>0.05, Fig. 7D).
Figure 6.
Expression of lncENST00000444102 and ABAT in myelodysplastic syndrome (MDS) samples and cell lines. (A) MDS and non-MDS control bone marrow samples were analyzed for lncENST00000444102 expression using RT-qPCR. (B) MDS cell lines, SKM-1 and THP-1, were analyzed for lncENST00000444102 expression using RT-qPCR. HEK-293 cells were used as a control. (C) MDS and control bone marrow samples were analyzed for ABAT expression using RT-qPCR. Data are presented as the means ± standard deviation. ****P<0.0001 and ***P<0.001 vs. the control. ABAT, reverse. 4-aminobutyrate aminotransferase.
Figure 7.
Overexpression of lncENST00000444102 and knockdown of ABAT expression in MDS SKM-1 and THP-1 cell lines. (A) SKM-1 and THP-1 cell lines were grown and stably infected with a lentivirus carrying lncENST00000444102 and then subjected to RT-qPCR analysis of lncENST00000444102 expression. (B) SKM-1 and THP-1 cells were grown and stably infected with a lentivirus carrying sh-ABAT and then subjected to RT-qPCR analysis of ABAT expression. (C) The stable lncENST00000444102-overexpressing SKM-1 and THP-1 cells were subjected to RT-qPCR analysis of ABAT mRNA expression level. (D) The stable ABAT-knockdown SKM-1 and THP-1 cells were subjected to RT-qPCR analysis of lncENST00000444102 expression level. Data are presented as the means ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. the controls; ns not significant. Control cells were not infected with a lentiviral vector, negative control cells were infected with a scramble lentivirus. sh/shRNA, short hairpin RNA. ABAT, 4-aminobutyrate aminotransfe.
Reduced cell viability and increase in apoptosis following lncENST00000444102 overexpression in MDS cells
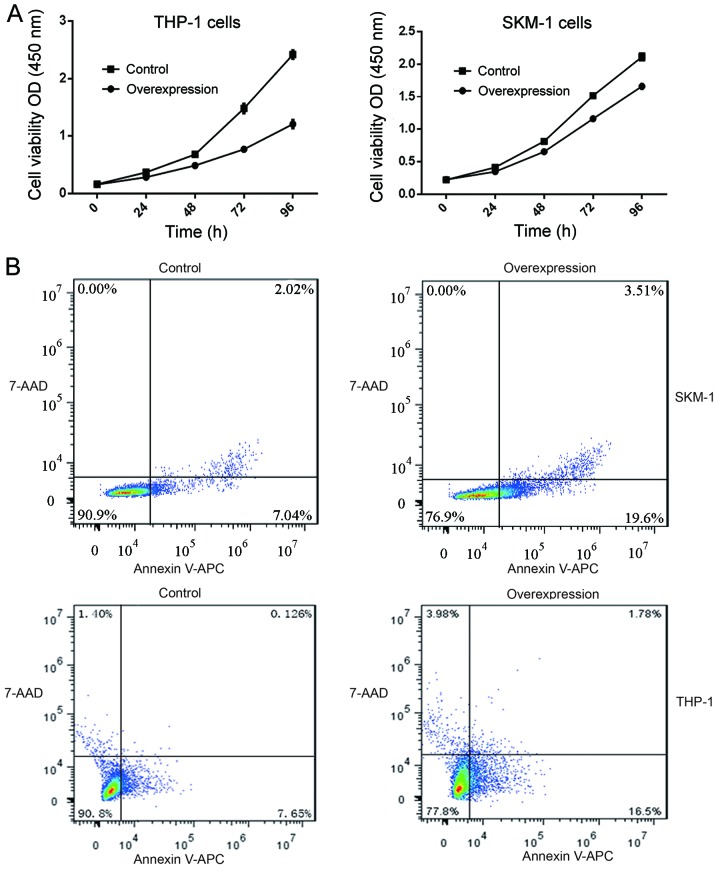
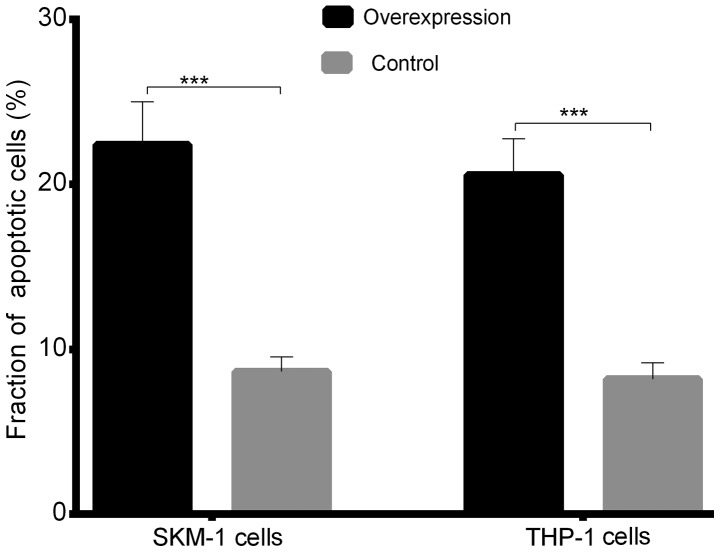
The cell viability CCK-8 assay showed that after 24 h in culture, the viability of SKM-1 and THP-1 cells with stable lncENST00000444102 overexpression started to decrease when compared with that of the control (P<0.05, Fig. 8A). The fraction of apoptotic cells was 22.41±2.596 in the lncENST00000444102-overexpressing SKM-1 cells, and 8.650±0.889 in the negative control; the fraction of apoptotic cells was 20.58±2.190 in the lncENST00000444102-overexpressing THP-1 cells and 8.192±0.997 in the negative control group (P<0.001, Fig. 8B). The flow cytometric apoptosis assay revealed that lncENST00000444102 overexpression promoted tumor cells to undergo apoptosis compared to control cells (P<0.001, Fig. 9). These data revealed that lncENST00000444102 overexpression reduced MDS cell viability by induction of MDS cell apoptosis.
Figure 8.
Reduction in MDS cell viability and induction of apoptosis after lncENST00000444102 overexpression. (A) Stable lncENST00000444102-overexpressing THP-1 and SKM-1 cells were subjected to cell viability CCK-8 assay. Data are presented as the means ± standard deviation. (B) Stable lncENST00000444102-overexpressing SKM-1 and THP-1 cells were subjected to flow cytometric apoptosis assay. Apoptosis of SKM-1 and THP-1 cells with lncENST00000444102 overexpression, as determined by Annexin V-APC/7-AAD staining at 48 h. MDS, myelodysplastic syndrome; 7-AAD, 7-amino-actinomycin D; APC, allophycocyanin; sh/shRNA, short hairpin RNA.
Figure 9.
