Abstract
3-hydroxybutyrate-co-3-hydroxyhexanoate (PHBHHx), which is naturally generated by biodegradable polyhydroxyalkanoates synthesized by bacteria, is an attractive material for drug delivery due to its controllable physical properties, non-toxicity, environmental friendliness, degradable properties and good biocompatibility. However, the intracellular trafficking network pathways, especially the autophagy mechanism of PHBHHx nanoparticles (NPs), have rarely been investigated. In this paper, we successfully prepared the NPs used solvent displacement method and investigated the autophagy pathways and other intracellular trafficking mechanisms based on NPs with the assistance of Rab proteins. We found that NPs were internalized in cells mainly via clathrin endocytosis and caveolin endocytosis. Beside the classical pathways, we discovered two new pathways: the micropinocytosis early endosome (EEs)-micropinocytosis-lysosome pathway and the EEs-liposome-lysosome pathway. NPs were delivered to cells through endocytosis recycling vesicles and GLUT4 exocytosis vesicles. Similar to other nanoparticles, NPs also induced intracellular autophagy and were then degraded via endolysosomal pathways. 3-MA and CQ were used as autophagy inhibitors to avoid the degradation of NPs through lysosomes by blocking endolysosomal pathways. Tumor volumes and weights were significantly decreased when autophagy inhibitors and chemical drugs packaged in NPs were cooperatively used.
Subject terms: Drug delivery, Drug delivery
Introduction
Application of nanoparticles to enhance the effectiveness of diagnostic agents and drugs is becoming a part of our medical armament1–4. More attention should be paid to novel nanoparticles or the diagnosis and treatment of different diseases5–8. Nanoparticles are used as tools to encapsulate and deliver traditional chemotherapeutic agents9. To enhance the therapeutic effects of traditional drugs, drug carriers are expected to have connections with the exterior of membranes to help deliver drugs to the cellular environmental10. Nano-sized features offer certain advantageous properties for nanoparticles on this journey and the intracellular uptake is the important step of the journey for nanoparticles to reach the bio-active site11. Obviously, in-depth understanding of the intracellular transport pathways is critical for the advancement of studies on nanotechnology and drug delivery.
3-hydroxybutyrate- co-3-hydroxyhexanoate (PHBHHx) is a copolymer naturally generated by biodegradable polyhydroxyalkanoates, which are synthesized by bacteria12. PHBHHx copolymer can dissolved into the common organic solvent such as methylene dichloride which make it a good candidate to apply to the biology and chemistry research field. Because it comes from microbes, PHBHHx NPs has unique characteristics due to its controllable physical and chemical properties, environmental friendliness, good degradable properties and biocompatibility when compared to other nanocarrier particles, including carbon nanotubes, liposome micelles, silica nanoparticles, dendrimers, solid lipid nanoparticles, anodic alumina nanotubes13–18. However, little research on the intracellular trafficking pathways of PHBHHx nanostructured materials has been made. In this article, we report a comprehensive investigation on the intracellular trafficking pathway of PHBHHx nanoparticles (NPs). 3-MA and CQ were used as autophagy inhibitors to avoid the degradation of PHBHHx NPs, and their implications for drug delivery are also explored. The volumes and weights of the tumors were significantly decreased when autophagy inhibitors and chemical drugs packaged in PHBHHx NPs were cooperatively used. Biological studies can provide supportive information on the understanding of other spherical nanostructured materials as well.
Rab GTPases are a family of proteins that act as coordinators of the vesicular trafficking pathways that are responsible for transporting the vast array of cellular cargo across membrane organelles19. Approximately 70 human Rab proteins have been discovered, most of which show close relationships with the trafficking of vesicles in and across intracellular trafficking vesica20. In this paper, Rab was used as a marker to explore the intracellular trafficking mechanism of PHBHHx NPs.
Results and Discussion
Preparation and characterization of PHBHHx NPs
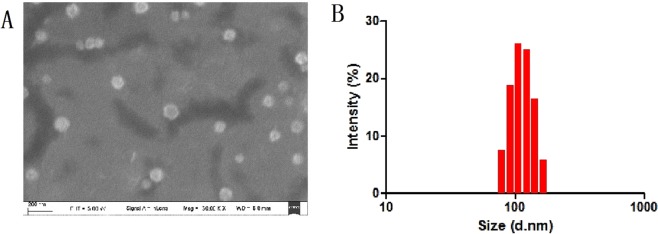
As shown in the SEM images in Fig. 1, the size of the PHBHHx NPs was approximately 110 nm, the size distribution analysis by DLS measurement (Fig. 1B) also shows good agreement with the SEM results. The zeta potential of the NPs was measured to be −10 ± 0.3 mV, indicating the negatively charged nature of the materials. The LC and EE of the paclitaxel-NPs were 7.41% and 68.36%, respectively.
Figure 1.
SEM characterization of PHBHHx NPs (A) DLS size distribution of PHBHHx NPs (B).
Endocytosis pathways of PHBHHx NPs
The endocytosis pathways are characterized by the engulfment of extracellular macromolecules, such as proteins, other foreign invader-like nanoparticles, or membrane constituents, via membrane invagination21. There are two classical endocytosis pathways: clathrin -dependent and clathrin-independent. Clathrin-dependent endocytosis can be subdivided into caveolin-dependent, caveolin-independent and macropinocytosis pathways based on the involvement of membrane proteins called caveolins. The caveolin-independent pathway can be further divided into Arf-6, RhoA-dependent, Cdc42 and flotillin pathways22. The classical endocytosis pathways of nanoparticles begin with their sequestration in vesicles, and then their delivery to EEs, LEs, and subsequently lysosomes as the final destination23.
