Abstract
Introduction
Exposure to ricin can be lethal and treatments that are under development have short windows of opportunity for administration after exposure. It is therefore essential to achieve early detection of ricin exposure to provide the best prognosis for exposed individuals. Ricin toxin can be detected in clinical samples via several antibody-based techniques, but the efficacy of these can be limited due to the rapid processing and cellular uptake of toxin in the body and subsequent low blood ricin concentrations. Other diagnostic tools that perform, in an orthogonal manner, are therefore desirable.
Objectives
To determine time-dependent metabolic changes in Sprague–Dawley rats following intravenous exposure to ricin.
Methods
Sprague–Dawley rats were intravenously exposed to ricin and multiple blood samples were collected from each animal for up to 48 h following exposure in two independent studies. Plasma samples were analysed applying HILIC and C18 reversed phase UHPLC–MS assays followed by univariate and multivariate analysis.
Results
In Sprague–Dawley rats we have demonstrated that metabolic changes measured in blood can distinguish between rats exposed intravenously to ricin and controls prior to the onset of behavioral signs of intoxication after 24 h. A total of 37 metabolites were significantly altered following exposure to ricin when compared to controls. The arginine/proline, bile acid and triacylglyceride metabolic pathways were highlighted as being important with two triacylglycerides at 8 h post exposure giving an AUROC score of 0.94. At 16 h and 24 h the AUROC score increased to 0.98 and 1.0 with the number of metabolites in the panel increasing to 5 and 7, respectively.
Conclusions
These data demonstrate that metabolites may be a useful tool to diagnose and detect ricin exposure, thus increasing the effectiveness of supportive therapy and future ricin-specific medical treatments.
Keywords: Ricin, Metabolomics, Arginine/proline metabolism, Bile acid metabolism, Triacylglyceride metabolism
Introduction
Ricinus communis, the castor bean plant, is indigenous to Eastern Africa, but is found commonly across the globe (Prince 2000). The castor oil obtained from the plant’s seeds is used in many products such as hydraulic fluid, engine lubricant and even in traditional medicine (Patel et al. 2016). The seeds also contain the toxic plant protein ricin (Schep et al. 2009) which belongs to the type 2 ribosome-inactivating protein (RIP) family and consists of two chains; Ricin Toxin A-chain (RTA) and Ricin Toxin B-chain (RTB) (Lord et al. 1994). The A-chain is a N-glycosidase enzyme responsible for the toxicity of ricin, while the B-chain facilitates entry into the cell by binding to cell surface glycolipids and proteins (Olsnes 2004). Once within the cell the RTA subunit is transported to the cytosol where it specifically targets a conserved adenine residue found in the 28S ribosomal RNA of the 60S subunit. This leads to inhibition of protein synthesis and ultimately cell apoptosis (May et al. 2013).
The LD50 of ricin is thought to be between 3 and 30 µg/kg in humans (Worbs et al. 2011) and in rodent models between 2 and 8 µg/kg (Roy et al. 2012) but can vary dependent on route of administration (i.e. intravenous vs. inhalation vs. ingestion) and dose. This high toxicity, along with the availability of the plant, its seeds, and relative ease of ricin extraction has lead ricin to be listed as a Category B threat agent by the United States Center for Disease Control (Khan et al. 2000; Rotz et al. 2002) and declared as a Schedule 1 agent by the Chemical Weapons Convention (CWC) (Kuca and Pohanka 2010). Indeed ricin has been used for assassination (Crompton and Gall 1980) and in several postal threats (Schier et al. 2007).
Currently, there are no specific treatments for ricin intoxication, however pre-clinical studies have demonstrated the utility of polyclonal or monoclonal ricin-neutralising antibodies (anti-toxin) to treat the intoxication (Gal et al. 2017; Van Slyke et al. 2016; Whitfield et al. 2017), although these are not yet licensed for clinical use. Data suggests that efficacy of these antibody therapies is significantly reduced if they are not given very early post exposure (Griffiths et al. 2007; Pratt et al. 2007). Other therapeutic strategies have been explored including “disease modifying” compounds which involve using inhibitors that target specific biochemical pathways such as the cytokine storm [Reviewed by D’Elia et al. (2013) and Gal et al. (2017)] to alleviate symptoms. The requirement for medical countermeasures for ricin intoxication to be administered early, combined with the short therapeutic window of pre-clinical therapies, highlights the requirement for early detection and diagnosis.
Early diagnosis of intoxication is essential to ensure that medical care and therapy are given in a timely manner as treatment efficacy can be reduced dramatically following the presentation of signs and symptoms. The generation of diagnostic tools, that can identify intoxication at the very early stages or even before changes in behavioral phenotypes, to stratify populations related to risk, are required to guide administration of therapy in the most effective way (Lukaszewski et al. 2008). This is of particular importance for acute diseases, including those caused by biological and toxin threat agents, where exposure can cause severe morbidity and mortality in days or even hours. This task is made even more difficult if the agent is unknown or is not detectable or cultureable (Patocka and Streda 2006; Ristanovic 2009).