The fraction of apoptotic cells in the SKM-1 and THP-1 cell lines with lncENST00000444102 overexpression. Data are presented as the means ± standard deviation. ***P<0.001 vs. the controls.
Discussion
Myelodysplastic syndromes (MDS) are a group of neoplastic bone marrow disorders characterized by abnormal blood cell morphology and can progress to acute myeloid leukemia (AML). In the present study, DELs and DEMs were profiled in MDS. Since our previous study demonstrated that expression of ABAT was reduced in MDS samples (8), we constructed an ABAT-DEL-DEM co-expression network in order to assess their role in MDS development. We found numerous DELs and DEMs in MDS samples compared with the controls. Our ABAT-DEL-DEM co-expression network identified six DELs that were co-expressed with ABAT in MDS, and function to regulate cell proliferation and morphogenesis, cell proliferation, and the enzyme-linked receptor protein signaling pathway. Furthermore, the expression of lncENST00000444102 and ABAT was significantly downregulated in MDS bone marrow samples and cell lines. In addition, lncENST00000444102 overexpression reduced tumor cell viability and increased apoptosis in MDS cell lines. This preliminary study links the ABAT-DEL-DEM co-expression network in MDS development. Future studies will further investigate how this network regulates MDS progression.
Indeed, in recent years since completion of the human genome project, research on the functional genome has become a popular research topic. Moreover, utilization of microarrays to profile DELs and DEMs in a disease vs. control is able to identify the causal relationship of altered gene expression; however, these data need to be i) validated in an independent cohort of samples; ii) functionally analyzed in cell lines, or bioinformatically; and iii) analyzed to identify a network or pathway. In the present study, we generated the transcriptomic data from MDS and hyperthyroid samples to construct the network consisting of ABAT, lncRNAs, and mRNAs to determine their role in MDS development. We used a multi-step approach to identify lncRNA-regulated mRNAs and pathways in MDS. We first generated DEL and DEM data using the Agilent human genome-wide gene expression 60K BeadChips in four cases of MDS patients vs. four cases of age-matched individuals with hypersplenism. We then identified 2705 DELs and 543 DEMs in MDS bone marrow samples vs. the controls using PCC analysis and thereafter, we used these data to construct the lncRNA and mRNA co-expression network (which contained six DELs related to ABAT and 135 co-expressed mRNAs). The network speculated that a particular mRNA could regulate a number of the targeting lncRNAs, while a single lncRNA can also regulate various mRNAs in MDS development. Furthermore, it is necessary to understand the potential functions of the lncRNA and mRNA co-expression network in MDS. Therefore, we performed the GO and KEGG gene pathway analyses. The gene enrichment analysis suggested that the co-expression network functions to regulate cell proliferation and morphogenesis, cell proliferation, and enzyme-linked receptor protein signaling pathways. The KEGG pathway analysis revealed that the co-expressed network was mainly involved in phagosome and metabolic pathways. However, further studies are needed to confirm these predictions.