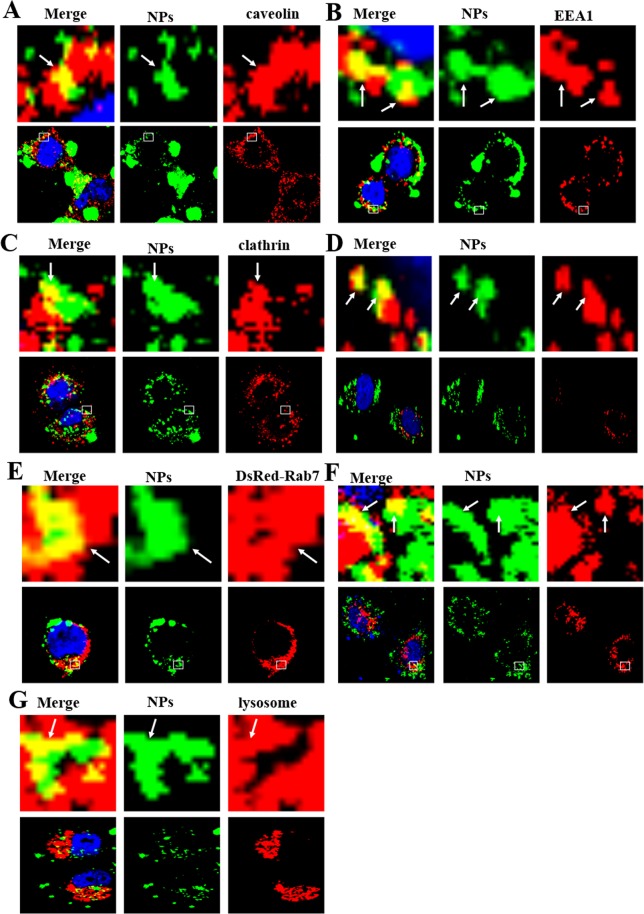
Coumarin-6-labeled NPs were prepared to identify the specific internalization pathways. MCF-7 cells were cultured with 1 mg/mL Coumarin-6-labeled NPs at 37 °C for 3 h, and the Coumarin-6-positive vesicles co-localized with caveolin-positive vesicles and clathrin-dependent positive vesicles (Figs 2A, S1A–D), rather than with other endocytosis-related vesicles (i.e., Arf-6, clathrin, flotillin, Cdc42, and RhoA-positive vesicles). This result clearly shows that cells internalize PHBHHx nanoparticles through clathrin-dependent pathways and caveolin-dependent endocytosis, a subtype of clathrin-independent pathway (Fig. 2A–C).
Figure 2.
Confocal images of endocytosis pathways. In MCF-7 cells, the NPs enter the cells through clathrin-dependent and caveolin-dependent endocytosis (A–C) after treatment with 1 mg/mL coumarin-6-labeled NPs for 3 h. Clathrin, caveolin and EEA1 were detected by primary antibodies against clathrin, caveolin and EEA1. (D–F) DsRed-Rab5, 7 and 9 cells were incubated with 1 mg/mL coumarin-6-labeled NPs for 3 h. (F) 1 mg/mL coumarin-6-labeled NPs transfected MCF-7 cells for 3 h and detected by Lyso-Tracker Red probes for 30 min. The above images are the enlarged ones in the white collar on the underside images. Scale bars: 10 μm.
A large array of Rab proteins has been identified, and they were confirmed to have a close relationship with vesicle trafficking and transport24. To validate the detailed endocytosis pathways of NPs, we use labeled Rab family proteins as markers. The first experiment carried out was a study on the relationship between NPs and two organelles (EEs and LEs) in classic endocytosis pathways using DsRed-Rab5, 7 and 9 proteins as probes.
The cells transfected by DsRed-Rab5, 7, and 9 were incubated with coumarin-6-labeled NPs at 37 °C for 3 h. As shown in Fig. 2B,D, Coumarin-6-containing vesicles merged with EEA1 and DsRed-Rab5, the markers of early endosomes. Meanwhile, coumarin-6-positive vesicles also fused with DsRed-Rab7 and DsRed-Rab9, the markers of late endosomes (Fig. 2E,F). These images revealed a classical endocytosis pathway involved in the process of NP endocytosis. This postulation can be further demonstrated by the observation of merged images of Coumarin-6-positive vesicles and lysosomes (Fig. 2G). These experiments provided an inside view of NP endocytosis pathways, indicating that the NPs were transported to EEs and LEs and degraded in lysosomes. This finding is identical to the classical endocytosis pathways.