The most common ricin detection methods rely on antibody recognition techniques such as enzyme linked immunosorbent assays (ELISA), flow cytometry and immuno-PCR (Kim et al. 2009; Men et al. 2010; Poli et al. 1994) to directly detect the toxin in clinical samples. ELISAs can be very sensitive being able to detect ricin at levels as low as 20 pg per 100 μl of tissue extract (Leith et al. 1988). They can also be highly adaptable showing efficacy in multiple clinical specimens and environmental samples including blood, urine and stool (Chen et al. 2014). The uptake of ricin into tissues is rapid (Moshiri et al. 2016). In addition, processing of ricin in the body differs depending on the route of exposure (Griffiths 2011). As a consequence, selecting the most appropriate sample type to test within the small window for detection of toxin after exposure has occurred remains a considerable challenge (Griffiths 2011). DNA analysis techniques have also been shown to be effective in detecting toxin exposure indirectly by exploiting the mechanism of action where ricin inhibits protein synthesis by removing an adenine from the 28S rRNA (Diakite et al. 2015; Pierce et al. 2011). More recently, “omic” techniques such as transcriptomics and proteomics, which are not only able to look at agent products, but also specific biomarkers of intoxication have been applied to models of intoxication. Indeed, a proteomic study where mice were exposed to aerosolised ricin highlighted that 6 mouse proteins were highly associated with ricin intoxication (Guo et al. 2014). The recent development of another “omic” technology, metabolomics, has the potential to be a complementary new technology (Dunn et al. 2011).
In the study reported here we sought to determine, in an intravenous rat model of ricin intoxication, whether a metabolomics approach could identify perturbed metabolic pathways prior to the onset of an adverse clinical phenotype. The discovery of putative biomarkers of exposure could then be validated in future investigations. Furthermore, such knowledge could increase the mechanistic understanding of ricin toxicity, beyond protein synthesis inhibition, thereby contributing to the development of an adverse outcome pathway (Hamm et al. 2017).
Materials and methods
Ricin stock preparation
Ricinus Communis Zanzibariensis toxin variant E was supplied by Dstl (batch 1124/29/1/1) at 2.7 mg/ml in Ca2+ and Mg2+-free phosphate buffered saline (PBS, GIBCO). The purity of this preparation was > 90% by SDS-PAGE and densitometry following Coomassie brilliant blue staining. Ricin was stored at − 80 °C prior to use.
Animals
All work was conducted under Home Office licence 30/3269. Sprague–Dawley rats were supplied by Charles River UK. Animals were pre-implanted with an identification/temperature chip. Animals were housed in the experimental animal house for 2 weeks prior to surgery with food and water ad libitum.
Surgery
All animals were surgically prepared with a vascular access button (VAB) connected to the carotid artery to enable blood sampling. Pre-medication analgesia was administered 1 h prior to surgery (Metacam 2 mg/ml s.c.). Anaesthesia was induced with 5% isoflurane in an induction box. The rats were moved to a nose cone as the surgical fields were close-clipped. They were then placed in a supine position on a heating pad and surgical fields were cleaned with chlorhexidine scrub solution. Anaesthesia was maintained with 2–5% Isoflurane (levels were determined by close monitoring of heart rate, respiration rate and level of response to external stimuli by a trained anaesthetist throughout the surgical procedure).
An incision (approximately 25 mm) was made on the ventral surface of the neck. A subcutaneous pocket was created by blunt dissection and then the left carotid artery was located and isolated. Two suture ligatures (Mersilk 6-0) were placed loosely around the blood vessel. The distal ligature was tied off and tension applied to the ligature in order to keep the vessel taut and straight during the cannulation. An atraumatic vessel clamp was used to prevent bleeding as the vessel was cannulated using a 2Fr polyurethane catheter pre-filled with 5 IU heparin in 0.9% saline. The catheter was secured in place with the pre-placed suture ligatures (above and between the suture rings on the catheter). The catheter patency was checked with a 5 IU heparin solution (< 10 µL). The catheter was then temporarily clamped and then capped with an appropriately sized pin. The catheter was tunnelled sub-cutaneously to the midline area on the dorsal surface, approximately 1 cm caudal to the base of the skull. The ventral incision was closed using 4/0 Vicryl suture. With the animal back in the prone position, the catheter was trimmed to the required length and connected to the VAB. 4/0 Mersilk was used to secure the catheter to the button. The VAB was then inserted under the skin and the wound closed with 4/0 Vicryl. Once surgery was complete, the line was checked, flushed and locked with a lock solution (500 IU heparin in 50% glucose in 0.9% saline). Rats received 5 ml fluids (Vetivex 18 s.c.) and were placed in a warm (29 °C) recovery box and monitored until fully recovered.
Study protocol
Following a recovery period of 6–7 days post-surgery in the home cage, rats were moved to the experimental room. A total of 24 rats over two independent experiments were studied for the metabolomics analysis (six rats in a discovery study (all ricin exposed) and sixteen in a validation study (8 ricin exposed vs. 8 PBS control, a complete set of samples could not be collected from one ricin-exposed rat due to a blocked catheter and this animal was omitted from the metabolomic analysis)). An additional 17 rats were challenged with ricin in a pilot study (n = 5) and for toxicokinetic measurements (n = 12) to determine the concentration of detectable ricin in the blood post exposure using ELISA (up to 24 h). Clinical signs data (bodyweight, temperature and clinical score) were collected for all 31 ricin-exposed rats and the 8 exposed to PBS. Statistical comparisons were made only to compare the PBS and the corresponding, time-matched, ricin-exposed animal data for weights and temperatures for the validation study (GraphPad Prism version 6.00 for Windows, GraphPad Software, La Jolla California USA). Animals from the pilot study and the discovery metabolomics study were used to follow the progression of intoxication up to a predetermined humane endpoint (see below). Rats in the toxicokinetic and validation metabolomics study were humanely killed at 24 and 32 h respectively. Only 10 out of 12 rat blood samples were used for ELISA analysis due to the vascular access buttons in 2 animals not functioning correctly. The total blood volume of each rat was estimated at the start of the experiment assuming a volume of 70 ml·kg−1 (Morton et al. 1993). A maximum of 15% total blood volume was taken from each rat over the course of the study.
