Abstract
Solar UV radiation is a major environmental risk factor for skin cancer. Despite decades of robust and meritorious investigation, our understanding of the mechanisms underlying UV-induced skin carcinogenesis remain incomplete. We previously performed comprehensive transcriptomic profiling in human keratinocytes following exposure to different UV radiation conditions to generate UV-specific gene expression signatures. In this study, we utilized Virtual Inference of Protein Activity by Enriched Regulon (VIPER), a robust systems biology tool, on UV-specific skin cell gene signatures to identify master regulators (MRs) of UV-induced transcriptomic changes. We identified multiple prominent candidate UV MRs, including forkhead box M1 (FOXM1), thyroid hormone receptor interactor 13 and DNA isomerase II alpha, which play important roles in cell cycle regulation and genome stability. MR protein activity was either activated or suppressed by UV in normal keratinocytes. Intriguingly, many of the UV-suppressed MRs were activated in human skin squamous cell carcinomas (SCCs), highlighting their importance in skin cancer development. We further demonstrated that selective inhibition of FOXM1, whose activity was elevated in SCC cells, was detrimental to SCC cell survival. Taken together, our study uncovered novel UV MRs that can be explored as new therapeutic targets for future skin cancer treatment.
We utilized robust systems biology tools to interrogate UV-specific gene expression signatures to identify prominent UV MRs. Our computational, experimental and clinical studies confirmed that FOXM1, a top-ranked MR, is closely involved with skin tumorigenesis.
Introduction
Skin cancer is the most common type of cancer in the United States and is estimated to affect one in every five Americans in their lifetime (1,2). Skin cancer encompasses both melanoma skin cancer and non-melanoma skin cancer, which includes basal cell carcinoma and squamous cell carcinoma (SCC). Genetic factors such as skin phototypes and family history contribute to an individual’s risk of developing skin cancer, as do environmental risk factors including chronic arsenic exposure, photosensitizing drugs, prolonged immunosuppression and ultraviolet radiation (UVR) (3–8). Solar UVR is a well-established carcinogen that can cause structural DNA damage, inflammation and immunosuppression, and can eventually lead to skin tumor formation (3–5). It is estimated that approximately 90% of non-melanoma skin cancers are associated with excessive exposure to UVR, and the incidence increases with age (9,10). Multiple signaling pathways and transcription factors (TFs) have been studied to probe the connection between UV exposure and skin carcinogenesis (4,5,11,12). Despite tremendous progress in the field of skin cancer research, the molecular mechanisms underlying UV-induced skin carcinogenesis remain incompletely understood.
Although ultraviolet B (UVB) accounts for approximately 5% of total solar UVR, UVB is responsible for the majority of sunburns and subsequent cancer incidence. UVB is mostly absorbed in the epidermis, therefore exerting a strong effect on epidermal keratinocytes. Upon skin exposure to UVR, two major forms of DNA photoproducts will occur: cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts (13–15). Cyclobutane pyrimidine dimers are more prevalent and less efficiently repaired than 6-4 pyrimidone photoproducts, and exert stronger carcinogenic effects (15). In addition to the effects of UVB, UVA can cause the formation of cyclobutane pyrimidine dimers or single-strand DNA breaks, or indirect DNA damage via the induction of reactive oxygen species and DNA-protein crosslinks (16,17).
Beyond DNA damage and mutations, UVR exposure has a profound impact on transcriptome stability. A single exposure of skin cells to UVR can cause substantial changes in gene expression, affecting thousands of genes (18,19). However, the regulatory pathways underlying this massive UV-induced gene expression dysregulation remain poorly defined. Traditional fold-change-based analysis of differential gene expression between basal and UVR-stimulated states is unable to distinguish between primary driver events and secondary associative events. Genome-wide reverse engineering approaches, such as the algorithm for the reconstruction of accurate cellular networks (ARACNe), have been developed to infer the direct targets of TFs and signal transduction proteins and construct transcriptional interaction networks (interactome) based on information theory (20). More recently, the Virtual Inference of Protein Activity by Enriched Regulon (VIPER) algorithm was developed to interrogate the interactome and identify enriched transcriptional targets of TFs with master regulator (MR) function that are causally linked with transcriptomic dysregulation in response to specific environmental stresses (21–23). These MR proteins are proposed to occupy gene regulation checkpoints, where multiple cellular signaling pathways converge (23,24).
To better define the molecular control of UV-induced global gene transcriptional dysregulation, we performed ARACNe and VIPER analyses on a large cohort of RNA-seq transcriptomic data sets representing UV-responsive gene signatures and human skin cancer gene profiles to identify UV and SCC MR genes, respectively. These analyses uncovered several prominent MRs of UVR that have important functional implications in skin cancer development. We also provided computational, experimental and clinical evidence supporting that forkhead box M1 (FOXM1), a top-ranked MR of UV response, is closely involved with skin tumorigenesis.