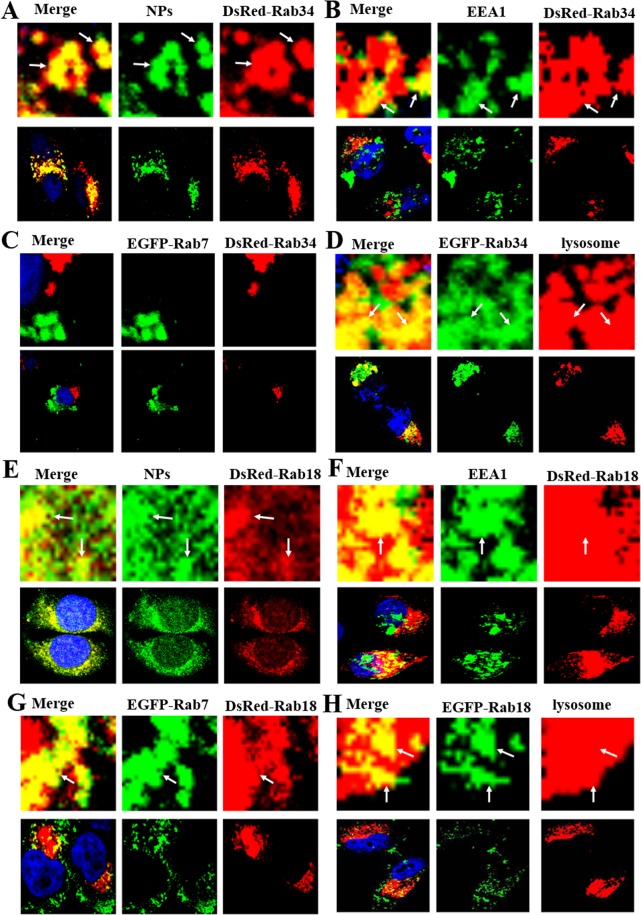
In addition to studies on EEs and LEs, other Rab proteins were used to analyze other pathways. Among Rab proteins, Rab18 is highly associated with the activity of lipid droplets, and Rab34 is a biomarker of the micropinocytosis process; Rab18 and Rab34 can therefore be used to illustrate the roles of lipid droplets and micropinocytosis, respectively, in NP endocytosis pathways25,26. DsRed-Rab18 and DsRed-Rab34 transfected MCF-7 cells were cultured at 37 °C for 3 h. Both Rab18 and Rab34 were observed to merge with NP-containing vesicles (Fig. 3A,E). To obtain more detailed information on the role of lipid droplets in the endocytosis pathways, the reactions between lipid droplets and markers for classical endocytosis paths (EEA1 and EGFP-Rab7) were also examined. Figure 3F–H shows the integration of Rab18-positive vesicles with EEA1, lysosomes, and EGFP-Rab7. This finding serves as proof of the participation of liposomes in the endocytosis pathways of NPs. This finding was also applied to studies on DsRed-Rab34, EGFP-Rab7 and lysosomes in the micropinocytosis process. To our surprise, Rab34 was found to merge with EEA1, lysosome but not EGFP-Rab7 (Fig. 3B–D). All this evidence points to the existence of a new endocytosis pathway: EEs (EEA1-positive)-macropinocytosis (Rab34 -positive)-lysosomes.
Figure 3.
Confocal microscopy pictures of endocytosis pathways. (A) MCF-7 cells were transfected by DsRed-Rab34 and then cultured with 1 mg/mL coumarin-6-labeled NPs for 3 h. (B) DsRed-Rab34 cells were cultured with 1 mg/mL NPs for 3 h; EEA1 was detected with a primary antibody against EEA1. (C) DsRed-Rab34 cells were co-cultured with EGFP-Rab7 and 1 mg/mL NPs for 3 h. (D) EGFP-Rab34 cells were cultured with 1 mg/mL NPs for 3 h and Lyso-Tracker Red probes for 30 min. (E–H) The same procedure was repeated, except that DsRed-Rab34 was replaced by DsRed-Rab18. The above images are the enlarged ones in the white collar on the underside images. Scale bars: 10 μm.
Recycling endosome pathways of NPs
The recycling endosome is an important organelle for the redelivery of protein receptors back to cell membranes from EEs27. According to previous reports, the action of recycling endosomes is facilitated by numerous accessory proteins. DsRed-Rab11 and DsRed-Rab35 participate in the process of slow endocytic recycling, and DsRed-Rab20 and DsRed-Rab25 are highly engaged in transportation between the apical recycling endosomes and apical plasma membrane28.
Herein, we report a study on intracellular trafficking pathways. MCF-7 cells were transfected with DsRed-Rab 11, 20, 25 and 35 and incubated with Coumarin-6-labeled NPs at 37 °C for 3 h. As shown in Figure S2A,B Rab11 and Rab35 marked slow recycling endosomes that fused with Coumarin -6-positive NPs, which also occurred on recycling endosomes marked with Rab20 and Rab25 (Figure S2C,D). As shown above, both slow and apical recycling endosome paths participate in the process of releasing NPs back to the cell membrane. The metabolic pathways of NPs started from clathrin -dependent pathways and caveolin-dependent endocytosis. Through these two processes, NPs were internalized in the cell and further transported to EEs, LEs, and lysosomes in classic endocytosis pathways. In addition, a new chain with the participation of EEs, macropinocytosis and lysosomes was discovered. Then, at the late stage of this metabolic pathway, both slow and apical recycling endosomes were observed in the inside-out process of NPs. Rab18-labeled liposomes were also observed in classical endocytosis pathways.
Exocytosis pathways of NPs
Secretory vesicles release their contents out of the cell through exocytosis29. This action is assisted by a group of Rab proteins. Recent studies on NPs showed that DsRed-Rab8, 10, and 14 regulate the flow of GLUT4 vesicles on Golgi20. To determine whether NPs can be transported out of the cell through pathways that the labeled proteins could monitor, DsRed-Rab8 and DsRed-Rab10 transfected MCF-7 cells were used. The transfected cells were further incubated with Coumarin -6-labeled NPs at 37 °C for 3 h. Figure S3A,B presents a merged image of Rab-positive vesicles (GLUT4 vesicle, marked with DsRed-Rab8 and Rab10) and Coumarin -6-positive vesicles. This evidence points to the involvement of Rab8- and Rab10-positive GLUT4 vesicle in the exocytosis process of NPs. Two critical procedures occur in the GLUT4-related pathways: retrieval of contents from early endosomes or LEs to the TGN, and formation of the vesicles derived from the TGN donor membranes.