Rats were placed in a heated chamber (39 °C) for up to 10 min before being restrained for dosing. Ricin was administered at a dose of 8 µg·kg−1 and a dose volume of 1 ml·kg−1 via the tail vein. This dose is expected to be ~ 1–2 LD50 (Franz 1997) and was chosen for this study to ensure the animals would have developed an adverse behavioural phenotype longer term. To confirm this, animals were monitored for signs of intoxication for up to 72 h after ricin administration. Rats were then moved into the metabolism cages and connected to an automated blood sampling system (ABS, Instech Solomon, USA). For the metabolomics studies samples were taken at − 16 h (pre-exposure baseline control), 0 h (time of exposure), 1 h, 8 h, 16 h, 24 h, 32 h and 48 h post exposure though later time points were not analysed because of severe intoxication and humane killing. For the toxicokinetic ELISA studies, samples were taken at 5 min, 20 min, 1 h, 5 h, 8 h and 24 h post exposure. Whole blood was used for ELISA analysis and plasma was prepared for metabolomics analysis. Rats were observed hourly and scored for signs of intoxication (behavioral phenotype). The indicators noted were condition of coat (e.g. ruffled), posture (hunched), mobility and pinching (drawing in of the abdomen). A score of 0 was assigned if signs were normal (no change to controls), 1 = minor alterations and 2 = severe signs. Body temperature (via an implanted identity chip) was also taken hourly. Any animal scored as severely intoxicated for all signs, i.e. a score of 8, was humanely killed by a schedule 1 method.
Toxicokinetics
Ricin ELISA kits (MP Biomedical) were kindly supplied by DST-G (Australia) and DSO National Laboratories (Singapore) for use in this work. The development of this assay is described in (Chen et al. 2014) and consists of a 32-well ELISA plate pre-coated with ricin-A chain specific monoclonal antibodies RA999 (Meridian Life Science, USA) and 7G12, (US Army Medical Research Institute of Infectious Diseases (Dertzbaugh et al. 2005). Blood samples from each exposed animal (30 μl) were added to 60 μl of Sample treatment buffer (STB) pre-aliquoted into the wells of a non-binding microwell plate. Control samples (kit-supplied positive and negative samples) were prepared similarly. Each ELISA plate was also used to analyse a range of known ricin concentrations prepared in naive whole blood. This methodology was used to generate standard curves to enable quantification of ricin within ex vivo samples. Samples containing known ricin concentrations were prepared by serial dilution of a known ricin standard and were treated in the same manner as described above.
The samples and controls were incubated at room temperature for 10 min. The biotinylated secondary antibody (kit-supplied rabbit polyclonal antibody raised against ricin toxoid, Toxin Technology, USA) was prepared by adding 1.6 ml of deionised water to the vial followed by gentle swirling to avoid foaming. The secondary antibody (25 μl) was then added to all wells of the ELISA plate, followed by 75 μl of the treated samples or controls from the non- binding plate. The loaded ELISA plate was then covered and incubated at 37 °C for 60 min. The plate was then washed 6 times with 250 μl of diluted wash buffer (kit-supplied) per well. A freshly made working solution of streptavidin peroxidase conjugate (Kit-supplied, 100 μl per well) was then added and the plate incubated at room temperature, for 10 min. The plate was washed again as above, and TMB reagent (kit-supplied, 100 μl) per well added. The plate was covered and incubated in the dark for 10 min. Stop buffer (2 M sulphuric acid; 50 μl/well) was added and the plate was gently agitated and read (within 10 min) at 450 nm and at a reference wavelength at 620 nm.
Readings taken at 620 nm were subtracted from readings at 450 nm. If the ELISA has performed well, the mean of the corrected absorbance readings of non-reactive controls should be equal to or below 0.1 units of OD. For the corrected reactive control values, these must be equal to or greater than 1.0 OD units. In addition, the cut off value for the assay, below which the sample data is excluded, is 2 × the mean of the nonreactive control.
Analysis of toxicokinetic data
Readings taken from the samples containing known concentrations of ricin were used to generate a standard curve. Data was background subtracted and fitted with a 4-parameter logistic non-linear curve fit (Eq. 1) using GraphPad Prism (Version 6 for Windows). No weighting was applied during curve fitting. Ricin concentrations in individual biological samples were interpolated from the curve fit using GraphPad Prism. Toxicokinetic data were analysed by fitting one and two phase exponential decays to the data and an extra sum of squares F-test was used to compare the fits (GraphPad Prism).
| 1 |
where Bottom = bottom plateau of curve fit, Top = top plateau of curve fit.
