ABSTRACT
Beyond being simply positive or negative, beneficial or inhibitory, microbial interactions can involve a diverse set of mechanisms, dependencies and dynamical properties. These more nuanced features have been described in great detail for some specific types of interactions, (e.g. pairwise metabolic cross-feeding, quorum sensing or antibiotic killing), often with the use of quantitative measurements and insight derived from modeling. With a growing understanding of the composition and dynamics of complex microbial communities for human health and other applications, we face the challenge of integrating information about these different interactions into comprehensive quantitative frameworks. Here, we review the literature on a wide set of microbial interactions, and explore the potential value of a formal categorization based on multidimensional vectors of attributes. We propose that such an encoding can facilitate systematic, direct comparisons of interaction mechanisms and dependencies, and we discuss the relevance of an atlas of interactions for future modeling and rational design efforts.
Keywords: microbial interactions, microbial communities, mutualism, microbiome, microbial ecology, systems biology of metabolism
Reports of inter-species interactions in microbial communities contain abundant information that defies simple ecological categorizations. Vectors of attributes can formally capture this knowledge, helping create interaction catalogues and predictive models.
INTRODUCTION
Microbes form complex ecosystems comprising up to hundreds or thousands of different species (Whitman, Coleman and Wiebe 1998; Ley et al. 2006; Walker and Pace 2007; Wilhelm and Matteson 2008; Brown et al. 2009; Qin et al. 2010; Welch and Huse 2011; Tecon and Or 2017). Increased exploration of these communities enabled by new technologies has yielded a wealth of information on their constituent organisms, as well as growing insight into the complex and rich web of relationships the organisms form between each other. As the study of microbial communities embraces a more systems-oriented approach (Raes and Bork 2008; Klitgord and Segrè 2011; Faust and Raes 2012; Fuhrman, Cram and Needham 2015; Poudel et al. 2016), more and more attention is being directed toward the myriad ways microbes interact, as well as toward the crucial roles these interactions play in defining community function. These relationships, which can range from mutualistic exchange of metabolic products (Imachi et al. 2000; Harcombe 2010), to antagonistic secretion of antibiotics (Jousset, Scheu and Bonkowski 2008; Cornforth and Foster 2015; Kelsic et al. 2015), to direct predation of individual organisms (Jurkevitch 2007; Chen et al. 2011; Kadouri et al. 2013), make up a vast space of relationships that are ubiquitous in the microbial world.
Despite the diversity and abundance of microbial life on the planet, we still face fundamental challenges in addressing broad, important questions pertaining to microbial interrelationships: are specific types of interactions more common among certain taxa than others? How prevalent is mutualism across all environments? How can we systematically compare interactions across individual studies? A first step in answering these questions is to categorize interactions based on common, recurrent properties. We may start with a widely-used classification system that is based on determining the ecological outcome each organism experiences in a pairwise interaction (Lidicker 1979; Keddy 2001) (Fig. 1A). These outcomes, on a very general level, can be either positive, neutral or negative and can encompass all possible pairs of effects: from mutualism (in which both participants experience a positive outcome) to competition (both participants experience a negative outcome). Between these two extremes are combinations of positive, neutral and negative outcomes such as amensalism, in which the actor experiences no benefit or detriment and the recipient experiences a negative outcome. Though this framework has formed the basis for a broad corpus of ecology research (Faust and Raes 2012), it is limited in that it cannot capture many nuances that are crucial in determining how individual interactions arise, change and affect a community (Margulis 1990; Smith 2001; Martin and Schwab 2013). Moreover, there exist inconsistencies in the very language used to describe these ecological outcomes, hindering comparison of intermicrobial behaviors between different disciplines and prompting efforts to standardize how interactions are described (West, Griffin and Gardner 2007; Smith et al. 2019; Tipton, Darcy and Hynson 2019).
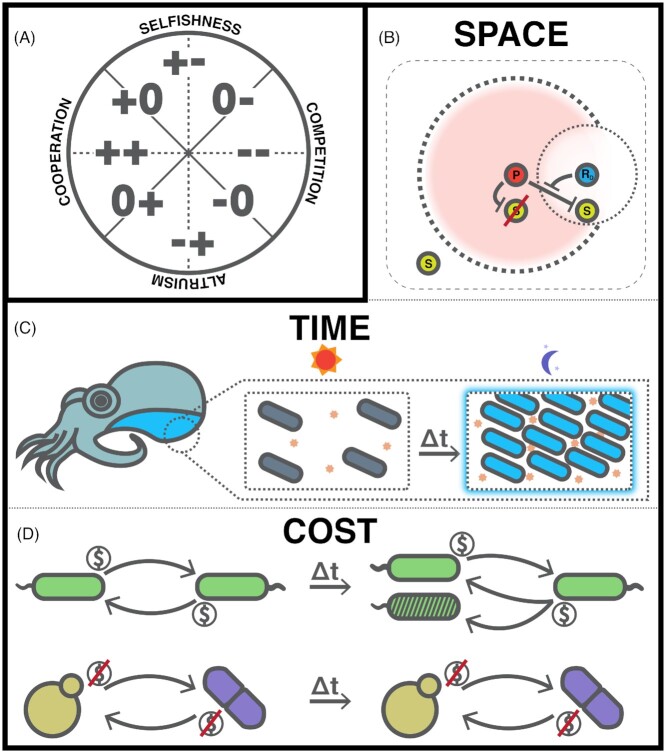
Figure 1.