As the contents could be retrieved from either EEs or LEs, they were studied separately. First, DsRed-Rab22 and DsRed-Rab31 were used as markers for the vesicles that transport from EEs to the TGN. MCF-7 cells transfected with Rab22 and 31 were further incubated with Coumarin-6-labeled NPs at 37 °C for 3 h. The merged confocal microscopic images produced from Rab-labeled vesicles (DsRed-Rab22 and -31 positive) and Coumarin-6-positive vesicles, as shown in Figure S3C,D, implied the interaction between EEs and TGN. Then, experiments on DsRed-Rab9 cells showed that DsRed-Rab9, 22, and 31 positive vesicles merged with Coumarin-6-positive vesicles. In line with this model, NPs were transmitted from LEs to the TGN. It is clearly established that the TGN and Golgi body could receive NPs from EEs and LEs, Rab8- and 10-positive GLUT4 transport vesicles paths are employed to move NPs (Figure S3E).
Autophagy pathways of NPs
Autophagy is a highly regulated process for eliminating a variety of intracellular materials, ranging from proteins to organelles, via lysosomal mechanisms30. Autophagy is a double-edged sword31. When tumors occur, cancer cells confer stress tolerance to stressors use autophagy, thereby maintaining tumor cell survival30. Autophagy is also activated when chemotherapeutic drugs invade32. When autophagy is induced, LC3-I is decreased, its C-terminal end is cut and LC3-II is generated from its precursor form33. LC3-I to LC3-II conversion is highly associated with the level of autophagosome formation, and the amount of LC3-II is an established marker of autophagy activity11.
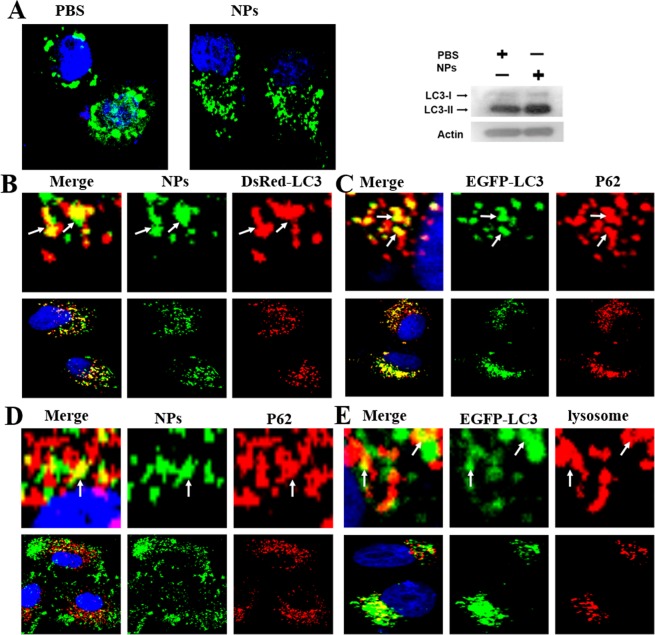
The EGFP-LC3 transfected MCF-7 cells were incubated with Coumarin-6-labeled NPs at 37 °C for 3 h. A large amount of autophagosomes appeared, and the level of LC3-II increased (Fig. 4A). The cells may regard the NPs as foreign invaders. To prove that NPs can induce autophagy, we used a red fluorescent probe (DsRed-LC3) to examine autophagy. The DsRed-LC3 cells were incubated with Coumarin-6-labeled NPs at 37 °C for 3 h (Fig. 4B). As we speculated, there were quantities of autophagosomes in cells. P62 is a marker of selective autophagy. We treated the cells with P62 and found that P62-positive vesicles could merge with Coumarin-6-positive vesicles (Fig. 4C,D). Therefore, NPs can induce selective autophagy. From the results above, we hypothesized that NPs was captured by P62 and transported to autophagosomes before being degraded in lysosomes (Fig. 4E).
Figure 4.
Confocal microscopy studies autophagy induced by NPs. (A) EGFP-LC3 cells were cultured with 1 mg/mL Coumarin-6-labeled NPs for 3 h. Western blotting of the cells cultured in (A). (B) DsRed-LC3 cells were cultured with 1 mg/mL Coumarin-6-labeled NPs for 3 h. (C) MCF-7 cells were cultured with 1 mg/mL NPs for 3 h, and P62 was detected with a primary antibody against P62. (D) The autophagosomes fused with lysosomes (arrows). (E) EGFP-LC3 cells were cultured with 1 mg/mL NPs for 3 h and Lyso-Tracker Red probes for 30 min. The above images are the enlarged ones in the white collar on the underside images. Scale bars: 10 μm.
Crosstalk among endocytosis, exocytosis and autophagy
Recent studies showed that autophagy is strongly correlated with Rab7, 8, 9, 11, 20, 32 and 33, which implies a relationship between endocytosis, exocytosis, and autophagy26. MCF-7 cells were co-transfected with EGFP-LC3 and DsRed-Rab proteins and incubated with Coumarin-6-labeled NPs at 37 °C for 3 h. Figure S4A–D shows that EGFP-LC3-positive vesicles merged with DsRed-Rab7, 34, 23 and 18. This finding demonstrated that autophagosomes might receive the NP vesicles from endocytosis pathways. Figure S4E,F shows that EGFP-LC3 positive vesicles merged with DsRed-Rab11 and 35, suggesting that autophagosomes might receive the NP vesicles from recycling endosome pathways. As EGFP-LC3-positive vesicles merged with DsRed-Rab8- and DsRed-Rab10-positive vesicles, we can also speculate that autophagosomes might receive the NP vesicles from GLUT4 vesicles (Figure S4G,H). Thus, there exists a complex mechanism among endocytosis, exocytosis and autophagy in cells.