Metabolomics study
Sample preparation
Plasma samples were extracted to separate low molecular weight metabolites from other biochemicals including proteins, RNA and DNA. Samples were thawed and extracted on ice. For the analysis of water-soluble metabolites, 180 µL of acetonitrile (LC–MS grade, LiChrosolv, Merck) was added to 60 µL of plasma followed by vortex mixing (15 s), centrifugation (13,000×g, 15 min) and transfer of the clear supernatant to a glass LC autosampler vial (VI-04-12-02RVG 300μL Plastic, Chromatography Direct, UK). For the analysis of lipid metabolites, 180 µL of isopropyl alcohol, IPA (LC-MS grade, LiChrosolv, Merck) was added to 60 µL of plasma followed by vortex mixing (15 s), centrifugation (13,000×g, 15 min) and transfer of the clear supernatant to a glass LC autosampler vial (VI-04-12-02RVG 300μL Plastic, Chromatography Direct, UK). A single pooled QC sample was prepared by combining 25 µL aliquots of all biological samples where adequate sample volume remained and vortex mixing (2 min). Aliquots (60 µL) of the pooled QC sample were extracted as defined above.
Ultra high performance liquid chromatography-mass spectrometry
The samples were analysed applying two Ultra High Performance Liquid Chromatography-Mass Spectrometry (UHPLC-MS) methods using a Dionex UltiMate 3000 Rapid Separation LC system (Thermo Fisher Scientific, MA, USA) coupled with a heated electrospray Q Exactive Focus mass spectrometer (Thermo Fisher Scientific, MA, USA). Polar extracts were analysed on a Accucore-150-Amide-HILIC column (100 × 2.1 mm, 2.6 μm, Thermo Fisher Scientific, MA, USA). Mobile phase A consisted of 10 mM ammonium formate and 0.1% formic acid in 95% acetonitrile/water and mobile phase B consisted of 10 mM ammonium formate and 0.1% formic acid in 50% acetonitrile/water. Flow rate was set for 0.50 ml·min−1 with the following gradient: t = 0.0, 1% B; t = 1.0, 1% B; t = 3.0, 15% B; t = 6.0, 50% B; t = 9.0, 95% B; t = 10.0, 95% B; t = 10.5, 1% B; t = 14.0, 1% B, all changes were linear with curve = 5. The column temperature was set to 35 °C and the injection volume was 2 μL. Data were acquired in positive and negative ionisation modes separately within the mass range of 70–1050 m/z at resolution 70,000 (FWHM at m/z 200). Ion source parameters were set as follows: Sheath gas = 53 arbitrary units, Aux gas = 14 arbitrary units, sweep gas = 3 arbitrary units, Spray Voltage = 3.5 kV, Capillary temp. = 269 °C, Aux gas heater temp. = 438 °C. Data dependent MS2 in ‘Discovery mode’ was used for the MS/MS spectra acquisition using following settings: resolution = 17,500 (FWHM at m/z 200); Isolation width = 3.0 m/z; stepped normalised collision energies (stepped NCE) = 25, 60, 100%. Spectra were acquired in three different mass ranges: 70–200 m/z; 200–400 m/z; 400–1000 m/z.
Non-polar extracts were analysed on a Hypersil GOLD column (100 × 2.1 mm, 1.9 µm; Thermo Fisher Scientific, MA, USA). Mobile phase A consisted of 10 mM ammonium formate and 0.1% formic acid in 60% acetonitrile/water and mobile phase B consisted of 10 mM ammonium formate and 0.1% formic acid in 90% propan-2-ol/water. Flow rate was set for 0.40 ml·min−1 with the following gradient: t = 0.0, 20% B; t = 0.5, 20% B, t = 8.5, 100% B; t = 9.5, 100% B; t = 11.5, 20% B; t = 14.0, 20% B, all changes were linear with curve = 5. The column temperature was set to 55 °C and the injection volume was 2μL. Data were acquired in positive and negative ionisation mode separately within the mass range of 150 – 2000 m/z at resolution 70,000 (FWHM at m/z 200). Ion source parameters were set as follows: Sheath gas = 50 arbitrary units, Aux gas = 13 arbitrary units, sweep gas = 3 arbitrary units, Spray Voltage = 3.5 kV, Capillary temp. = 263 °C, Aux gas heater temp. = 425 °C. Data dependent MS2 in ‘Discovery mode’ was used for the MS/MS spectra acquisition using following settings: resolution = 17,500 (FWHM at m/z 200); Isolation width = 3.0 m/z; stepped normalised collision energies (stepped NCE) = 20, 50, 80%. Spectra were acquired in three different mass ranges: 200–400 m/z; 400–700 m/z; 700–1500 m/z.
A Thermo ExactiveTune 2.8 SP1 build 2806 was used as instrument control software in both cases and data were acquired in profile mode. Quality control (QC) samples were analysed as the first ten injections and then every seventh injection with two QC samples at the end of the analytical batch. Two blank samples were analysed, the first as the 6th injection and then the second at the end of each batch.