The multifaceted nature of microbial interactions. (A), Axes commonly used to classify interactions, adapted from Lidicker 1979. A single interaction can be represented as a point within the axes, which quantify the ecological outcomes experienced by the participants and the strength of the interaction. For example, a point close to the bottom of the axes (corresponding to −+) represents an altruistic scenario in which one participant experiences a net negative outcome and the second participant receives a positive one. (B-D), Examples of attributes observed in interactions that resist straightforward, benefit-oriented classification. Each of the interactions displayed feature some kind of mutualistic outcome, but exhibit crucial dependencies that impact the nature of the interaction. (B), An interaction that confers differing benefits on its participants based on their spatial configuration, reported by Kelsic et al. (Kelsic et al. 2015). Here, a colony of Streptomyces “P” produces an antibiotic that kills sensitive E. coli “S” within a given radius. If, however, an antibiotic-degrading Streptomyces population “RD” is placed within this radius, E. coli is able to survive within its immediate vicinity. Therefore, depending on its location, E. coli can either experience a neutral or negative effect from the antibiotic-producing Streptomyces. (C), Time-dependent intraspecies interaction between Vibrio fischeri cells within the light organ of the Hawaiian bobtail squid. During the day, the squid releases the majority of V. fischeri, diminishing their concentration within the organ. As the V. fischeri population regrows, individuals secrete signaling molecules that, upon reaching a critical concentration, lead to the expression of luminescence genes. In this way, the symbionts allow the squid to bioluminesce at night. The day–night cycle therefore drives this transition through its effects on squid physiology, signaling molecule concentration, and bacterial cell density. (D), Two mutualistic interactions that impose differing metabolic costs on participants. Top: Intraspecies interactions within Pseudomonas fluorescens populations reported by Rainey and Rainey (Rainey and Rainey 2003). Initially, cooperating individuals secrete an adhesive polymer to form a biofilm. This process occurs at a metabolic cost to individual organisms. Over time, defecting individuals stopped producing the polymer but continued benefitting from the collective production within the group. This ‘cheating’ diminished the viability of the community in the short term, leading to more complex interaction dynamics over longer timescales. Bottom: A simplified schematic of a mutualistic interaction based on non-costly overflow metabolism demonstrated by Ponomarova et al. Here, Saccharomyces cerevisiae uses overflow metabolism to secrete amino acids which allow for the growth of Lactococcus lactis. Lactococcus lactis, in turn, provides glucose and galactose to the yeast through lactose hydrolysis, yielding a stable symbiotic relationship (Ponomarova et al. 2017). Though these two mutualisms are fundamentally different, neither represents the sole possible outcome of costly or non-costly interactions. Previous work has shown how cheating could in fact stabilize mutualisms (Foster and Kokko 2006), and that cheating itself may pose less of a threat to community collapse as commonly thought (Frederickson 2017).
In light of these limitations, we ask if it is possible to use additional properties to enhance the vocabulary we use to describe and compare interactions. It has indeed become possible to capture more and more fine details of interactions through recently-developed technologies and computational tools, such as measurements of individual exchanged molecules using metabolomics (Tang 2011; Chamoun, Aliferis and Jabaji 2015; Bilyk and Luzhetskyy 2016), inference of entire co-occurrence networks based on metagenomic sequencing (Qin et al. 2010; Steele et al. 2011; Barberán et al. 2012; Gilbert et al. 2012; Lloyd-Price et al. 2017), and direct detection of interspecies synergy using microfluidics (Park et al. 2011; Hsu et al. 2019). Unfortunately, as this wealth of data is often reported on an observation-by-observation basis, it remains difficult to comprehensively classify interactions and systematically compare them across studies. As a way of facing this challenge, we may therefore look for inspiration in integrative categorizations of complex phenomena in other areas of biology (e.g. metabolic networks (Kanehisa and Goto 2000; Caspi et al. 2016), protein interaction networks (Szklarczyk et al. 2017), or 16s sequences (DeSantis et al. 2006)), which have become valuable standardized repositories of knowledge, as well as starting points for data-driven analyses that would have been otherwise impossible (Barberán et al. 2012; Magnúsdóttir et al. 2016; Goldford et al. 2018).
In this mini-review, we compile a diverse list of known microbial interactions and comment on the limitations of describing and classifying them based solely on simple ecological outcomes. We then provide an example of how one can instead embrace the multifaceted nature of microbial relationships by explicitly ascribing numerical variables to different properties of the interacting partners. In doing so, we propose a framework for encoding interaction data in a way that enables quantitative analysis and comparison across studies. This formalization could be viewed as one of many incremental steps needed to better understand these interactions, as well as a way to encourage conversations on how to structure future methodical comparisons, data-mining efforts, and data-driven analyses.
MICROBIAL INTERACTIONS ARE MULTIDIMENSIONAL, DYNAMIC PHENOMENA
Examples of interactions that can be best understood by more explicitly accounting for different attributes include those that change significantly based on spatial configurations. An interesting example is that of Aggregatibacter actinomycetemcomitans (Aa) and Streptococcus gordonii (Sg), bacteria isolated from the human oral cavity. Sg has been shown to secrete lactate, the preferred carbon source of Aa, as a metabolic byproduct (Brown and Whiteley 2007; Tong, Zeng and Burne 2011). Lactate is taken up by Aa, which experiences growth benefits as a result (Ramsey, Rumbaugh and Whiteley 2011). It has been shown in a murine infection model, however, that while this metabolic exchange benefits the virulence capabilities of Aa, Sg also secretes high amounts of hydrogen peroxide, a potent antimicrobial. Stacy et al. demonstrated that Aa can adopt a twofold detoxification-dispersion response to this challenge, which allows it to spatially position itself at an optimal distance from Sg and detoxify the hydrogen peroxide to consume lactate (Stacy et al. 2014). This response is also thought to yield benefits to Sg, enhancing the overall fitness of the community and the strength of the infection. This interaction could be termed mutualistic using a simple interpretation of ecological outcome since, despite a clear antagonistic action by one of its participants, it results in a net beneficial relationship. However, only adopting such an interpretation risks abstracting away key nuances within the interaction that detail paradoxical, yet important, mechanisms for cooperation and competition. A different example of spatially dependent interactions was investigated by Kelsic et al. using a synthetic community made up of Escherichia coli and two Streptomyces strains isolated from soil (Kelsic et al. 2015). In this study, E. coli were exposed to an antibiotic-producing Streptomyces strain on an agar plate. As the E. coli population was sensitive to the antibiotic produced, it was only able to grow outside the radius formed by the antagonistic Streptomyces. However, if an antibiotic-degrading Streptomyces strain was placed within this killing radius, E. coli were shown to grow in an area around the detoxifying strain (Fig. 1B). As such, the spatial configuration of the interaction directly determined the growth outcomes of its participants. Recent modeling efforts have also demonstrated how the nature of an interaction can change when spatial information is considered (Armitage and Jones 2019), underscoring the importance of capturing these dependencies.
As with spatial organization, time-dependent sharing of nutrients or toxin sequestration has also been shown to affect the nature of microbial interactions. For example, in the same study mentioned above, Kelsic et al. used dynamical modeling to show how four organisms with varying degrees of antibiotic production and degradation capabilities could stably coexist in various temporal modes (stable equilibrium, limit cycles, or chaotic oscillations) without spatial separation (Kelsic et al. 2015). In nature, the importance of temporal dynamics on interactions is particularly evident in host-microbe symbioses, where circadian cycles impact gene regulation and metabolic processes in the host and among their symbionts (Roden and Ingle 2009; Liang, Bushman and FitzGerald 2015; Heath-Heckman 2016; Staley et al. 2017). A classic example of such a scenario is the intraspecies signaling patterns shown within the Vibrio fischeri communities living within the squid Euprymna scolopes. V. fischeri colonize the light organ of the squid, allowing it to bioluminesce at night. This result arises from a sequence of biological processes, all ultimately driven by the day–night cycle (Fig. 1C) (Verma and Miyashiro 2013). As the nature of these interactions vary drastically over different temporal scales, it is important to consider this attribute as a crucial factor in classification.