3-MA and CQ as inhibitors of autophagy
As a double-edged sword, autophagy can not only prevent the formation of cancer but also can provide the endurance of survival for the cancer cells once the tumor formed. With the development of chemistry and biology technologies, many chemotherapeutic drugs are synthesized for different cancers. However, most can induce autophagy, which decreases the effect of drugs. We designed an experiment to test this hypothesis. Paclitaxel, doxorubicin, 5-FU and cyclophosphamide are representative chemotherapeutic drugs. MCF-7 cells were cultured with EGFP-LC3, then incubated with DMSO, paclitaxel, doxorubicin, 5-FU or cyclophosphamide at 37 °C for 24 hour. As expected, a large amount of autophagosomes was induced (Figures S5A and S6A).
3-MA and CQ are two commonly used autophagy inhibitors31. 3-MA inhibits autophagy via the inhibition of type III phosphatidylinositol 3-kinases (PI-3K), CQ prevents autophagy by blocking autophagosome fusion and degradation31. MCF-7 cells were further co-cultured with the autophagy inhibitors 3-MA and CQ and the chemotherapeutic drugs. Figures S5B–D and S6B show that there were fewer autophagosomes in the 3-MA group and more autophagosomes in the CQ group compared to the control group, indicating that 3-MA prevented the formation of autophagosomes and CQ inhibited autophagosome fusion with lysosomes.
Inhibiting autophagy blocks the degradation of NPs in auto-lysosome pathways
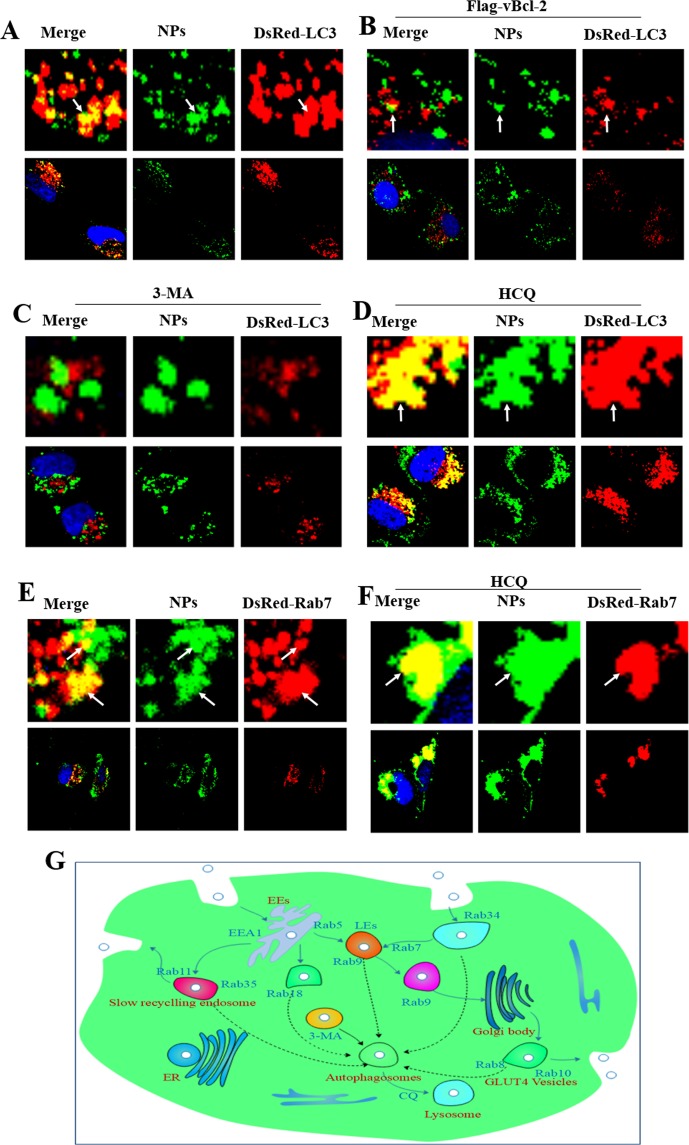
Because NPs can induce autophagy and nanoparticle degradation in lysosomes, autophagy inhibitors should block the degradation of NPs through the auto-lysosome pathway. A comparative study of autophagy inhibition by 3-MA and CQ was performed. The DsRed-LC3 transfected MCF-7 cells were incubated with Coumarin-6-labeled NPs and 3-MA at 37 °C for 3 h. DsRed-LC3-positive vesicles did not merge with Coumarin -6-positive vesicles (Fig. 5A–C). Therefore, 3-MA possessed the function of inhibiting autophagy induced by NPs. In the same way, we found Coumarin-6-positive vesicles aggregated in LEs (Fig. 5D). CQ can interrupt the paths of NPs from LEs to lysosomes. We then incubated the DsRed-LC3 transfected cells with Coumarin-6-labeled NPs, CQ and DsRed-Rab7 at 37 °C for 3 hour. The control group was in the same condition but lacked CQ (Fig. 5E,F). Figure 5 shows that the result was as expected. In summary, chemical drugs such as 3-MA and CQ can not only inhibit autophagy and endocytosis pathways, but can also block NPs degraded in lysosome pathways (Fig. 5G).
Figure 5.
Confocal microscopy pictures of inhibiting autophagy of NPs. (A,B) DsRed-LC3 cells were cultured with 1 mg/mL Coumarin-6-labeled NPs for 3 h; DsRed-LC3 cells were co-treated with Flag-vBcl-2 and 1 mg/mL Coumarin-6-labeled NPs for 3 h. (C,D) The DsRed-LC3 cells were cultured with 10 μM 3-MA or 30 μM CQ. (E,F) DsRed-Rab7 cells were cultured with 1 mg/mL Coumarin-6-labeled NPs for 3 h; the DsRed-Rab7 cells were cultured with 30 μM CQ. (G) Schematic diagram of NP degradation pathways in the intracellular network. The above images are the enlarged ones in the white collar on the underside images.