As known gene profiling generates an enormous amount of data and it is impossible to handle all of the data in a single study. Thus, in the present study, we chose to focus on lncENST00000444102 and ABAT in MDS by first assessing their expression levels in MDS samples and cell lines, and then by overexpressing this particular lncRNA and knocking down ABAT expression in MDS cell lines. We found that their expression was both downregulated in MDS samples and cell lines. lncENST00000444102 overexpression upregulated ABAT expression but reduced tumor cell viability by inducing apoptosis in vitro. However, knockdown of ABAT expression had no effect on the expression of lncENST00000444102 in MDS cell lines, indicating that lncENST00000444102 should indirectly regulate ABAT expression in MDS cells, although our present study did not identify how lncENST00000444102 affects ABAT expression. Our bioinformatic analysis did reveal various lncRNA targeting genes and our future studies will investigate these genes that may potentially regulate ABAT expression.
The present study profiled DELs and DEMs in MDS bone marrow samples vs. controls and our data analyses were preliminary. We also did not compare the DELs and DEMs to findings in the literature to confirm whether they are indeed altered in MDS. The expression level of lncRNA-targeting genes was not confirmed in MDS using RT-qPCR or their interactions in cells using a luciferase reporter assay. Thus, there are many limitations in the present study, e.g. the small sample size as one of the limitations of the study. If we had included more cases, particularly lower risk cases, we would have obtained different results. In this research, we focused on the differences between MDS and non-MDS, and in future research we need to explore the differences between low-risk and hight-risk MDS. We also need to explore the differences between MDS and AML, CMML (chronic myelomonocytic leukemia) and MDS. A growing number of in vivo lncRNA studies have reported discrepancies with phenotypes observed in cultured cell lines. Thus, we initially validated the function of long non-coding RNAs in vitro and then we will try to illuminate the discrepancies in vivo in subsequent research.
lncRNAs are involved in a variety of biological processes and regulate gene expression in cis or trans through diverse mechanisms. We found that lncENST00000444102 plays a role in MDS development, but how it functions and what changes in gene expression will occur are issues that must be explored and addressed in subsequent research. Moreover, further studies are needed to investigate how this network regulates MDS progression and to assess the molecular biologic role of lncENST00000444102 in MDS. In conclusion, our present study provides novel insight to better understand MDS development and a methodology for future data analysis was hereby proposed.
Supplementary Material
Acknowledgements
The authors would like to thank all patients and physicians who participated in this study.
Funding
The present study was supported in part by grants from the Science and Technology Commission of Shanghai Municipality (grant no. 17ZR1403600) and from the Three-Year Action Plan on Public Health, Phase IV, Shanghai, China (grant no. 15GWZK0801).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YC and GZ performed the experiments, analyzed the data and wrote the manuscript. YC and GZ contributed to sample collection. NL and ZL performed part of the research and collected the data. XW and JG designed the research study and contributed to the drafting of manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Huashan Hospital, School of Medicine, Fudan University. All participants provided a written informed consent form before being enrolled into this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that the authors have no competing interests.