Materials and methods
Collection and processing of gene expression datasets
RNA-seq gene expression data files were obtained from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) for a total of 21 UV-irradiated and corresponding non-irradiated control samples and five pairs of human SCC samples with matched adjacent normal skin (Gene Expression Omnibus accession number GSE85443). Gene expression signatures representing each UV or SCC sample were obtained through differential gene expression analysis using the DESeq2 R software package (25). We also downloaded RNA-seq data from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) for approximately 470 cutaneous melanoma samples. This data was transformed using a variance-stabilizing transformation method and normalized using DESeq2.
Assembly of skin cancer-specific interactome and MR analyses
To generate a skin cancer-specific protein interactome, we first normalized gene expression values in the RNA-seq data downloaded from the TCGA using the DESeq2 package. We then used ARACNe to compute the mutual information between each transcriptional regulator and its target genes. Regulator–target gene interactions with P-value < 1e-7 were used to construct a skin cancer-specific interactome that contained 5701 regulators, including both TFs and signal transduction proteins, which regulate the expression of 20 500 genes through 872 810 putative interactions. We subsequently used this interactome to infer UVR MR proteins with the msVIPER function in the VIPER R package. VIPER analysis transformed the UV gene signatures into protein activity signatures by computing enrichment of the regulator’s target genes in the gene signature of interest. To identify the most conserved and robust set of UV MRs, we averaged regulator activity signatures across all 21 UV signatures. Proteins whose activities were significantly altered (Z-score > 1.96 or <−1.96, P-value < 0.05) were designated as UV MRs. We selected significantly activated or repressed regulators that represent candidate MRs of UV response. We identified MRs of SCC carcinogenesis through similar VIPER analyses using SCC signatures that we reported previously (25).
Gene set enrichment analysis and survival analysis
We performed gene set enrichment analysis (GSEA) to test mutual enrichment between the UV MR set and the SCC MR set, as reported previously (25). To assess if the activity of UV MRs or SCC MRs could predict patient survival, we carried out survival analysis using the Cox proportional hazard regression model based on VIPER-inferred protein activities in patient samples. To do so, we downloaded the RNA-seq data of skin cancer patients from TCGA and inferred protein activities based on the skin cancer interactome. Survival status and survival time for the same patient cohort were also obtained from TCGA.
Cells and reagents
Primary human keratinocytes and SCC13 tumor cells were established as previously described through the Columbia University Skin Disease Research Center tissue culture core facility (19,26). The protocol was exempt by our Institutional Review Board. Cells were cultured in 154CF medium supplemented with human keratinocyte growth supplement (Life Technologies, Carlsbad, CA). Thiostrepton, an inhibitor of FOXM1 transcriptional activity, was purchased from Sigma-Aldrich (St. Louis, MO). Thiostrepton was dissolved in DMSO at a final concentration of 10 mM as a stock solution. The working concentration of thiostrepton was 5 μM throughout this study (27).
FOXM1 functional validation studies
To determine FOXM1 protein expression status, we performed immunofluorescence (IF) staining on tissue sections from human SCC and adjacent non-tumor skin, and from UV-induced mouse SCC tissue and adjacent non-tumor skin (25,28). In addition, we performed IF staining experiments on cultured human SCC cells (SCC13) and normal keratinocytes. Anti-FOXM1 antibody used in these studies was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All images were acquired using a Zeiss fluorescence confocal microscope.
CRISPR/Cas9-mediated deletion of FOXM1 in SCC13 cells
To deplete FOXM1 in SCC13 cells, single-guide RNA (sgRNA) sequences were selected to flank the genomic region surrounding the human FOXM1 gene (CGGCCACCCTACTCTTACA and CTTCGAGACCATCAGCGTCC) using the CRISPR Design Tool (29). FOXM1-targeting sgRNA was cloned into the pXPR-based LentiCRISPR lentivirus vector expression system following a protocol reported previously (30). FOXM1-depleted stable SCC13 clones were generated following puromycin selection. Viability of control SCC13 cells, thiostrepton-treated SCC13 cells and FOXM1-depleted SCC13 cells were assessed using the EasyProbes Cell Viability Imaging Kit following the manufacturer’s protocol (GeneCopoeia, Rockville, MD).
Results
Network analysis identified UV-specific and SCC-specific MRs
In previous studies, we used RNA-seq to generate a large transcriptomic dataset from primary keratinocytes exposed to various UVR conditions (18). UV-specific transcriptional signatures were identified by differential gene expression analysis between cells exposed to UVB and nonexposed control cells. Using pathway analysis, we found that the UV signature was enriched for genes involved in pathways associated with DNA repair, cell growth and regulation of gene expression, consistent with established effects of UV radiation (18). To better understand UV-induced gene regulatory patterns, we performed advanced network analyses; namely, we transformed UV transcriptional signatures into a protein activity matrix using two well-established network algorithms, VIPER and ARACNe. VIPER computes the enrichment of regulator-controlled genes (regulon) inferred by ARACNe from the specific phenotype signature. Regulators whose target genes were significantly enriched in the signature were selected as candidate UV MRs.