Autophagy inhibitor increased the tumor suppression effect of paclitaxel by inhibiting autophagy in vivo
From the previous cell experiments, we found that autophagy inhibitors such as CQ can block NPs degraded in lysosome pathways. We thus speculated that the use of autophagy inhibitors together with drugs in the nanoparticle drug delivery platform can increase the effectiveness of the chemical drugs.
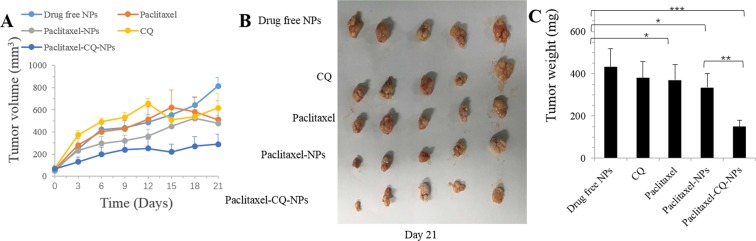
The xenograft SCID mouse model is used to verify the curative effects. We treated the mice with drug-free NPs, paclitaxel, paclitaxel-NPs, CQ and paclitaxel-CQ-NPs every three days for seven continuous cycles. After the intraperitoneal injections, the tumor sizes were measured. The control group was injected with normal saline. Paclitaxel-NPs had an inhibitory effect on tumor growth. Paclitaxel- and CQ-NPs had similar effects on tumor growth. The paclitaxel without NPs may have been almost degraded in the lysosome. The sizes of the tumors treated with paclitaxel-CQ-NPs were markedly lower than those of the other groups, demonstrating that the cooperative use of autophagy inhibitor and chemical drugs packaged in NPs can remarkably increase the curative effect (Fig. 6).
Figure 6.
Autophagy inhibitor CQ increased the tumor suppression effect of drugs by inhibiting autophagy in vivo. Tumor growth curve of the SCID mice bearing MCF-7 cell xenografts after injection with drug-free NPs, paclitaxel, paclitaxel-NPs, CQ and paclitaxel-CQ-NPs. (A) Morphology of the tumors of each group removed from the sacrificed mice at the study end point. (B) Tumor weight at the study end point. (C) Data are shown as the means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared to controls.
Conclusions
In this study, we found that NPs were internalized in cells mainly by clathrin endocytosis and caveolin endocytosis. NP movement into cells followed the classical endocytosis paths: EEs-LEs-lysosomes. We also discovered two new paths: the micropinocytosis EEs-micropinocytosis- lysosomes paths and the EEs-liposome- lysosomes paths. Endocytosis recycling and GLUT4 exocytosis vesicles were the paths through which NPs were delivered from cells. Similar to nanoparticles, NPs also induced intracellular autophagy and were then degraded via the endolysosomal pathways. Autophagy inhibitors, such as 3-MA and CQ, were able to block the endolysosomal pathways to avoid the degradation of NPs in lysosomes. The co-delivery of CQ and paclitaxel in NPs dramatically prevented the growth of tumors in vivo. These new intracellular network traffic mechanisms will provide new ideas for exploring the cellular behavior of nanoparticles. A new enduring and efficient drug delivery system, such as the co-delivery of an autophagy inhibitor and chemical drugs in the same nanoparticle, may be developed to help cure cancer.
Materials and Methods
Animals
Xenograft SCID mouse (6−8week sold) used inthe experiments were obtained from the Guang dong MedicalLaboratory Animal Center (Guangdong,China). All animal experiments followed the animal protocols approved by the Animal Care and Use Committee of Tsinghua University. Animals received care following the NSFC regulations concerning the use of experimental animals.
Materials
PHBHHx is kindly provided by BluephaLab, China. Phosphoric acid (H3PO4), ethanol (C2H5OH), coumarin-6 and hydrochloric acid (HCl) were bought from Shenzhen Tianxiang chemical glass instrument trading company (China). Polyvinyl alcohol(PVA), dimethyl sulphoxide (DMSO), acetone (C3H6O), Chloroquine (CQ), bovine serum albumin and 3-methyladenine (3-MA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). N-acryloxysuccinimide (NAS), acrylamide (AAm), N, N, N′, N′-tetramethylethylenediamine (TEMED), N, N-methylene bisacrylamide (BIS), ammonium persulphate (APS), were obtained from Aladdin Industrial Co. LTD. (Shanghai, China). Antibodies against LC3, Arf-6, RhoA, Flotillin, Caveolin, Cdc42, P62, EEA1, Clathrin, were from Cell Signaling Technology. Lyso-Tracker Red and N-(3-Aminopropyl) methacrylamide hydrochloride were from Beyotime Biotechnology (Shanghai, China) and Polymer Science, Inc., respectively.
Preparation and characterization of PHBHHx NPs
PHBHHx nanoparticles (NPs) were formulated via a solvent displacement method34,35. Briefly, 5 mg PHBHHx was dissolved in 1 mL of dichloromethane, followed by intense stirring. Then, the mixture was added dropwise into 10 mL of 0.6% polyvinyl alcohol (PVA) under magnetic stirring at 800 rpm, and then stirred for 10 min. The system was left in the fume hood overnight under magnetic stirring to eliminate the dichloromethane. The NPs were collected by centrifugation at 12,000 rpm for 15 min at room temperature and washed twice. Then, the particles were dispersed in ultra-pure water and freeze-dried for at least 23 h. The freeze-dried powder was kept in the freezer until use.
The method of Coumarin-6-loaded NPs and paclitaxel-NPs was the same as described above except that 5 mg mL−1 and 50 µg mL−1 of coumarin-6 and 10 mg paclitaxel powders were added instead of using pure PHBHHx.