Raw data processing
Raw data acquired in each analytical batch were converted from the instrument-specific format to the mzML file format applying the open access ProteoWizard software (Kessner et al. 2008). Deconvolution was performed with XCMS software according to the following settings of Min peak width (4 for HILIC and 6 for lipids); max peak width (30); ppm (12 for HILIC and 14 for lipids); mzdiff (0.001); gapInit (0.5 for HILIC and 0.4 for lipids); gapExtend (2.4); bw (0.25); mzwid (0.01) (Smith et al. 2006). A data matrix of metabolite features (m/z-retention time pairs) vs. samples was constructed with peak areas provided where the metabolite feature was detected for each sample. Putative annotation of metabolites or metabolite groups was performed by applying the PUTMEDID-LCMS workflows operating in the Taverna workflow environment (Brown et al. 2011). We applied 5 ppm mass error and a retention time range of 2 s in feature grouping and molecular formula and metabolite matching. As different metabolites can be detected with the same accurate m/z (for example, isomers with the same molecular formula), multiple annotations could be observed for a single detected metabolite feature. Also, a single metabolite could be detected as multiple molecules, particularly as a different type of ion (e.g., protonated and sodiated ions). Throughout this article, the term “metabolite” refers to either single metabolites or groups of molecules with the same retention time and the same accurate m/z. All molecules were annotated according to guidelines for reporting of chemical analysis results, specifically to Metabolomics Standards Initiative level 2 (Sumner et al. 2007).
QC filtering and multivariate data analysis
The data for pooled QC samples were applied to perform QC filtering. For each metabolite feature detected QC samples 1–8 were removed and the relative standard deviation and percentage detection rate were calculated. Metabolite features with a RSD > 30% and a percentage detection rate < 60% were deleted from the dataset. Missing values in the data were replaced by values applying k nearest neighbour (kNN) missing value imputation followed by normalisation to sample total peak area (as a percentage), glog transformation and Pareto scaling prior to data analysis. Principal Components Analysis (PCA) was then performed to assess the technical variability (measured by the replicate analysis of a pooled QC sample) and biological variability as part of the quality control process. Areas under the Receiver Operator Curves (AUROC) were calculated in MetaboAnalyst with multiple metabolites combined (Xia et al. 2015).
Univariate data analysis
Data were normalised to sample total peak area (as a percentage) and were defined as normalized concentration (%), no missing value imputation or scaling was performed and glog transformation was performed prior to data analysis. For the initial discovery study, paired within-subject t-tests were applied to define changes in metabolite concentrations over time (up to 48 h). For the validation study which included unexposed control animals, ‘Within-subjects repeated measures ANOVA’ was applied to investigate time-specific statistically significant changes (up to 32 h), phenotype-specific statistically significant changes and time × phenotype interactions which were statistically significant, in the software MetaboAnalyst (Guo et al. 2016). All statistical analyses are reported following correction for multiple testing applying the Benjamin-Hochberg method. All metabolite features which were statistically significant in both the discovery and validation datasets were passed forward for further interpretation with all other metabolite features being removed from the datasets. Following this, multiple replicates of the same metabolite features were filtered to leave one replicate with the p value furthest from 0.05.
Results
Model development