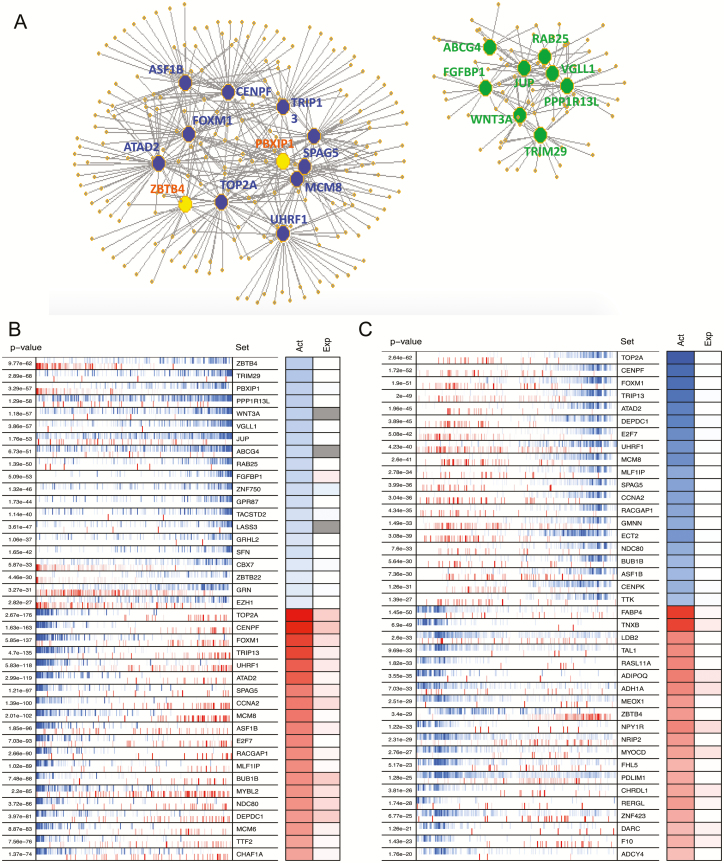
We used 21 transcriptomic profiles representing various UVR conditions and used the VIPER algorithm to superimpose UV transcriptional signatures onto a regulatory network, identifying regulators whose target genes were enriched in these signatures (Figure 1A). We selected the top 20 significantly activated and inactivated proteins (P < 0.05) as candidate MRs of UV response (Figure 1B). Through a similar analysis on RNA-seq data from five pairs of SCCs and adjacent normal skin, we also identified the top 20 activated and inactivated proteins in SCC signatures as potential MRs of SCC tumorigenesis (Figure 1C). Several of the identified MRs, such as FOXM1 and centromere protein F (CENPF), have been previously implicated in other human cancers (31), but their roles in skin cancer development have not been elucidated.
Figure 1.
(A) Network maps illustrating the regulons of top-ranked UV-responsive MRs identified by VIPER analysis. The network to the left represents MRs (large dots) and their corresponding inferred targets (small dots) involved in cell cycle, chromosome segregation, sister chromatid cohesion and DNA repair. The network to the right represents MRs and their targets involved in keratinocyte differentiation, skin barrier function, protein sumoylation and epidermis development. Edges in the network represent the regulatory relationship between each MR and its inferred targets. Blue: decreased MR activity; yellow and green: increased MR activity. (B) Heat map showing the inferred protein activity (Act) and differential gene expression (Exp) status of top-ranked UV MRs. (C) Heat map showing the inferred protein activity (Act) and differential gene expression (Exp) status of top-ranked SCC MRs. In both B and C, the left panel shows the enrichment P-value of each MR’s regulon set in the UV signature or SCC signature, respectively. The right panel shows the color scale of each MR protein activity (Act) and their corresponding differential mRNA expression (Exp) level. Blue: increased MR activity or mRNA expression after UV irradiation; red: decreased MR activity or mRNA expression after UV irradiation. Protein activity was quantitatively inferred by VIPER and differential mRNA expression was determined by DESeq2.
Correlation between altered MR protein activity and mRNA expression
We evaluated data from the top 40 UV MRs to test whether any correlation exists between changes in MR protein activity (as predicted by the VIPER program) and mRNA expression following UVR (based on the RNA-seq data). Increased MR protein activity showed no correlation with changes in mRNA expression following UVR (Supplementary Table 1, available at Carcinogenesis Online). Therefore, the increased activities of these MRs may be related to post-transcriptional regulation events. In contrast, approximately 80% of MRs with decreased protein activities displayed significant reductions in mRNA expression (Supplementary Table 2, available at Carcinogenesis Online). This positive correlation suggests that decreases in mRNA level reduced protein expression and thus diminished the activity of the MRs.