The drug loading content (LC) and drug encapsulation efficiency (EE) were calculated like the previously report36. Briefly, under vigorous vortexing, paclitaxel-NPs were dissolved in 1 mL of dichloromethane. The solution was transferred to 5 mL of mobile phase consisting of deionized water and acetonitrile (50:50,v/v). A nitrogen stream was introduced to evaporate dichloromethane for about 15 min, and then a clear solution was obtained for HPLC analysis (LC 1200, Agilent Technologies, Santa Clara, CA, USA). The measurement was performed in triplicate.
Plasmid and transfection
DsRed-Rab5 and DsRed-Rab7 were obtained from Addgene. KSHV Flag-vBcl-2 plasmid was kindly provided by Professor Beth Levine from Department of Medicine, University of Texas. Rab family genes in the T Vector and sub-cloned into EGFP-C1 and DsRed-C1 were kindly provided by Professor Jiahuai Han’s Lab. All the plasmids were confirmed by automated DNA sequencing. And cells were transfected with the plasmids by Lipofectamine 2000 (Invitrogen).
Characterization of NPs
The shape and structure of NPs were characterized by a scanning electron microscope (ZEISS, SUPRA-55, SAPPHIRE). Zeta potential was measured with a Malvern particle sizer Nano-ZS.
Cell culture
The MCF-7 cells were incubate in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS).
Cellular uptake of NPs
Non-transfected or DsRed-Rab family gens were cultured with 1 mg/mL coumarin-6-label NPs at 37 °C for 3 h. Cells were incubated with Lyso-Tracker Red for 0.5 h for lysosome detection. After washing with PBS for three times, the cells were fixed with 4% paraformaldehyde for 10 min, stained with DAPI for 5 min and washed with PBS for three times. The Confocal microscopy was performed with a FLUO-VIEW laser scanning confocal microscope (Olympus, FV1000, Olympus Optical, Japan) under sequential scanning mode using a 60~100 × objective37.
Autophagy assays
Cells were transfected with EGFP-LC3 under the conditions described above. Confocal microscopic images were used to calculate the EGFP-LC3 translocation. An anti-LC3 antibody was quantified by Level of LC3II protein38.
Immunoblotting
Immunoblotting analysis was similar to the ref.39 In short, cell lysates were dissolved in 12% SDS-PAGE by immunoblotting with a LC3 antibody. An ECL detection system (Thermo Fisher Scientific, Schwerte, Germany) was used in this process.
Immunofluorescence assay
Primary antibodies, EEA1, P62, Caveolin, Cdc42, Clathrin, LC3, Flotillin, Arf-6 were used to incubate the cells.
Statistical methodology
All results are reported as the mean ± S.E.M. of three independent experiments. Comparisons were performed using a two-tailed paired Student’s t test. (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary information
Acknowledgements
This work was financially supported by National Natural Science Foundations of China (Grants 21573124 and 21874082) and Innovation Foundation for Science and Technology of Shenzhen (Grants JCYJ20170413104646428 and JCYJ20170817172150505).
Author Contributions
S. Sun and X. Jin conceived the project and designed the experiments. X. Sun, C. Cheng and J. Zhang was primarily responsible for experimental operation, data collection, analysis and discussions. L. Mei and L. Huang revised the manuscript. X. Sun prepared figures and wrote the main manuscript text. S. Sun supervised the whole research. All authors contributed to experimental design, data analysis and discussions.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiangyu Sun, Cheng Cheng and Jinxie Zhang contributed equally.
Contributor Information
Xing Jin, Email: jinxing70@163.com.
Shuqing Sun, Email: sun.shuqing@sz.tsinghua.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45632-y.
References
- 1.Esfandiari, N., Arzanani, M. K. & Koohi-Habibi, M. The study of toxicity and pathogenicity risk of Potato Virus X/Herceptin nanoparticles as agents for cancer therapy. Cancer Nanotechnol. 9, 1 (13 pp.)-11 (13 pp.), 10.1186/s12645-018-0036-6 (2018).
- 2.Hao J, et al. Temporary suppression the sequestrated function of host macrophages for better nanoparticles tumor delivery. Drug Deliv. 2018;25:1289–1301. doi: 10.1080/10717544.2018.1474965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang XD, Gu XQ, Wang HM, Yang JY, Mao SR. Enhanced delivery of doxorubicin to the liver through self-assembled nanoparticles formed via conjugation of glycyrrhetinic acid to the hydroxyl group of hyaluronic acid. Carbohydr. Polym. 2018;195:170–179. doi: 10.1016/j.carbpol.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 4.Li F, et al. Facile Synthesis of pH-sensitive Germanium Nanocrystals with High Quantum Yield for Intracellular Acidic Compartment Imaging. Small. 2015;11:1954–1961. doi: 10.1002/smll.201402743. [DOI] [PubMed] [Google Scholar]
- 5.Guo XL, et al. Multi-functionalized chitosan nanoparticles for enhanced chemotherapy in lung cancer. Carbohydr. Polym. 2018;195:311–320. doi: 10.1016/j.carbpol.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Bush X, Yan BF, Chen JA. Gemcitabine nanoparticles promote antitumor immunity against melanoma. Biomaterials. 2019;189:48–59. doi: 10.1016/j.biomaterials.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li GH, Zhu L, He YH, Tan H, Sun SQ. Digital triplex DNA assay based on plasmonic nanocrystals. Anal. Bioanal.Chem. 2017;409:3657–3666. doi: 10.1007/s00216-017-0307-9. [DOI] [PubMed] [Google Scholar]
- 8.Li GH, et al. Digital Concentration Readout of DNA by Absolute Quantification of Optically Countable Gold Nanorods. Anal. Chem. 2016;88:10994–11000. doi: 10.1021/acs.analchem.6b02712. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Santos A, Kaur G, Evdokiou A, Losic D. Structurally engineered anodic alumina nanotubes as nano-carriers for delivery of anticancer therapeutics. Biomaterials. 2014;35:5517–5526. doi: 10.1016/j.biomaterials.2014.03.059. [DOI] [PubMed] [Google Scholar]