Previous data were utilised to determine the most appropriate i.v. ricin dose for rats for a metabolomics study. Depending on the dose of ricin, morbidity and mortality onset varied between hours and days (data not shown). It was decided that a dose where the majority of rats showed clinical or behavioural signs of ricin intoxication, but did not succumb to intoxication within 48 h of exposure would be suitable. This dose was estimated to be 8 µg·kg−1. This progression of intoxication would allow for multiple samples to be taken.
In-vivo signs of intoxication
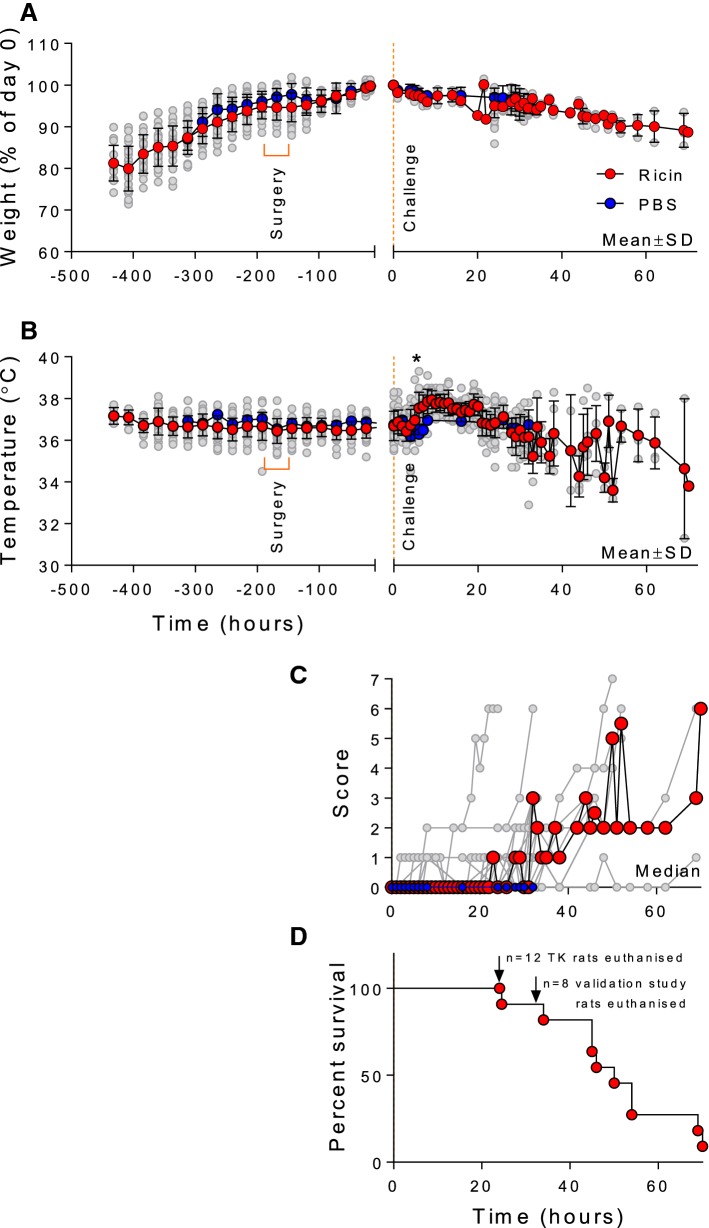
Data relating to signs of intoxication are reported. All rats gradually gained weight during their acclimatisation to the housing in the experimental animal house; they showed a small decrease, on average, following surgery but then gained weight steadily until challenge with ricin. Following exposure, rats in both the PBS and ricin-exposed groups gradually lost weight. There was no statistically significant difference between the two groups and weight loss at this point is likely to be due to stresses of handling for exposure and blood sampling. Ricin-exposed animals in the pilot and discovery metabolomics study continued to lose weight until the humane end point or the end of the study (Fig. 1). Following intoxication, temperature readings were taken hourly and increased, on average, by 1 °C from 6 h post challenge in all ricin exposed rats (Fig. 1B). Comparison of PBS and corresponding ricin-exposed animals from the validation study shows that this change is statistically significant at the 6 and 8 h time points, (p < 0.001 and p < 0.01 respectively). From ~ 30 h after intoxication, temperatures became quite variable and dropped to below normal ranges in some animals (Fig. 1B). This is also consistent with the appearance of behavioral signs of intoxication which started as minor observed signs in the majority of animals between 20 and 30 h after intoxication (Fig. 1C). Clinical signs of intoxication in the remaining animals gradually increased in severity and animal numbers drop during this time course as the rats reached the pre-determined humane endpoint of severe intoxication showing that the ricin dose of 8 µg·kg−1 causes a lethal intoxication in Sprague–Dawley rats (Fig. 1D).
Fig. 1.
Effect of ricin exposure on bodyweight, temperature, signs of intoxication and survival. Data from up to 31 ricin-exposed rats are shown as mean ± SD (panel A and B) and median for Panel C (red data points) and for individual rats (grey data points in all panels) for bodyweight (panel A), body temperature (panel B) and clinical score (panel C). Data for PBS-exposed rats are shown as blue data points as mean ± SD and median only. Bodyweight data are normalised to t = 0, the time-point at which the ricin (8 µg·kg−1 i.v.) challenge was administered. Clinical scores data (panel C) are calculated from those rats alive at each time point. On reaching the humane endpoint (score = 8) or the end of the study (see panel D) rats are excluded from this analysis. Survival data for all animals in the study are shown in panel D, planned euthanasia at the experiment end-point is indicated and these animals were censored from the analysis. *Statistically significant difference between PBS and corresponding ricin-exposed animals in the validation study, p < 0.001 and p < 0.01 at 6 and 8 h respectively, 2-way ANOVA and Sidak’s multiple comparisons test
Ricin toxicokinetics
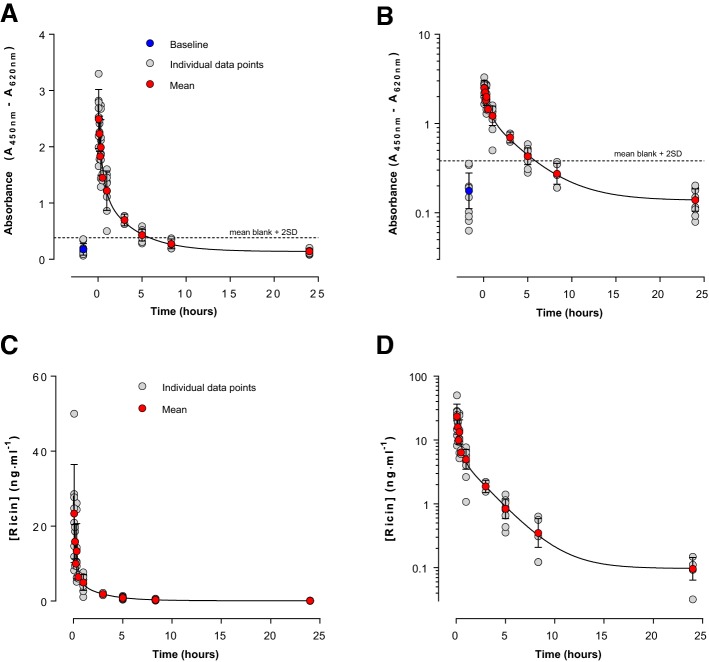
Using standard ELISA techniques the concentration of ricin in blood following i.v. ricin exposure was determined. Data demonstrated that free ricin was rapidly removed from the blood in a biphasic manner (Fig. 2 and Table 1). Ricin levels in the blood were below the limit of detection of this ELISA (n = 10 rats) at ~ 5 h post exposure (Fig. 2). This is prior to the appearance of behavioral signs of intoxication (Fig. 1), which started to become apparent in the majority of rats administered this dose of ricin between about 20 h and 30 h after exposure in this study albeit at a minor level of clinical score initially (Fig. 1C). A lack of observable behavioral signs of intoxication at this 5 h point means there is a window of opportunity for metabolic biomarkers to provide an additional route to identifying whether individuals have been exposed to ricin, to complement direct toxin detection methods.