The classic axes commonly used to describe ecological interactions (Fig. 1A) are also limited in the sense that specific points on this graph (e.g. ++) group together interactions that may be based on very different mechanisms and costs of metabolite production and exchange. For example, two organisms that benefit metabolically from each other (thus falling in the ++ category) may do so in two fundamentally different ways: a first possibility is an interaction mediated by evolved secretion of metabolically costly products (Harcombe 2010; Wintermute and Silver 2010; Celiker and Gore 2012; Mee et al. 2014; Zomorrodi and Segrè 2017). A second type of interaction could be the outcome of secreted byproducts that are not costly to the producer, and that are not ‘intended’ to specifically benefit any other particular organism. Such non-costly secretions are also described as byproduct benefits (Sachs et al. 2004), and can lead to the emergence of mutually beneficial interactions through selfish actions by individual organisms. Examples of this type of metabolite-mediated exchange include overflow metabolism and secretion of incompletely reduced carbon sources (Molenaar et al. 2009; Basan et al. 2015), which are secretions that can be strongly dependent on environmental context (Klitgord et al. 2010; Chamberlain, Bronstein and Rudgers 2014; Pacheco, Moel and Segrè 2019). Importantly, the cost of metabolite production and secretion can change the interaction's susceptibility to cheating phenotypes (Hamilton 1964; West et al. 2006; D'Souza et al. 2018), which in turn can have implications for the long-term stability of the relationship (Fig. 1D). Additionally, the strong environmental dependence of interactions is especially problematic when studying ‘uncultivable’ organisms or when designing synthetic microbial communities, whose members may have metabolic dependencies that are difficult to satisfy experimentally (Rappé and Giovannoni 2003; Vartoukian, Palmer and Wade 2010; Stewart 2012). The formation of an interaction often requires the fulfillment of particular environmental conditions, such as the presence of specific carbon or nitrogen sources or a particular pH range, which might be strongly modulated by spatial or temporal dynamics.
For some interactions, the ecological outcomes experienced by their participants remain unclear. Nonetheless, other properties can be clearly identified to yield insight into the mechanisms of the relationship, as exemplified by the interaction between two archaea: Igniococcus hospitalis and Nanoarchaeum equitans. Both species are able to form a stable co-culture, but such a partnership is only necessary for the survival of N. equitans, and not for that of I. hospitalis (Huber et al. 2003). In co-culture experiments, N. equitans appears to rely on H2S, the primary metabolic end product of I. hospitalis, as well as on amino acids and lipids provided by I. hospitalis (Jahn et al. 2004, 2008). This interaction would therefore appear to be parasitic. However, this relationship results in no detriment to the I. hospitalis population and there even appear to be evolved structural features that allow for a tight physical connection between both organisms. This strong coupling suggests that there is some benefit given by N. equitans to I. hospitalis, though experiments have so far been unable to identify such a mechanism, making any definitive labeling of an interaction type elusive. This interaction exhibits other important properties, such as contact-dependence, that are not captured in a framework based solely on ecological outcomes.
Finally, interactions that involve three or more species present further classification challenges. It is often possible to assign clear benefits when the participants in a multispecies interaction can be divided into two distinct roles, as in, for example, an interaction observed between Myxococcus xanthus and a consortium of prey bacteria (Berleman et al. 2006). Here, the predatory M. xanthus can be assigned a positive outcome and its prey bacteria can collectively be assigned a negative, detrimental one. Additionally, if an interaction confers effects of the same sign to all recipients (e.g. a tripartite symbiosis in which all organisms benefit (Lőrincz et al. 2010) it is possible to place it on an ecological outcome axis (Fig. 1A). If, however, different members within a higher-order interaction are influenced by effects of different signs, as is likely to occur within complex communities, it becomes less clear how to classify the interaction without reducing it to a set of pairwise interactions. This limitation has important implications for understanding the structure and function of natural microbial ecosystems. Though pairwise relationships can be used to gain insights into overall community features (Guo and Boedicker 2016; Friedman, Higgins and Gore 2017; Venturelli et al. 2018), their predictive power can vary depending on the model used or on interaction mechanisms (Carrara et al. 2015; Momeni, Xie and Shou 2017). As such, the way in which features of higher-order interactions are represented must be carefully considered.
A MULTI-DIMENSIONAL FRAMEWORK FOR DESCRIBING MICROBIAL INTERACTIONS
The above examples pose the question of whether it is possible to move beyond individual narratives and formally encode multiple features of observed microbial interactions, extending the classical ecological outcome axis. If some of these features are ubiquitous across microbial communities, one could formalize the multidimensional nature of microbial relationships and devise an expanded list of attributes that can classify interactions at a higher resolution and in a manner that better highlights their diverse, dynamic qualities. In this section, we introduce a possible scheme for capturing some of these attributes, and exemplify the application of this framework to a compendium of interactions found in the literature. The attributes we will consider here are: the molecular vehicle (if present) that mediates the interaction, whether the interaction is specific to a particular recipient, whether there is a fitness cost to engaging in the interaction, the site (cytoplasm, membrane or extracellular) in which the interaction takes place, the biome or habitat in which the interaction has been observed, whether the interaction was observed to depend on specific spatial configurations, temporal dynamics or direct physical contact, as well as the sign of the ultimate ecological outcome incurred by the participants. It is our hope that incorporating these attributes, further formalized in Table 1, into descriptions of interactions will enhance our understanding of the landscape of known microbial relationships.
Table 1.
Definitions of key interaction attributes. We describe microbial interactions using a number of attributes, each of which is assigned a numerical value based on experimental observations. Each attribute defined here corresponds to a column in our interaction catalog (Supplementary Table 1). We quantify most features in a binary way: using a ‘0’ if the interaction does not exhibit a certain attribute and a ‘1’ if it does. For example, if an interaction involved the exchange of a peptide, it would contain a ‘1’ in the ‘peptides’ column. Costs and ecological outcomes are specific to the organisms in the interactions, that is, there are columns for costs and outcomes for each of the participants. In a pairwise commensal interaction, for instance, there would be a ‘0’ in the column corresponding to the outcome gained by participant 1 and a ‘1’ in the column corresponding to the outcome gained by participant 2.