To understand the molecular functions of the identified MRs, we queried the GeneCards® Human Gene Database, which provides genomic, proteomic and functional information. As summarized in Table 1, most UV-activated MRs are involved in cell growth and differentiation, whereas UV-repressed MRs are mainly involved in DNA repair and cell cycle regulation. As expected, many of the identified MRs are TFs, including both activated MRs (TRIM29, ZNF750 and GRHL2) and repressed MRs (FOXM1, TRIP13 and UHRF). To further understand the cellular functions and biological processes in which affected MRs are involved, we performed pathway analysis using DAVID on the regulons of the top 20 MRs. As depicted in the heatmap in Figure 2, most regulons were involved in cell cycle regulation, chromosome separation and keratinocyte differentiation.
Table 1.
Molecular function of the identified MRs
| Activated MRs | Function(s) | Suppressed MRs | Function(s) |
|---|---|---|---|
| ZBTB4 | Mammalian epigenetic regulator | TOP2A | DNA topoisomerase that controls DNA topologic states during transcription |
| TRIM29 | Zinc finger and leucine zipper transcription factor | CENPF | Regulating DNA synthesis and cell cycle progression |
| PBXIP1 | Co-factor of PBX1 homeodomain protein | FOXM1 | Transcription factor involved in cell proliferation |
| PPP1R13L | Inhibitor of tumor suppressor p53 | TRIP13 | Transcriptional activator involved in cell proliferation |
| WNT3A | Signaling molecule | UHRF1 | Transcription factor involved in cell cycle regulation |
| VGLL1 | Co-factor of TEA domain transcription factor | ATAD2 | Chaperone-like protein involved in estrogen-induced cell proliferation |
| JUP | Cadherin binding protein | SPAG5 | Regulator of mitotic spindles |
| ABCG4 | Membrane transporter protein | CCNA2 | Regulator of cell cycles |
| RAB25 | Involved in membrane trafficking and cell survival | MCM8 | Regulator of DNA replication |
| FGFBP1 | Carrier protein of signaling molecule FGF | ASF1B | Modulating nucleosome structure of chromatin |
| ZNF750 | Transcription factor involved in epidermis differentiation | E2F7 | Transcription factor with sequence-specific DNA binding |
| GPR87 | Needed for p53/TP53-dependent survival response to DNA damage | RACGAP1 | Mediator during cell cycle cytokinesis |
| TACSTD2 | Growth factor receptor | MLF1IP | Component of CENPA-NAC complex required for mitotic progression |
| LASS3 | Required for synthesis of long-chain ceramides in the epidermis | BUB1B | Essential component of mitotic checkpoint |
| GRHL2 | Transcription factor involved in epithelial development | MYBL2 | Transcription factor for cell survival, proliferation and differentiation |
| SFN | Activator of p53 | NDC80 | Required for chromosome segregation |
| CBX7 | Affecting the lifespan of cells | DEPDC1 | Possible involvement in transcription regulation |
| ZBTB22 | Involved in transcriptional regulation | MCM6 | Part of the MCM2-7 complex to ensure once per cell cycle DNA replication |
| GRN | cytokine-like activity, linked to tumorigenesis | TTF2 | Transcription termination factor |
| EZH1 | Mediating methylation of histones | CHAF1A | Component of Chromosome Assembly Factor 1 complex |
Figure 2.
Heatmap showing significantly enriched pathways based on regulons of the top 20 MRs. Most regulons were involved in cell cycle, chromosome separation and keratinocyte differentiation.
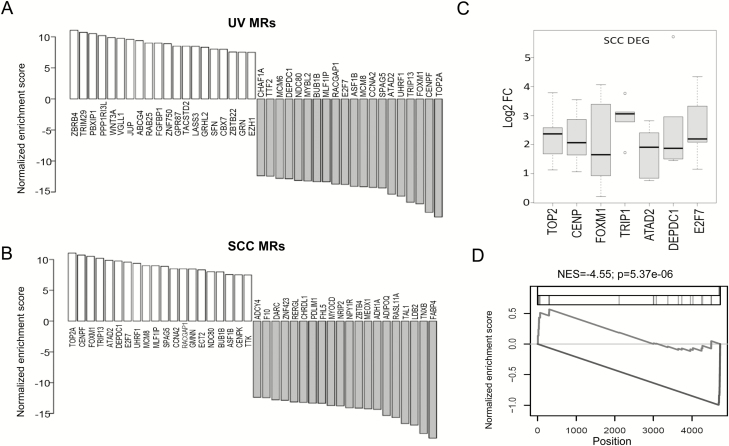
Activities of UV MRs and SCC MRs are inversely regulated
The predicted activities of UV and SCC MR proteins are schematically illustrated in Figure 3A and B. In support of the role of UV MRs in skin cancer, many were significantly upregulated in human SCCs compared to adjacent normal skin (Figure 3C, n = 5). We then asked if there was any similarity between UV MRs and SCC MRs. To test this, we performed GSEA to compare the mutual enrichment of these two MR groups. As shown in Figure 3D, there was no enrichment of UV-activated MRs in the SCC MR set. However, UV-repressed MRs shared an intriguingly significant similarity with MRs that were activated in SCCs. This finding highlights the distinctive roles of these MRs in regulating UV response and skin cancer development.