Fig. 2.
Ricin toxicokinetics following administration of 8 µg·kg−1 ricin i.v. Data are shown as mean ± SD (red or blue points) from up to 10 individual animals (grey points) at each time point (the vascular access buttons of 2 animals were not patent and no samples could be collected from these animals). In the upper panels, A and B, a limit of detection could be estimated from the blank absorbance plus 2 times its SD (horizontal line). Whilst this is indicative of a detection limit it is based on relatively few determinations and for quantitative analysis of the ricin toxicokinetics all ricin concentrations that could be interpolated from the standard curve were analysed (lower panels C, D). Curve fits shown are for 2 phase exponential decays, which in all cases provided statistically better fits to the data than a one phase model
Table 1.
Kinetic parameters describing disappearance of ricin from blood following in vivo administration of 8 µg kg−1 i.v. in rats
| Assay | C0 (ng ml−1) | t½ fast (h) | t½ slow (h) | % fast |
|---|---|---|---|---|
| ELISA | 28.1 (15.5–40.7) | 0.20 (0.11–1.25) | 1.78 (1.31–2.77) | 79.6 (66.1–93.0) |
Values and 95% confidence intervals are shown in parentheses
Metabolic response to ricin exposure
An untargeted metabolomics approach applying UHPLC-MS to measure water-soluble and lipid metabolites was performed to investigate the dynamic response to ricin exposure for greater than 1000 metabolites in rat plasma. Two independent studies were performed to enable the discovery and subsequent validation of metabolic changes to increase the reliability of the results reported. 163 and 363 metabolites were statistically significant in the discovery and validation studies, respectively. Statistical significance between different timepoints is reported for ricin-treated rats in the discovery study and statistical significance between control and ricin-treated rats is reported for the validation study. Following comparison of the results for these two studies, 37 metabolites were identified as statistically significant in both studies (Table 2).
Table 2.
Annotated metabolites showing statistical significance in both the discovery and validation studies
| Metabolites | p value (discovery) | p value (validation) | Metabolite class |
|---|---|---|---|
| Ornithine | 1.60E−03 | 7.23E−06 | Arginine and proline metabolism |
| Arginine | 3.35E−05 | 1.10E−14 | Arginine and proline metabolism |
| β-Guanidinopropionic acid and/or Creatine | 1.80E−03 | 8.71E−06 | Arginine and proline metabolism |
| 3-Indolepropionic acid and/or Indole-3-methyl acetate | 3.00E−03 | 9.04E−17 | Aromatic metabolism |
| Varanic acid | 1.10E−03 | 4.86E−09 | Bile acid metabolism |
| Taurallocholic acid and/or Tauro-b-muricholic acid and/or Taurocholic acid and/or Taurohyocholate and/or Taurohyocholic acid and/or Tauroursocholic acid | 1.69E−05 | 4.25E−11 | Bile acid metabolism |