| Attribute | Definition | Quantification |
|---|---|---|
| Specificity | The reported mechanism of interaction is deployed in a manner specific to the recipient (e.g. signaling molecules specific to one species vs. nonspecific secretion of waste products). | Binary |
| Cost | Engagement in the reported interaction (e.g. secreting a metabolite) imposes a fitness burden on a participant (i.e. the individual fitness/growth rate of an organism would initially have been greater had it not been involved in the interaction). | Binary |
| Ecological outcome | The ultimate ecological effect the interaction confers on each participant. Combining these values for both participants in a pairwise interaction yields its overall ecological outcome (e.g. 1,-1 corresponds to selfishness; 1,1 corresponds to mutualism, etc.). |
|
| Contact dependence | Reported interaction features organisms engaging in direct physical contact. | Binary |
| Time dependence | Reported relationship features organisms interacting according specific temporal frames (e.g. occurring only at one point in a circadian cycle). | Binary |
| Spatial dependence | Reported interaction features organisms displaying particular spatial configurations (e.g. colonies separated by some distance on an agar plate as opposed to interacting in mixed cultures). | Binary |
| Site | The site, relative to the microbes involved, in which the interaction is reported to take place: extracellular (e.g. signaling molecule release or metabolic exchange), membrane (e.g. protein docking or conjugation), or cytoplasm (e.g. direct predation). | Binary value for each site |
| Habitat | The biome(s) in which the interaction or participating organisms have been observed: aquatic, biofilm, food product, multicellular host, soil, synthetic, or ubiquitous. | Binary value for each habitat |
| Compounds involved | The type of molecule that mediates the interaction: small molecules (e.g. carbohydrates or metabolic intermediates, but not secondary metabolites), nucleic acids (e.g. DNA), peptides (e.g. amino acids), or secondary metabolites (e.g. quorum sensing molecules). | Binary value for each compound type |
The set of interactions to which we applied the proposed framework consists of 74 microbial relationships sourced from the literature, which together make up a ‘catalog’ in which each interaction is characterized by a ‘barcode’ of quantifiable attributes (Table S1, Supporting Information). As we focused almost exclusively on collecting interactions that had sufficient data to assign values to each of our attributes, our catalog is limited to a small set of well-characterized observations. Nonetheless, it is intended to represent a cross-section of all known interactions, featuring a wide variety of biomes, mechanisms, taxa and dependencies. We also note that our assignment of features is specific to the experiments reported in each study. As a result, the attributes we report here should not be considered universal to all interactions between the reported taxa. Instead, they serve to encode the complex nature of observed microbial interactions in a manner that is compatible with current methods of data collection. Nonetheless, the gradual establishment of a standardized set of attributes to be measured in newly designed experiments could lead to large compendia of these multi-dimensional traits that may enable more generalizable insights.
A majority of the interactions that we compiled occurred between two distinct microbial species. However, we have also included some interactions that have been studied at lower taxonomic resolutions (e.g. at the genus or phylum level). Some of these relationships list multiple species of a particular genus — or even relatively undefined consortia of organisms – as individual participants. Although different species within a large taxonomic grouping will undoubtedly display varying interaction properties individually, studies may group several organisms together to highlight their collective performance of a function of interest. For example, a set of nitrogen-fixing archaea was framed as a single entity that engages in an interaction with organisms of the bacterial genus Desulfosarcina (catalog entry 6, (Dekas, Poretsky and Orphan 2009)). In grouping these varied organisms together, Dekas et al. highlighted the cooperative behavior of the archaeal consortium as it applied to its bacterial symbionts. Since identifying relationships of interest in complex natural microbial systems sometimes necessitates such levels of abstraction, our catalog reflects the taxonomic groupings for each interaction as they were reported.
As an example of how a specific interaction can be translated into our proposed multi-dimensional classification, we will analyze here in detail a predatory relationship between two opportunistic pathogens: Pseudomonas aeruginosa and the filamentous form of the fungus Candida albicans, initially identified by Hogan and Kolter ((Hogan and Kolter 2002), catalog entry 48). Here, P. aeruginosa is reported to form a biofilm on the filaments of C. albicans, which kills the fungus. By eliminating a competitor, this action allows P. aeruginosa to more effectively consume nutrients. We quantify the ecological outcomes of this interaction by assigning a positive outcome to P. aeruginosa and a negative one to C. albicans. The study also described mechanisms displayed by P. aeruginosa, that were necessary to initiate the interaction, chief among which was an ability to differentiate the filamentous form of the fungus from the yeast form. We therefore deem this interaction to be targeted towards a particular organism and phenotype, as the bacterium did not attack yeast-form C. albicans. The authors described a dependence of the interaction on quorum sensing pathways, indicating the probable role of secondary metabolites. Since these molecules are exchanged between organisms, we record the extracellular environment as the primary site for this relationship. As the activation pathways that lead to the formation of biofilms and the secretion of secondary metabolites are metabolically costly, we assign to P. aeruginosa a positive metabolic cost of initiating predation. Since the interaction results in the death of C. albicans, we also deem the interaction to impose a fitness cost on the fungus. Lastly, the interaction features direct contact between both species and depends on their spatial proximity, so we list contact and space as key dependencies. In all, we describe this interaction with 22 different variables, which collectively make up its interaction ‘barcode.’
Though this ‘barcoding’ constitutes a rudimentary formalization of the complex attributes an interaction can exhibit, it has some immediate ramifications. Firstly, it allows for identification of interaction attributes that may not be intuitively correlated. For example, are interactions involving signaling molecules more likely than others to also exhibit spatial dependence? In which environments could such correlations hold? Using our limited dataset, for instance, we calculated a Spearman correlation coefficient of 0.41 between the column detailing spatial dependence and the column containing secondary metabolite data (compared to −0.24, −0.04 and −0.04 for small molecules, nucleic acids and peptides respectively). Though modest, this significant correlation (P = 0.0003) hints at possible relationships between spatial dependence and secondary metabolite use for interactions in our dataset. By performing similar analyses on a greater number of interactions, it may be possible to infer stronger, additional correlation patterns emerging from the data. For example, it would be possible to ask if all ‘barcode’ combinations tend to occur, or if there are specific attribute combinations that are either very frequent or rare. Moreover, it would be possible to highlight the discovery of new types of interactions that exhibit unique attribute combinations not previously observed. Such a framework could also be useful for identifying commonalities among interactions from different biomes, and for comparing seemingly unrelated interactions to each other.
An ability to answer the general questions discussed above would undoubtedly require more data than the small collection of interactions we have compiled here. Moreover, annotating newly discovered interactions to the level of detail we propose is not a trivial task, as generating a single interaction ‘barcode’ likely requires deployment of a variety of different experimental and computational methods (Raes and Bork 2008; Faust and Raes 2012) in addition to manual curation of the catalog itself. Because of these challenges, some mechanistic attributes remain unknown even for the relatively well-characterized set of interactions that we have compiled here. For instance, the presence and strength of a microbial interaction can also be determined by environmental constraints such as temperature (Price and Sowers 2004; Dell, Pawar and Savage 2014; Lin et al. 2016) and pH (López-Lara, Sohlenkamp and Geiger 2003; Jin and Kirk 2018; Ratzke and Gore 2018), as well as by the chemical composition of the environments (Klitgord et al. 2010; Chamberlain, Bronstein and Rudgers 2014; Ponomarova et al. 2017; Ziesack et al. 2018; Pacheco, Moel and Segrè 2019). Moreover, microbial interactions may change based on properties of the populations themselves such as relative population size (Kong et al. 2018; Venturelli et al. 2018), order of colonization (Mazumdar, Amar and Segrè 2013; Dang and Lovell 2016), or stochasticity in community assembly (Zhou et al. 2013; Vega and Gore 2017; von Bronk et al. 2017). Future cataloguing and analysis efforts would benefit from the reporting of as many of these attributes as possible in individual studies, as well as from improved data mining methods to incorporate existing data into numerical frameworks.