Figure 3.
(A and B) Changes in UV MR protein activity (A) or SCC MR protein activity (B) based on VIPER analyses. Blue: increased activity; red: decreased activity. (C) Box plot showing differential expression of selected UV MR genes (DEG) in human SCCs compared to adjacent normal skin (n = 5). Y-axis represents the log2 fold change (Log2FC) in selected MR gene expression between each SCC and adjacent normal skin pair. (D) Enrichment plot showing a significant enrichment of repressed UV MR set in SCC MR activities. VIPER-inferred SCC protein activities were sorted from the highest (left) to the lowest (right) scores. The blue line indicates activated UV MR sets and the red line indicates repressed UV MR sets. NES: normalized enrichment score.
To further elucidate the role of the shared MRs in skin cancer development, we used available skin cancer data from TCGA to perform survival analysis, correlating patient survival time and MR activities. Skin melanoma patient survival status and survival time were analyzed using the Cox proportional hazard regression model to evaluate the significance of MR activities in predicting patient survival. We found that the activities of FGFBP1, RAB25, ABCG4, JUP, VGLL1, TRIM29, FOXM1, TRIP13, UHRF1, SPAG5, CCNA2 and ASF1B are significantly associated with decreased patient survival time (P < 0.05) (Table 2) and predict poor prognosis.
Table 2.
Cox proportional hazard (CoxPH) analysis of the correlation between patient survival time and the activity of each MRs in melanoma patients
| MR | UV_NESa | CoxPH_Exp(coeff)b | CoxPH_P value |
|---|---|---|---|
| FOXM1 | −16.88764 | 1.44 | 3.95E-07 |
| SPAG5 | −14.35344 | 1.32 | 0.00256 |
| TRIP13 | −16.64743 | 1.28 | 0.000496 |
| ASF1B | −14.09895 | 1.2 | 0.0191 |
| CCNA2 | −14.24951 | 1.19 | 0.0304 |
| UHRF1 | −15.66974 | 1.17 | 0.0146 |
| JUP | 9.609899 | 1.14 | 0.00778 |
| CENPF | −18.26369 | 1.13 | 0.0866 |
| ABCG4 | 9.398624 | 1.12 | 0.00417 |
| PPP1R13L | 10.20518 | 1.11 | 0.0711 |
| VGLL1 | 9.764145 | 1.1 | 0.000185 |
| TRIM29 | 10.73277 | 1.09 | 2.15E-06 |
| RAB25 | 9.010712 | 1.09 | 3.02E-05 |
| TOP2A | −19.00923 | 1.09 | 0.245 |
| FGFBP1 | 9.003473 | 1.06 | 0.00171 |
| WNT3A | 9.88639 | 1.05 | 0.105 |
| ATAD2 | −15.34317 | 1.01 | 0.91 |
| MCM8 | −14.15302 | 0.982 | 0.835 |
| ZBTB4 | 11.05652 | 0.938 | 0.542 |
| PBXIP1 | 10.51629 | 0.894 | 0.197 |
aAveraged NES (normalized enrichment scores) among 21 UV radiation signatures as determined by VIPER analysis.
bThe exp (coefficients) of the cox model indicates how probably the activity of a specific MR will increase (<1) or decrease (>1) the patient survival. For example, the exp (coefficients) of FOXM1 is 1.44, which indicates that skin cancer patients with higher FOXM1 activity will have poorer survival probability.
Elevated expression of FOXM1 in skin SCC
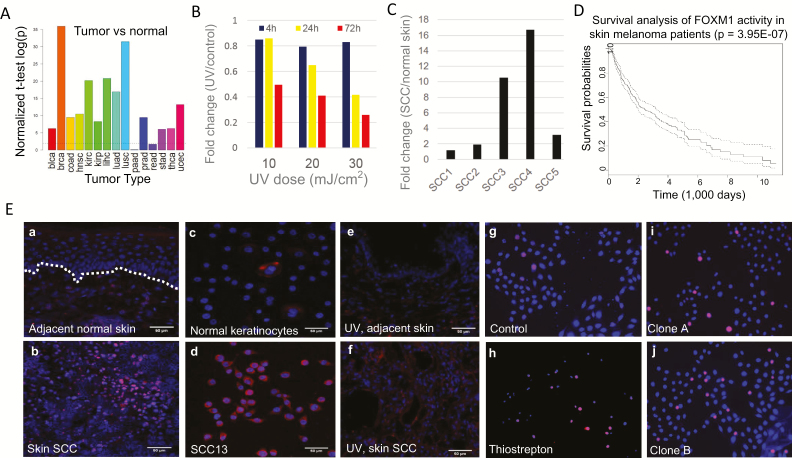
Among the identified MR proteins, FOXM1 displayed the most significant change in activity following UVR. FOXM1 encodes a forkhead domain transcription factor that has been shown to play important roles in various cancers (32). Analysis of RNA-seq data from multiple cancer types in TCGA has demonstrated that FOXM1 mRNA expression is significantly increased in the majority of human cancers (Figure 4A). However, its function in skin cancer development remains elusive. To elucidate the role of FOXM1 in UV-induced skin cancer development, we analyzed the UV and SCC RNA-seq cohorts and found that FOXM1 mRNA expression was repressed by UV, but increased in human SCCs (Figure 4B and C). Survival analysis using the Cox proportional hazard regression model confirmed a significant inverse correlation between increased FOXM1 activity and melanoma cancer patient survival time (Figure 4D).