- 10.Afsharzadeh M, Hashemi M, Mokhtarzadeh A, Abnous K, Ramezani M. Recent advances in co-delivery systems based on polymeric nanoparticle for cancer treatment. Artif. Cells Nanomed. Biotechnol. 2018;46:1095–1110. doi: 10.1080/21691401.2017.1376675. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Enhancing therapeutic effects of docetaxel-loaded dendritic copolymer nanoparticles by co-treatment with autophagy inhibitor on breast cancer. Theranostics. 2014;4:1085–1095. doi: 10.7150/thno.9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y-C, et al. A specific drug targeting system based on polyhydroxyalkanoate granule binding protein PhaP fused with targeted cell ligands. Biomaterials. 2008;29:4823–4830. doi: 10.1016/j.biomaterials.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chemical Society Reviews. 2012;41:2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye C, et al. PHB/PHBHHx scaffolds and human adipose-derived stem cells for cartilage tissue engineering. Biomaterials. 2009;30:4401–4406. doi: 10.1016/j.biomaterials.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Peng Q, Zhang ZR, Gong T, Chen GQ, Sun X. A rapid-acting, long-acting insulin formulation based on a phospholipid complex loaded PHBHHx nanoparticles. Biomaterials. 2012;33:1583–1588. doi: 10.1016/j.biomaterials.2011.10.072. [DOI] [PubMed] [Google Scholar]
- 16.Croissant JG, Fatieiev Y, Khashab NM. Degradability and Clearance of Silicon, Organosilica, Silsesquioxane, Silica Mixed Oxide, and Mesoporous Silica Nanoparticles. Advanced Materials. 2017;29:1–51. doi: 10.1002/adma.201604634. [DOI] [PubMed] [Google Scholar]
- 17.Martins JP, et al. Engineered Multifunctional Albumin-Decorated Porous Silicon Nanoparticles for FcRn Translocation of Insulin. Small. 2018;14:1–11. doi: 10.1002/smll.201800462. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, et al. Systematic investigation of intracellular trafficking behavior of one-dimensional alumina nanotubes. Journal of Materials Chemistry B. 2019 doi: 10.1039/c8tb03349h. [DOI] [PubMed] [Google Scholar]
- 19.Jordens I, Marsman M, Kuijl C, Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–1077. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 20.Bhuin T, Roy JK. Rab proteins: the key regulators of intracellular vesicle transport. Exp. Cell Res. 2014;328:1–19. doi: 10.1016/j.yexcr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 21.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J. Controlled Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huotari J, Helenius A. Endosome maturation. The EMBO journal. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peruchetti DDB, Silva-Aguiar RP, Siqueira GM, Dias WB, Caruso-Neves C. High Glucose Reduces Megalin-Mediated Albumin Endocytosis in Renal Proximal Tubule Cells through Protein Kinase B O-GlcNAcylation. J Biol Chem. 2018;29:11388–11400. doi: 10.1074/jbc.RA117.001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozeki S, et al. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 26.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nature reviews Molecular cell biology. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 27.Allgood Samual C., Neunuebel M. Ramona. The recycling endosome and bacterial pathogens. Cellular Microbiology. 2018;20(7):e12857. doi: 10.1111/cmi.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akinc A, Battaglia G. Exploiting endocytosis for nanomedicines. Cold Spring Harbor Perspect. Biol. 2013;5:a0169801–a01698024. doi: 10.1101/cshperspect.a016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruinsma S, et al. Small molecules that inhibit the late stage of Munc13-4-dependent secretory granule exocytosis in mast cells. Journal of Biological Chemistry. 2018;293:8217–8229. doi: 10.1074/jbc.RA117.001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Mei L, Zhang XD, Feng SS. Autophagy inhibition strategy for advanced nanomedicine. Nanomedicine. 2014;9:377–380. doi: 10.2217/nnm.13.218. [DOI] [PubMed] [Google Scholar]
- 32.Szatmári Z, Sass M. The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy. 2014;10:1154–1166. doi: 10.4161/auto.29395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penaloza JP, et al. Intracellular trafficking and cellular uptake mechanism of PHBV nanoparticles for targeted delivery in epithelial cell lines. J Nanobiotechnology. 2017;15:1–15. doi: 10.1186/s12951-016-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, et al. Enhanced anti-tumor efficacy by co-delivery of doxorubicin and paclitaxel with amphiphilic methoxy PEG-PLGA copolymer nanoparticles. Biomaterials. 2011;32:8281–8290. doi: 10.1016/j.biomaterials.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Zeng XW, et al. Cholic acid-functionalized nanoparticles of star-shaped PLGA-vitamin E TPGS copolymer for docetaxel delivery to cervical cancer. Biomaterials. 2013;34:6058–6067. doi: 10.1016/j.biomaterials.2013.04.052. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J. et al. Doxorubicin-loaded star-shaped copolymer PLGA-vitamin E TPGS nanoparticles for lung cancer therapy. Journal of Materials Science-Materials in Medicine26, 10.1007/s10856-015-5498-z (2015). [DOI] [PubMed]
- 38.Hoyer-Hansen M, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Molecular Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Liang X, et al. pH-Triggered burst intracellular release from hollow microspheres to induce autophagic cancer cell death. Journal of Materials Chemistry B. 2015;3:9383–9396. doi: 10.1039/c5tb00328h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.