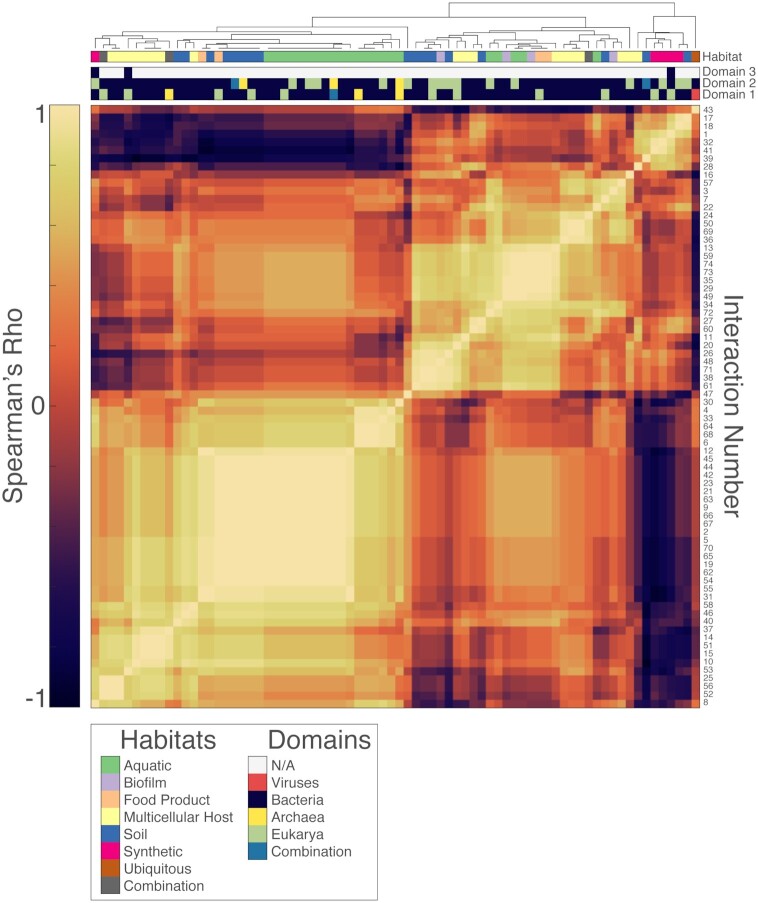
Despite these challenges, we will outline how a quantitative method of comparing interactions may be approached using the dataset we have compiled. Here, we used our annotation framework to calculate distance metrics between each interaction. These distances consider, in an unbiased way, all of the attributes we have annotated for each interaction. In this manner, we are able to generate a hierarchical tree and heatmap (Fig. 2) to visualize which interactions are most similar to each other. As our encoding of interaction properties yielded numerical vectors that are difficult to parse and intuitively compare by eye, such a tree (which likely bears no resemblance to evolutionary or phylogenetic trees) is a useful way to visually interpret similarities or differences between interactions. Our clustering yielded several key observations: for example, the case of prophage-bacteria mutualism reported by Barondess and Beckwith ((Barondess and Beckwith 1995), catalog entry 43) is, perhaps intuitively, grouped furthest away from all other organisms. This grouping occurred independently of the large relative taxonomic difference between viruses and bacteria, archaea and eukaryotes, as taxonomic information was not considered for the clustering. Instead, its grouping is likely due to the ubiquity of this interaction in terms of habitat – it is generally not limited to a particular environment, which differentiates it from other interactions in our dataset. As data on more and more interactions becomes available, we may begin to use such clustering techniques to gain more universal insight on the general global distributions of certain interaction types.
Figure 2.
Hierarchical clustering of microbial interactions, numbered according to their catalog entry in Table S1 (Supporting Information). Numerical values for interaction attributes (specificity, costs, ecological outcomes, dependencies, site, habitat and compounds involved) were normalized from 0 to 1. Multi-column attributes (i.e. those that contain specific values for individual participants, such as ecological outcome and cost) were additionally encoded into single unique values using the Cantor pairing function. Unknown values (comprising 2.1% of the dataset) were imputed with the mean of each column to enable all interactions to be compared. The normalized values were used to calculate pairwise distances between each interaction using Spearman's rho. Hierarchical clustering was then performed to generate a tree based on the resulting distance matrix. Taxonomic information (which was not used to generate clustering), as well as habitat information is displayed for all interactions.
In addition, this analysis revealed proximity between some interactions based on mechanism of action. For example, siderophore-mediated signaling interactions reported separately by Lamont et al. in Pseudomonas ((Lamont et al. 2002), catalog entry 49) and Guan et al. in Vibrio ((Guan, Kanoh and Kamino 2001), catalog entry 73) are grouped within the same cluster. Interactions not mediated by any molecular exchange (Catalog entries 28, 32, 40, 41 and 46) are also largely grouped separately from those that require a molecular vehicle. In addition, observations of direct amino acid exchange ((Accolas, Veaux and Auclair 1971; Gobbetti, Corsetti and Rossi 1994; Holguin and Bashan 1996; Rikhvanov et al. 1999; Garcia et al. 2015), catalog entries 31, 12, 54, 55 and 9) display close proximity despite having been observed in very different habitats. This grouping shows that our clustering is sensitive to mechanism of action, and may provide a tool for identifying similar interactions across very different ecosystems.
Another feature of this multi-dimensional distance analysis is that it does not necessarily group interactions within the same benefit/detriment category together (e.g. all commensal interactions or all mutualistic interactions), as shown in the close groupings of a Burkholderia-Rhizopus toxin-dependent interaction (Partida-Martinez et al. 2007) and a eukaryote-bacteria protein-based relationship ((Schulz et al. 2016), catalog entries 17 and 18). These interactions, despite involving very different mechanisms, are similar in a less straightforward way: they are both mutualistic and both spatially and contact-dependent. Their close grouping therefore stems from this combination of features, which is rare in the dataset and places them at a greater distance from most other interactions. Such a use of a combination of factors to determine proximity is essential for more complete comparisons of interactions, and can also provide clues as to the types of interactions that may be rare in nature or have yet to be observed.