Figure 4.
(A) Illustration of the average fold change in FOXM1 mRNA expression between human cancers and their respective normal control tissue. (B) Time- and dose-dependent changes in FOXM1 mRNA expression following UVR in primary human keratinocytes. (C) Fold change in FOXM1 mRNA expression between human SCCs and adjacent normal skin. (D) Inverse correlation between FOXM1 expression and melanoma patient survival time based on TCGA data. (E) FOXM1 protein expression in human SCC tumor tissue and adjacent normal skin from the same patient (a and b); FOXM1 protein expression in human SCC13 tumor cells and normal keratinocytes (c and d); FOXM1 protein expression in UV-induced mouse SCC tumor and adjacent control skin (e and f); Viability staining of control SCC13 cells (g), SCC13 treated with 5 μM thiostrepton (h) or with FOXM1-targeting CRISPR/Cas9 (i and j). Red: FOXM1 expression (a–f) or apoptotic cells (g–j). Blue: DAPI nuclei staining. The dotted white line in image 4E-a indicates the basement membrane between the epidermis and dermis.
By means of IF staining, we compared the level of FOXM1 protein expression in human SCC tumor tissue with that in adjacent normal skin tissue. We found that while FOXM1 protein expression was barely detectable in normal skin epidermis, it was aberrantly upregulated in SCC tumor tissue (Figure 4E, a and b). Similarly, we found the FOXM1 protein to be highly expressed in human SCC tumor derived SCC13 cancer cells but not in normal human keratinocytes (Figure 4E, c and d). Importantly, IF analysis of UV-induced mouse SCC tumors also confirmed upregulation of FOXM1 protein expression compared to non-tumor bearing adjacent skin (Figure 4E, e and f).
To test the role of elevated FOXM1 expression in SCC cell survival and proliferation, we treated human SCC13 cells with thiostrepton, a potent inhibitor of FOXM1 transcriptional activity, and found that it caused significant SCC cell death (Figure 4E, g and h). Furthermore, CRISPR-mediated depletion of FOXM1 in SCC13 cells increased cell death as well, although the effect appeared to be less pronounced than thiostrepton treatment (Figure 4E, i and j). Taken together, these results demonstrate that FOXM1 activity is closely involved in SCC development and cell survival.
Discussion
Skin cancer is a major and growing public health problem. Improved understanding of the molecular control of skin cancer pathogenesis has the potential to profoundly improve cancer prevention and treatment. In this study, we employed ARACNe to analyze a large-scale skin cancer RNA-seq gene expression data set obtained from TCGA to construct a transcriptional regulatory network (interactome), which represents the genome-wide repertoire of regulator–target gene interactions in skin cancer cells. We then applied VIPER on expression data from UV-irradiated normal human keratinocytes or SCC tissues to interrogate this interactome and identify candidate MRs. We identified a set of UV MRs that are located at the top of transcriptional regulatory hierarchies responsive to UVR. Approximately 47% of UV-activated MRs and 67% of UV-suppressed MRs are TFs and have been previously implicated in cancer development promoters (33–39). However, most have not been studied in the specific context of skin cancer.
When comparing the predicted protein activity of UV MRs with their mRNA expression data following UVR, we found that for UV-activated MRs, protein activity does not depend on increased mRNA expression. In contrast, there was a strong correlation between the activity of UV-suppressed MRs and their mRNA expression following UVR. It is interesting to note that other studies have shown many of the UV-suppressed MRs to be cancer promoters (33–38). This is consistent with our GSEA analysis, which found that UV-suppressed MRs are activated in SCCs (Figure 3D). It is possible that upon exposure to UVR, normal skin keratinocytes can activate anti-cancer pathways by suppressing the activity of tumor-promoting MRs. During skin cancer initiation and progression, however, these cancer-promoting MRs eventually become aberrantly activated to facilitate tumor growth and development. Elucidating the mechanisms underpinning the upregulation of these tumor-promoting MRs will significantly improve our understandings of skin cancer pathogenesis.