References
- 1.Germing U, Kobbe G, Haas R, Gattermann N. Myelodysplastic syndromes: Diagnosis, prognosis, and treatment. Dtsch Arztebl Int. 2013;110:783–790. doi: 10.3238/arztebl.2013.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsley RC, Ebert BL. Molecular pathophysiology of myelodysplastic syndromes. Annu Rev Pathol. 2013;8:21–47. doi: 10.1146/annurev-pathol-011811-132436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Issa JP. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood. 2013;121:3811–3817. doi: 10.1182/blood-2013-02-451757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glenthøj A, Ørskov AD, Hansen JW, Hadrup SR, O'Connell C, Grønbæk K. Immune mechanisms in myelodysplastic syndrome. Int J Mol Sci. 2016;17:E944. doi: 10.3390/ijms17060944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varney ME, Melgar K, Niederkorn M, Smith M, Barreyro L, Starczynowski DT. Deconstructing innate immune signaling in myelodysplastic syndromes. Exp Hematol. 2015;43:587–598. doi: 10.1016/j.exphem.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gañán-Gómez I, Wei Y, Starczynowski DT, Colla S, Yang H, Cabrero-Calvo M, Bohannan ZS, Verma A, Steidl U, Garcia-Manero G. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 2015;29:1458–1469. doi: 10.1038/leu.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O'Keefe C, Sekeres M, Saunthararajah Y, Maciejewski JP. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Yang F, Li S, Liu M, Ying S, Jia X, Wang X. CpG island methylator phenotype of myelodysplastic syndrome identified through genome-wide profiling of DNA methylation and gene expression. Br J Haematol. 2014;165:649–658. doi: 10.1111/bjh.12811. [DOI] [PubMed] [Google Scholar]
- 9.Zhao G, Li N, Li S, Wu W, Wang X, Gu J. The high methylation of 4-aminobutyrate aminotransferase gene predicts a poor prognosis in patients with myelodysplastic syndrome. Int J Oncol. 2019;54:491–504. doi: 10.3892/ijo.2018.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina-Kauwe LK, Tobin AJ, De Meirleir L, Jaeken J, Jakobs C, Nyhan WL, Gibson KM. 4-Aminobutyrate aminotransferase (GABA-transaminase) deficiency. J Inherit Metab Dis. 1999;22:414–427. doi: 10.1023/A:1005500122231. [DOI] [PubMed] [Google Scholar]
- 11.Jaeken J, Casaer P, de Cock P, Corbeel L, Eeckels R, Eggermont E, Schechter PJ, Brucher JM. Gamma-aminobutyric acid-transaminase deficiency: A newly recognized inborn error of neurotransmitter metabolism. Neuropediatrics. 1084;15:165–169. doi: 10.1055/s-2008-1052362. [DOI] [PubMed] [Google Scholar]
- 12.Wegerer M, Adena S, Pfennig A, Czamara D, Sailer U, Bettecken T, Müller-Myhsok B, Modell S, Ising M. Variants within the GABA transaminase (ABAT) gene region are associated with somatosensory evoked EEG potentials in families at high risk for affective disorders. Psychol Med. 2013;43:1207–1217. doi: 10.1017/S0033291711002923. [DOI] [PubMed] [Google Scholar]
- 13.Maguire SE, Rhoades S, Chen WF, Sengupta A, Yue Z, Lim JC, Mitchell CH, Weljie AM, Sehgal A. Independent effects of γ-Aminobutyric acid transaminase (GABAT) on metabolic and sleep homeostasis. J Biol Chem. 2015;290:20407–20416. doi: 10.1074/jbc.M114.602276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP, International Molecular Genetics Study of Autism Consortium Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: Evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jirholt J, Asling B, Hammond P, Davidson G, Knutsson M, Walentinsson A, Jensen JM, Lehmann A, Agreus L, Lagerström-Fermer M. 4-Aminobutyrate aminotransferase (ABAT): Genetic and pharmacological evidence for an involvement in gastro esophageal reflux disease. PLos One. 2011;6:E19095. doi: 10.1371/journal.pone.0019095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budczies J, Brockmöller SF, Müller BM, Barupal DK, Richter-Ehrenstein C, Kleine-Tebbe A, Griffin JL, Orešič M, Dietel M, Denkert C, Fiehn O. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: Alterations in glutamine and beta-alanine metabolism. J Proteomics. 2013;94:279–288. doi: 10.1016/j.jprot.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Jansen MP, Sas L, Sieuwerts AM, Van Cauwenberghe C, Ramirez-Ardila D, Look M, Ruigrok-Ritstier K, Finetti P, Bertucci F, Timmermans MM, et al. Decreased expression of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer and endocrine therapy resistance in advanced disease. Mol Oncol. 2015;9:1218–1233. doi: 10.1016/j.molonc.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Malavé NI, Rao DS. Long noncoding RNAs in hematopoietic malignancies. Brief Funct Genomics. 2016;15:227–238. doi: 10.1093/bfgp/elv047. [DOI] [PubMed] [Google Scholar]
- 21.Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, Georgiou I, Bourantas KL. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34:148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, Bloomfield CD. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Huang Da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin MF, Jungreis I, Kellis M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prüss M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geiling B, Vandal G, Posner AR, de Bruyns A, Dutchak KL, Garnett S, Dankort D. A modular lentiviral and retroviral construction system to rapidly generate vectors for gene expression and gene knockdown in vitro and in vivo. PLos One. 2013;8:e76279. doi: 10.1371/journal.pone.0076279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Will S, Jabbari H. Sparse RNA folding revisited: Space-efficient minimum free energy structure prediction. Algorithms Mol Biol. 2016;11:7. doi: 10.1186/s13015-016-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.