Lastly, this framework can be extended to analyzing interactions that involve more than two organisms. For example, the ‘ecological outcome’ and ‘cost’ categories can have as many columns as there are partners within any given interaction. In this way, it is possible to assign corresponding values to each participant within the entire multispecies relationship. To compare these attributes with those of other interactions, we used a pairing function to collapse these multivariable characteristics into single unique numerical values. In this way, for example, we can directly compare a synthetic tripartite symbiosis between Azotobacter, Alternaria and Chlamydomonas ((Lőrincz et al. 2010), catalog entry 8) with all others in our dataset. This example clustered very closely to a relationship between yeast and Acinetobacter ((Smith, Des Etages and Snyder 2004), catalog entry 52), allowing us to appreciate similarities in their mechanisms (both interactions involved the exchange of small molecules and have spatially dependent components) despite each having a different number of individual participants. If, however, an interaction involving more than two organisms can be divided into two clear roles, it is still possible to analyze it under a pairwise framework. For example, in the relationship involving Thioploca, a genus of marine sulfur bacteria, and a set of anaerobic ammonium oxidizing (anammox) bacteria, Thioploca is defined as the first participant as it was observed to unidirectionally provide small molecules to the anammox consortium, which is defined collectively as the second participant ((Scholten et al. 2007), catalog entry 68). We can then analyze such an interaction in the same way as we would those involving two distinct organisms. As a result, we notice that this interaction is grouped closely to other marine bacterial metabolic exchanges, showing that interactions with similar attributes can cluster together independently of the number of organisms involved.
The specific encoding structure we have proposed here is flexible, as different attributes can be highlighted using variants of the framework and refined as additional cases and data become available. In addition, our particular clustering analysis is highly sensitive to the distance metrics used and to the numerical inputs that were used to quantify individual attributes. Therefore, further research could address the question of whether alternative metric schemes should be used or different methods for dimensionality reduction could be employed. Regardless of the comparison techniques employed, however, we believe that a continued effort to formally encode interaction properties as we have done here could facilitate comparisons of diverse inter-microbial networks, especially as data from multiple microbial ecosystems are increasingly made available. While the formalization exemplified here is limited to a numerical representation of known attributes, a framework like it could help implement more comprehensive mathematical models for microbial ecosystem dynamics, applicable to understanding complex natural communities. For example, different interaction attributes could translate to specific terms in appropriate differential equations that describe community function (Carrara et al. 2015; Momeni, Xie and Shou 2017; Hart et al. 2019) enabling quantitative predictions of population dynamics. If available, data providing deeper information on interaction mechanisms, such as gene expression, binding affinities, and reaction rates could be also systematically encoded in future compendia. Such an encoding would need to be much more complex than the simple framework we have proposed here in order to enable comparison across interactions, but it would have the potential to facilitate quantitative community modeling efforts. For example, transporter parameters such as Km and Vmax values can be directly incorporated into metabolic cross-feeding simulations using genome-scale models (Khandelwal et al. 2013; Harcombe et al. 2014). Additionally, the design of synthetic microbial consortia could greatly benefit from the availability of a curated list of interaction properties. One could imagine selecting candidate organisms based on their known interactions in certain contexts, potentially expediting the process of assembling multispecies communities with desired phenotypes.
SUMMARY
Descriptions of microbial interactions range from those that report individual symbioses in exquisite detail, to an increasing number of large-scale measurements of pairwise interactions enabled by new technologies. As we try to make sense of these interactions with the aid of network analyses and computer simulations, an emerging challenge is how to categorize these different types of relationships and formally encode their underlying mechanisms and attributes to enable comparison across datasets. In this mini-review, we provided examples of well-characterized microbial interactions to highlight their complex nature, and we illustrated the limitations of a broadly used ecological classification system. At the same time, we showed how one can in principle distill recurrent interaction characteristics that can be translated into multi-dimensional profiles. In an effort to embrace these details and characterize interactions in a more unified framework, we proposed a list of quantifiable attributes that can more fully capture the multidimensional nature of these phenomena. We compiled a collection of diverse interactions and quantitatively compared them according to these attributes, showing how such analysis can serve as a stepping stone towards more comprehensive quantitative frameworks for addressing important questions in microbial ecology.
As more data becomes available for a greater number of interactions, we may begin to incorporate additional attributes that can impact their interpretation, such as dependence on environmental substrates, pH or strain abundances. In addition, if non-mechanistic data from methods such as network inference are also to be incorporated into future iterations of this framework, it will be important to add identifiers to denote the experimental and computational techniques used to infer the resultant interactions. These considerations are crucial, as variability in different methods of inferring interactions is likely to have an impact on which data are available to collect and compare.
We anticipate that efforts similar to the one proposed here could grow into large databases of microbial relationships based on detailed observations, phenotypic measurements and ecosystem-level sequencing efforts. As we continue to gather data about microbial interactions and learn which of their attributes are most useful to encode, a standard format for these knowledge repositories (e.g. SBML for systems biology (Hucka et al. 2003) or SBOL for synthetic biology (Galdzicki et al. 2014)) may emerge to further codify reporting methods and facilitate comparative analyses. We therefore hope that, by compiling, classifying and analyzing this small collection of microbial relationships, our effort can motivate further efforts and conversations on how to gather, formalize and mine multi-dimensional data on microbial interactions.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank members of the Segrè lab for inspiring conversations.
FUNDING
This work was supported by the Defense Advanced Research Projects Agency (purchase request no. HR0011515303, contract no. HR0011-15-C-0091), the Army Research Office under the Multidisciplinary University Research Initiatives (MURI) award W911NF-12-1-0390, the United States Department of Energy (DE-SC0012627), the National Institutes of Health (T32GM100842, 5R01DE024468, R01GM121950 and Sub_P30DK036836_P&F), the National Science Foundation (1457695 and NSFOCE-BSF 1635070), the Human Frontiers Science Program (RGP0020/2016) and the Boston University Interdisciplinary Biomedical Research Office. A.R.P. is supported by a Howard Hughes Medical Institute Gilliam Fellowship and a National Academies of Sciences, Engineering and Medicine Ford Foundation Predoctoral Fellowship.
Conflicts of interests. None declared.