Some of the MRs, including CENPF and FOXM1, have been shown previously to function as MRs in regulating prostate cancer development (31). Importantly, co-silencing FOXM1 and CENPF with shRNAs has a greater effect on reducing cancer cellular proliferation than silencing either alone, suggesting that FOXM1 partners with other MRs in driving and sustaining cancer development and growth. In this study, we found that FOXM1 is aberrantly upregulated in skin SCC tissues and SCC-derived tumor cells (Figure 4E). Cancer survival analysis confirmed that elevated FOXM1 activity is inversely correlated with melanoma patient survival (Figure 4D). Due to that cutaneous SCCs are rarely fatal, currently there is no SCC survival database available to perform similar analysis to exam the relationship between FOXM1 activity and skin SCC patient survival. CRISPR-mediated depletion or pharmacological Inhibition of FOXM1 activity effectively decreased the viability of SCC cells. Nevertheless, genetic depletion of FOXM1 seems to be less potent than the proteasome inhibitor thiostrepton. This may be attributable to the fact that thiostrepton, which is a potent inhibitor of FOXM1 transcriptional activity (40–42), may exert additional FOXM1-independent effects on cancer cell survival. The anti-cancer effect of thiostrepton is proposed to occur through stabilizing a negative regulator of FOXM1, which binds to FOXM1 and inhibits its transcriptional activity in cancer cells (43). Additional studies are warranted to identify other FOXM1-interacting proteins that may co-regulate skin tumorigenesis.
In summary, by means of ARACNe and VIPER analyses, we have identified novel MRs that regulate skin UV response and SCC tumorigenesis. MRs identified using this unbiased analysis of genome-wide regulatory networks can enhance our understanding of the molecular control of human skin cancer development. MRs are often highly conserved in different tumor subtypes, which makes them promising therapeutic targets. In addition to FOXM1, other MRs identified in this study are key regulators of cancer development, such as thyroid hormone receptor interactor 13 (TRIP13) and DNA isomerase II alpha (TOP2A) (36,44–46). Following appropriate validation studies, future research will focus on synergistic effects among these MRs in skin cancer development, which will ultimately facilitate the development of novel targeted approaches for skin cancer prevention and treatment.
Funding
This work was supported in part by NIH/NIAMS grant K01AR064315, the Columbia University Herbert Irving Comprehensive Cancer Center (P30 CA013696), the Columbia University Skin Disease Research Center (P30AR44535) and the Center for Environmental Health in Northern Manhattan (P30 ES009089).
Supplementary Material
Acknowledgements
We thank Dr. Angela Christiano for her generous support. We thank the excellent technical assistance from Rong Du, Kevin Sun and Tao Su with cell culture, tissue procurement and RNA preparation. We are also grateful to the support from the Molecular Pathology and Genomics Technologies Core Facilities at the Herbert Irving Cancer Research Center at Columbia University Medical Center.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ARACNe
algorithm for the reconstruction of accurate cellular networks
- FOXM1
forkhead box M1
- GSEA
gene set enrichment analysis
- IF
immunofluorescence
- MR
master regulator
- SCC
squamous cell carcinoma
- TCGA
The Cancer Genome Atlas
- TFs
transcription factors
- UVB
ultraviolet B
- UVR
ultraviolet radiation
- VIPER
Virtual Inference of Protein Activity by Enriched Regulon
References
- 1. Stern R.S. (2010) Prevalence of a history of skin cancer in 2007: results of an incidence-based model. Arch. Dermatol., 146, 279–282. [DOI] [PubMed] [Google Scholar]
- 2. Guy G.P., Jr, et al. ; Centers for Disease Control and Prevention (CDC). (2015) Vital signs: melanoma incidence and mortality trends and projections - United States, 1982-2030. MMWR. Morb. Mortal. Wkly. Rep., 64, 591–596. [PMC free article] [PubMed] [Google Scholar]
- 3. Sahu R.P., et al. (2012) The environmental stressor ultraviolet B radiation inhibits murine antitumor immunity through its ability to generate platelet-activating factor agonists. Carcinogenesis, 33, 1360–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kemp M.G., et al. (2017) Insulin-like growth factor 1 receptor signaling is required for optimal atr-chk1 kinase signaling in ultraviolet B (UVB)-irradiated Human Keratinocytes. J. Biol. Chem., 292, 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lewis D.A., et al. (2010) The IGF-1/IGF-1R signaling axis in the skin: a new role for the dermis in aging-associated skin cancer. Oncogene, 29, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mayer J.E., et al. (2016) Arsenic and skin cancer in the USA: the current evidence regarding arsenic-contaminated drinking water. Int. J. Dermatol., 55, e585–e591. [DOI] [PubMed] [Google Scholar]
- 7. Robinson S.N., et al. (2013) Photosensitizing agents and the risk of non-melanoma skin cancer: a population-based case-control study. J. Invest. Dermatol., 133, 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcovich S., et al. (2017) Skin cancer epidemics in the elderly as an emerging issue in geriatric oncology. Aging Dis., 8, 643–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christophers A.J. (1998) Melanoma is not caused by sunlight. Mutat. Res., 422, 113–117. [DOI] [PubMed] [Google Scholar]
- 10. Chitsazzadeh V., et al. (2016) Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat. Commun., 7, 12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nickoloff B.J., et al. (2002) Life and death signaling pathways contributing to skin cancer. J. Investig. Dermatol. Symp. Proc., 7, 27–35. [DOI] [PubMed] [Google Scholar]