REFERENCES
- Accolas J, Veaux M, Auclair J. Étude des interactions entre diverses bactéries lactiques thermophiles et mésophiles, en relation avec la fabrication des fromages à pâte cuite. Lait. 1971;51:249–72. [Google Scholar]
- Armitage DW, Jones SE. How sample heterogeneity can obscure the signal of microbial interactions. bioRxiv. 2019:520668. DOI: 10.1101/520668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Bates ST, Casamayor EOet al.. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barondess JJ, Beckwith J. bor gene of phage lambda, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J Bacteriol. 1995;177:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basan M, Hui S, Okano Het al.. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Chumley T, Cheung Pet al.. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol. 2006;188:5888–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyk O, Luzhetskyy A. Metabolic engineering of natural product biosynthesis in actinobacteria. Curr Opin Biotechnol. 2016;42:98–107. [DOI] [PubMed] [Google Scholar]
- von Bronk B, Schaffer SA, Götz Aet al.. Effects of stochasticity and division of labor in toxin production on two-strain bacterial competition in Escherichia coli. Balaban N (ed). PLOS Biol. 2017;15:e2001457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M V, Philip GK, Bunge JAet al.. Microbial community structure in the North Pacific ocean. ISME J. 2009;3:1374–86. [DOI] [PubMed] [Google Scholar]
- Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara F, Giometto A, Seymour Met al.. Inferring species interactions in ecological communities: a comparison of methods at different levels of complexity. Rees M (ed). Methods Ecol Evol. 2015;6:895–906. [Google Scholar]
- Caspi R, Billington R, Ferrer Let al.. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiker H, Gore J. Competition between species can stabilize public-goods cooperation within a species. Mol Syst Biol. 2012;8:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions?. Etienne R (ed). Ecol Lett. 2014;17:881–90. [DOI] [PubMed] [Google Scholar]
- Chamoun R, Aliferis KA, Jabaji S. Identification of signatory secondary metabolites during mycoparasitism of Rhizoctonia solani by Stachybotrys elegans. Front Microbiol. 2015;6:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Athar R, Zheng Get al.. Prey bacteria shape the community structure of their predators. ISME J. 2011;5:1314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Foster KR. Antibiotics and the art of bacterial war. Proc Natl Acad Sci. 2015;112:10827–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza G, Shitut S, Preussger Det al.. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep. 2018;35:455–88. [DOI] [PubMed] [Google Scholar]
- Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80:91–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science (80-). 2009;326:422–6. [DOI] [PubMed] [Google Scholar]
- Dell AI, Pawar S, Savage VM. Temperature dependence of trophic interactions are driven by asymmetry of species responses and foraging strategy. Humphries M (ed). J Anim Ecol. 2014;83:70–84. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen Net al.. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–50. [DOI] [PubMed] [Google Scholar]
- Foster KR, Kokko H. Cheating can stabilize cooperation in mutualisms. Proceedings Biol Sci. 2006;273:2233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson ME. Mutualisms are not on the verge of breakdown. Trends Ecol Evol. 2017;32:727–34. [DOI] [PubMed] [Google Scholar]
- Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1:0109. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM. Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol. 2015;13:133–46. [DOI] [PubMed] [Google Scholar]
- Galdzicki M, Clancy KP, Oberortner Eet al.. The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat Biotechnol. 2014;32:545–50. [DOI] [PubMed] [Google Scholar]
- Garcia SL, Buck M, McMahon KDet al.. Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol Ecol. 2015;24:4449–59. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JGet al.. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbetti M, Corsetti A, Rossi J. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of carbohydrates. Appl Microbiol Biotechnol. 1994;41:456–60. [DOI] [PubMed] [Google Scholar]
- Goldford JE, Lu N, Bajić Det al.. Emergent simplicity in microbial community assembly. Science (80-). 2018;361:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LL, Kanoh K, Kamino K. Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Appl Environ Microbiol. 2001;67:1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Boedicker JQ. The contribution of high-order metabolic interactions to the global activity of a four-species microbial community. Reed JL (ed). PLOS Comput Biol. 2016;12:e1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. [DOI] [PubMed] [Google Scholar]
- Harcombe W. Novel cooperation experimentally evolved between species. Evolution (N Y). 2010;64:2166–72. [DOI] [PubMed] [Google Scholar]
- Harcombe WR, Riehl WJ, Dukovski Iet al.. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SFM, Mi H, Green Ret al.. Uncovering and resolving challenges of quantitative modeling in a simplified community of interacting cells. Sanchez A (ed). PLOS Biol. 2019;17:e3000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath-Heckman EAC. The metronome of symbiosis: interactions between microbes and the host circadian clock. Integr Comp Biol. 2016;56:776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Kolter R. Pseudomonas-candida interactions: an ecological role for virulence factors. Science (80-). 2002;296:2229–32. [DOI] [PubMed] [Google Scholar]
- Holguin G, Bashan Y. Nitrogen-fixation by Azospirillum brasilense Cd is promoted when co-cultured with a mangrove rhizosphere bacterium (Staphylococcus sp.). Soil Biol Biochem. 1996;28:1651–60. [Google Scholar]
- Hsu RH, Clark RL, Tan JWet al.. Rapid microbial interaction network inference in microfluidic droplets. bioRxiv. 2019:521823. DOI: 10.1101/521823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Stetter KOet al.. The phylum Nanoarchaeota: Present knowledge and future perspectives of a unique form of life. Res Microbiol. 2003;154:165–71. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HMet al.. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. [DOI] [PubMed] [Google Scholar]
- Imachi H, Sekiguchi Y, Kamagata Yet al.. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl Environ Microbiol. 2000;66:3608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn U, Gallenberger M, Paper Wet al.. Nanoarchaeum equitans and Ignicoccus hospitalis: New insights into a unique, intimate association of two archaea. J Bacteriol. 2008;190:1743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn U, Summons R, Sturt Het al.. Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I. Arch Microbiol. 2004;182:404–13. [DOI] [PubMed] [Google Scholar]
- Jin Q, Kirk MF. pH as a primary control in environmental microbiology: 1. thermodynamic perspective. Front Environ Sci. 2018;6:21. [Google Scholar]
- Jousset A, Scheu S, Bonkowski M. Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria. Funct Ecol. 2008;22:714–9. [Google Scholar]
- Jurkevitch E(ed). Predatory Prokaryotes. Berlin Heidelberg: Springer, 2007. [Google Scholar]
- Kadouri DE, To K, Shanks RMQet al.. Predatory bacteria: a potential ally against multidrug-resistant Gram-negative pathogens. PLoS One. 2013;8:e63397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28. DOI: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddy PA. Competition. Dordrecht Netherlands: Springer, 2001. [Google Scholar]
- Kelsic ED, Zhao J, Vetsigian Ket al.. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal RA, Olivier BG, Röling WFMet al.. Community flux balance analysis for microbial consortia at balanced growth. Vera J (ed). PLoS One. 2013;8:e64567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgord N, Segrè D. Ecosystems biology of microbial metabolism. Curr Opin Biotechnol. 2011;22:541–6. [DOI] [PubMed] [Google Scholar]