- 12. Rodust P.M., et al. (2009) UV-induced squamous cell carcinoma–a role for antiapoptotic signalling pathways. Br. J. Dermatol., 161 (suppl. 3), 107–115. [DOI] [PubMed] [Google Scholar]
- 13. Pfeifer G.P., et al. (2005) Mutations induced by ultraviolet light. Mutat. Res., 571, 19–31. [DOI] [PubMed] [Google Scholar]
- 14. Brash D.E. (2015) UV signature mutations. Photochem. Photobiol., 91, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfeifer G.P., et al. (2012) UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem. Photobiol. Sci., 11, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khan A.Q., et al. (2018) Roles of UVA radiation and DNA damage responses in melanoma pathogenesis. Environ. Mol. Mutagen., 59, 438–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rünger T.M. (2013) Much remains to be learned about how UVR induces mutations. J. Invest. Dermatol., 133, 1717–1719. [DOI] [PubMed] [Google Scholar]
- 18. Shen Y., et al. (2016) Transcriptome analysis identifies the dysregulation of ultraviolet target genes in human skin cancers. PLoS One, 11, e0163054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun X., et al. (2016) Distinctive molecular responses to ultraviolet radiation between keratinocytes and melanocytes. Exp. Dermatol., 25, 708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Margolin A.A., et al. (2006) Reverse engineering cellular networks. Nat. Protoc., 1, 662–671. [DOI] [PubMed] [Google Scholar]
- 21. Yang J., et al. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell, 117, 927–939. [DOI] [PubMed] [Google Scholar]
- 22. De D., et al. (2014) Inhibition of master transcription factors in pluripotent cells induces early stage differentiation. Proc. Natl. Acad. Sci. U. S. A., 111, 1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alvarez M.J., et al. (2016) Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet., 48, 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan S.S.-K., et al. (2013) What is a master regulator? J. Stem Cell Res. Ther., 3, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen Y., et al. (2017) Epigenetic and genetic dissections of UV-induced global gene dysregulation in skin cells through multi-omics analyses. Sci. Rep., 7, 42646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L., et al. (2014) Inhibition of p38 MAPK signaling augments skin tumorigenesis via NOX2 driven ROS generation. PLoS One, 9, e97245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siraj A.K., et al. (2018) FoxM1 is an independent poor prognostic marker and therapeutic target for advanced Middle Eastern breast cancer. Oncotarget, 9, 17466–17482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim H., et al. (2012) Loss of hairless confers susceptibility to UVB-induced tumorigenesis via disruption of NF-kappaB signaling. Plos One, 7, e39691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ran F.A., et al. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc., 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shalem O., et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aytes A., et al. (2014) Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell, 25, 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gemenetzidis E., et al. (2010) Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res., 70, 9515–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chung V.Y., et al. (2016) GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep., 6, 19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raychaudhuri P., et al. (2011) FoxM1: a master regulator of tumor metastasis. Cancer Res., 71, 4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qiu F., et al. (2015) TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int. J. Clin. Exp. Pathol., 8, 5053–5061. [PMC free article] [PubMed] [Google Scholar]
- 36. Kurita K., et al. (2016) TRIP13 is expressed in colorectal cancer and promotes cancer cell invasion. Oncol. Lett., 12, 5240–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang F., et al. (2012) UHRF1 promotes cell growth and metastasis through repression of p16(ink4a) in colorectal cancer. Ann. Surg. Oncol., 19, 2753–2762. [DOI] [PubMed] [Google Scholar]
- 38. Kanehira M., et al. (2007) Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene, 26, 6448–6455. [DOI] [PubMed] [Google Scholar]
- 39. Thurlings I., et al. (2017) Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene, 36, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radhakrishnan S.K., et al. (2006) Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res., 66, 9731–9735. [DOI] [PubMed] [Google Scholar]
- 41. Bhat U.G., et al. (2009) Thiazole antibiotics target FoxM1 and induce apoptosis in human cancer cells. PLoS One, 4, e5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hegde N.S., et al. (2011) The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem., 3, 725–731. [DOI] [PubMed] [Google Scholar]
- 43. Gartel A.L. (2011) Thiostrepton, proteasome inhibitors and FOXM1. Cell Cycle, 10, 4341–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Banerjee R., et al. (2014) TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat. Commun., 5, 4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lan J., et al. (2014) TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol., 35, 179–187. [DOI] [PubMed] [Google Scholar]
- 46. Schaefer-Klein J.L., et al. (2015) Topoisomerase 2 Alpha cooperates with androgen receptor to contribute to prostate cancer progression. PLoS One, 10, e0142327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.