- Klitgord N, Segrè D, Hattori Met al.. Environments that induce synthetic microbial ecosystems. Papin JA (ed). PLoS Comput Biol. 2010;6:e1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Meldgin DR, Collins JJet al.. Designing microbial consortia with defined social interactions. Nat Chem Biol. 2018;14:821–9. [DOI] [PubMed] [Google Scholar]
- Lamont IL, Beare PA, Ochsner Uet al.. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2002;99:7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox Jet al.. Unexpected diversity and complexity of the guerrero negro hypersaline microbial mat. Appl Environ Microbiol. 2006;72:3685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. 2015;112:10479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidicker WZ. A clarification of interactions in ecological systems. Bioscience. 1979;29:475–7. [Google Scholar]
- Lin Q, He G, Rui Jet al.. Microorganism-regulated mechanisms of temperature effects on the performance of anaerobic digestion. Microb Cell Fact. 2016;15:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard Get al.. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol Plant-Microbe Interact. 2003;16:567–79. [DOI] [PubMed] [Google Scholar]
- Lőrincz Z, Preininger É, Kósa Aet al.. Artificial tripartite symbiosis involving a green alga (Chlamydomonas), a bacterium (Azotobacter) and a fungus (Alternaria): Morphological and physiological characterization. Folia Microbiol (Praha). 2010;55:393–400. [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir S, Heinken A, Kutt Let al.. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2016, DOI: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- Margulis L. Words as battle cries: symbiogenesis and the new field of endocytobiology. Bioscience. 1990;40:673. [PubMed] [Google Scholar]
- Martin BD, Schwab E. Current usage of symbiosis and associated terminology. Int J Biol. 2013;5. DOI: 10.5539/ijb.v5n1p32 [Google Scholar]
- Mazumdar V, Amar S, Segrè D. Metabolic proximity in the order of colonization of a microbial community. PLoS One. 2013;8:e77617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee MT, Collins JJ, Church GMet al.. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar D, van Berlo R, de Ridder Det al.. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol. 2009;5:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni B, Xie L, Shou W. Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. Elife. 2017;6:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Moel M, Segrè D. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat Commun. 2019;10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kerner A, Burns MAet al.. Microdroplet-enabled highly parallel co-cultivation of microbial communities. Herrera-Estrella A (ed). PLoS One. 2011;6:e17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez LP, Monajembashi S, Greulich K-Oet al.. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol. 2007;17:773–7. [DOI] [PubMed] [Google Scholar]
- Ponomarova O, Gabrielli N, Sévin DCet al.. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017;5:345–357.. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel R, Jumpponen A, Schlatter DCet al.. Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology. 2016;106:1083–96. [DOI] [PubMed] [Google Scholar]
- Price PB, Sowers T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci USA. 2004;101:4631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes Jet al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–9. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. Nature. 2003;425:72–4. [DOI] [PubMed] [Google Scholar]
- Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. M. Sullam P (ed). PLoS Pathog. 2011;7:e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–94. [DOI] [PubMed] [Google Scholar]
- Ratzke C, Gore J. Modifying and reacting to the environmental pH can drive bacterial interactions. Pal C (ed). PLOS Biol. 2018;16:e2004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhvanov EG, Varakina NN, Sozinov DYet al.. Association of bacteria and yeasts in hot springs. Appl Environ Microbiol. 1999;65:4292–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden LC, Ingle RA. Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell. 2009;21:2546–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JL, Mueller UG, Wilcox TPet al.. The evolution of cooperation. Q Rev Biol. 2004;79:135–60. [DOI] [PubMed] [Google Scholar]
- Scholten JC, Culley DE, Brockman FJet al.. Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: involvement of an ancient horizontal gene transfer. Biochem Biophys Res Commun. 2007;352:48–54. [DOI] [PubMed] [Google Scholar]
- Schulz F, Martijn J, Wascher Fet al.. A Rickettsiales symbiont of amoebae with ancient features. Environ Microbiol. 2016;18:2326–42. [DOI] [PubMed] [Google Scholar]
- Smith DC. Symbiosis research at the end of the millenium. Hydrobiologia. 2001;461:49–54. [Google Scholar]
- Smith MG, Des Etages SG, Snyder M. Microbial synergy via an ethanol-triggered pathway. Mol Cell Biol. 2004;24:3874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NW, Shorten PR, Altermann Eet al.. The classification and evolution of bacterial cross-feeding. Front Ecol Evol. 2019;7:153. [Google Scholar]
- Stacy A, Everett J, Jorth Pet al.. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci. 2014;111:7819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C, Ferrieri AP, Tfaily MMet al.. Diurnal cycling of rhizosphere bacterial communities is associated with shifts in carbon metabolism. Microbiome. 2017;5:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia Let al.. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook Het al.. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. Microbial metabolomics. Curr Genomics. 2011;12:391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Or D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol Rev. 2017;41:599–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton L, Darcy JL, Hynson NA. A developing symbiosis: enabling cross-talk between ecologists and microbiome scientists. Front Microbiol. 2019;10:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Zeng L, Burne RA. The EIIABMan phosphotransferase system permease regulates carbohydrate catabolite repression in Streptococcus gordonii. Appl Environ Microbiol. 2011;77:1957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of “unculturable” bacteria. FEMS Microbiol Lett. 2010;309:1–7. [DOI] [PubMed] [Google Scholar]
- Vega NM, Gore J. Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine. Cadwell K (ed). PLOS Biol. 2017;15:e2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli OS, Carr AC, Fisher Get al.. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14:e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Miyashiro T. Quorum sensing in the squid-Vibrio symbiosis. Int J Mol Sci. 2013;14:16386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JJ, Pace NR. Endolithic microbial ecosystems. Annu Rev Microbiol. 2007;61:331–47. [DOI] [PubMed] [Google Scholar]
- Welch DB, Huse SM. Microbial Diversity in the Deep Sea and the Underexplored “Rare Biosphere”. In: de Bruijn FJ(ed). Handbook of Molecular Microbial Ecology II. 2011. DOI: doi:10.1002/9781118010549.ch24 [Google Scholar]
- West SA, Griffin AS, Gardner Aet al.. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J Evol Biol. 2007;20:415–32. [DOI] [PubMed] [Google Scholar]
- Whitman W, Coleman D, Wiebe W. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm SW, Matteson AR. Freshwater and marine virioplankton: a brief overview of commonalities and differences. Freshw Biol. 2008;53:1076–89. [Google Scholar]
- Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu W, Deng Yet al.. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. MBio. 2013;4:e00584–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesack M, Gibson T, Shumaker AMet al.. Inducible cooperation in a synthetic gut bacterial consortium introduces population balance and stability. bioRxiv. 2018:426171. DOI: 10.1101/426171 [DOI] [Google Scholar]
- Zomorrodi AR, Segrè D. Genome-driven evolutionary game theory helps understand the rise of metabolic interdependencies in microbial communities. Nat Commun. 2017;8:1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.