| 3a,7b,12a-Trihydroxyoxocholanyl-Glycine and/or Glycocholic acid | 3.90E−03 | 1.82E−10 | Bile acid metabolism |
| 1,3,12-Trihydroxycholan-24-oic acid and/or 1b,3a,12a-Trihydroxy-5b-cholanoic acid or isomer | 2.10E−03 | 7.74E−10 | Bile acid metabolism |
| Sulfolithocholylglycine | 4.00E−05 | 3.55E−12 | Bile acid metabolism |
| Taurochenodesoxycholic acid and/or Taurodeoxycholic acid and/or Tauroursodeoxycholic acid | 4.50E−05 | 6.10E−12 | Bile acid metabolism |
| 3a,7b,12a-Trihydroxyoxocholanyl-Glycine and/or Glycocholic acid | 3.90E−03 | 4.44E−09 | Bile acid metabolism |
| GalCer(d18:0/22:0) and/or GlcCer(d18:0/22:0) | 1.60E−03 | 4.54E−15 | Ceramides |
| GalCer(d18:1/24:1) and/or GlcCer(d18:1/24:1) | 1.40E−03 | 3.00E−16 | Ceramides |
| GalCer(d18:1/16:0) and/or GlcCer(d14:1/20:0) and/or GlcCer(d16:1/18:0) and/or GlcCer(d18:1/16:0) | 1.00E−05 | 3.00E−16 | Ceramides |
| PC[38:2] | 4.60E−03 | 3.28E−06 | Glycerophospholipid |
| PC[34:2] and/or PE-NMe[36:2] | 8.00E−05 | 2.13E−13 | Glycerophospholipid |
| PC[34:1] and/or PE-NMe2(O-16:0/O-16:0) | 2.85E−05 | 2.70E−11 | Glycerophospholipid |
| PC(16:0/5:0(CHO)) and/or LysoPC[20:2] and/or PC(O-18:2/2:0) | 1.90E−03 | 6.52E−07 | Glycerophospholipid |
| PA[30:4] and/or PA[28:1] | 1.90E−03 | 4.25E−05 | Glycerophospholipid |
| LysoPE[22:1] | 2.10E−03 | 4.48E−06 | Lysoglycerophospholipid |
| LysoPC[22:1] | 1.80E−03 | 1.86E−05 | Lysoglycerophospholipid |
| LysoPC[20:2] and/or PC(O-18:2/2:0) | 3.60E−03 | 3.17E−06 | Lysoglycerophospholipid |
| LysoPC[24:1] | 3.00E−05 | 1.80E−03 | Lysoglycerophospholipid |
| Nicotinamide N-oxide and/or Urocanic acid | 6.98E−06 | 7.70E−15 | Mixed classes |
| 3-Pyridinebutanoic acid and/or Norsalsolinol | 7.27E−05 | 9.15E−15 | Mixed classes |
| Beta-Tyrosine and/or Threo-3-Phenylserine and/or Tyrosine | 1.60E−03 | 9.18E−06 | Mixed classes |
| 1-(2-Furanylmethyl)-1H-pyrrole | 1.40E−03 | 9.15E−15 | Other class |
| Isoleucyl-Valine and/or Leucyl-Valine | 1.30E−03 | 1.31E−08 | Peptide |
| SM(d18:2/24:1) and/or SM(d18:0/22:3) and/or SM(d16:1/22:0) and/or SM(d18:1/20:0) and/or | 1.40E−03 | 1.28E−06 | Sphingolipids |
| Cholesterol sulfate | 6.00E−05 | 6.92E−08 | Sterol and steroid (including Vitamin D) metabolism |
| 3-Deoxyvitamin D3 | 1.10E−03 | 3.17E−07 | Sterol and steroid (including Vitamin D) metabolism |
| TG[60:9] | 1.50E−03 | 1.20E−05 | Triacylglyceride |
| TG[60:10] | 8.00E−05 | 5.00E−05 | Triacylglyceride |
| TG[54:6] and/or TG[53:6] and/or TG[51:3] and/orTG[49:0] | 8.10E−05 | 3.90E−03 | Triacylglyceride |
| TG[53:8] and/or TG[51:5] and/or TG[50:2] and/or TG[47:1] | 2.0E−03 | 5.50E−05 | Triacylglyceride |
| TG[58:4] | 3.16E−05 | 4.80E−03 | Triacylglyceride |
| TG[49:3] | 1.80E−03 | 2.15E−05 | Triacylglyceride |
The p value (following correction for multiple testing) and designated metabolite class/metabolic pathway are included to demonstrate enhancement of specific metabolite class/metabolic pathway
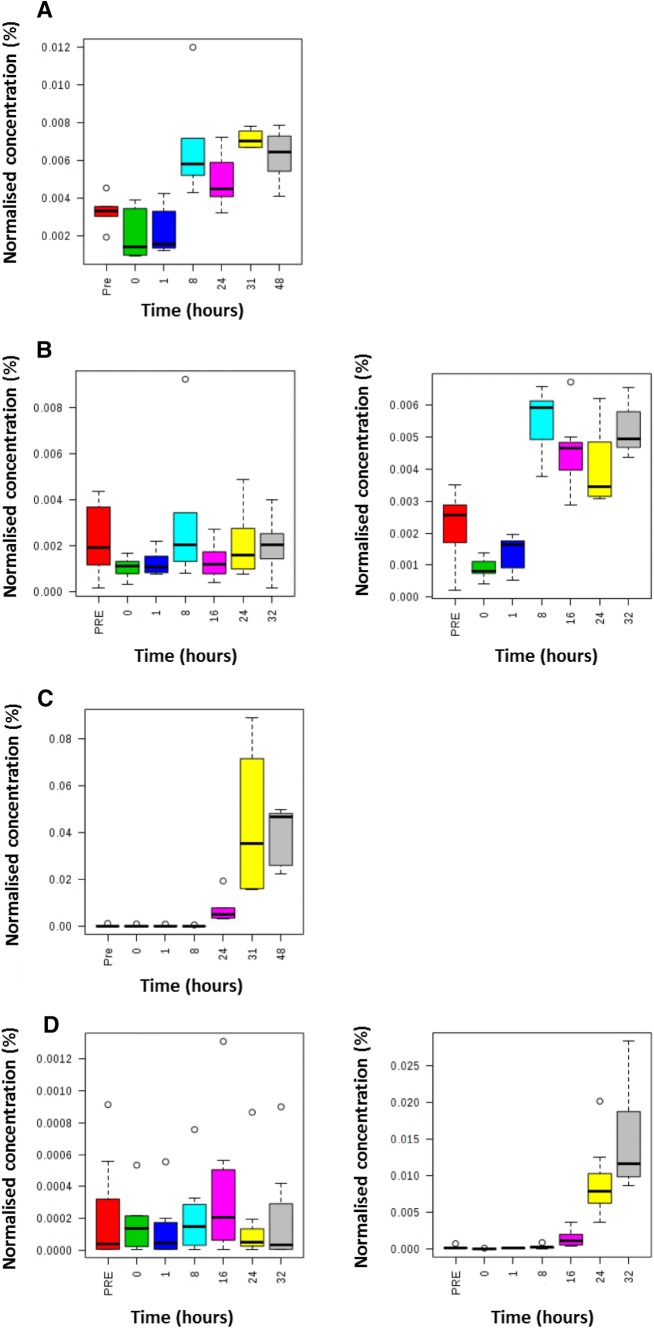
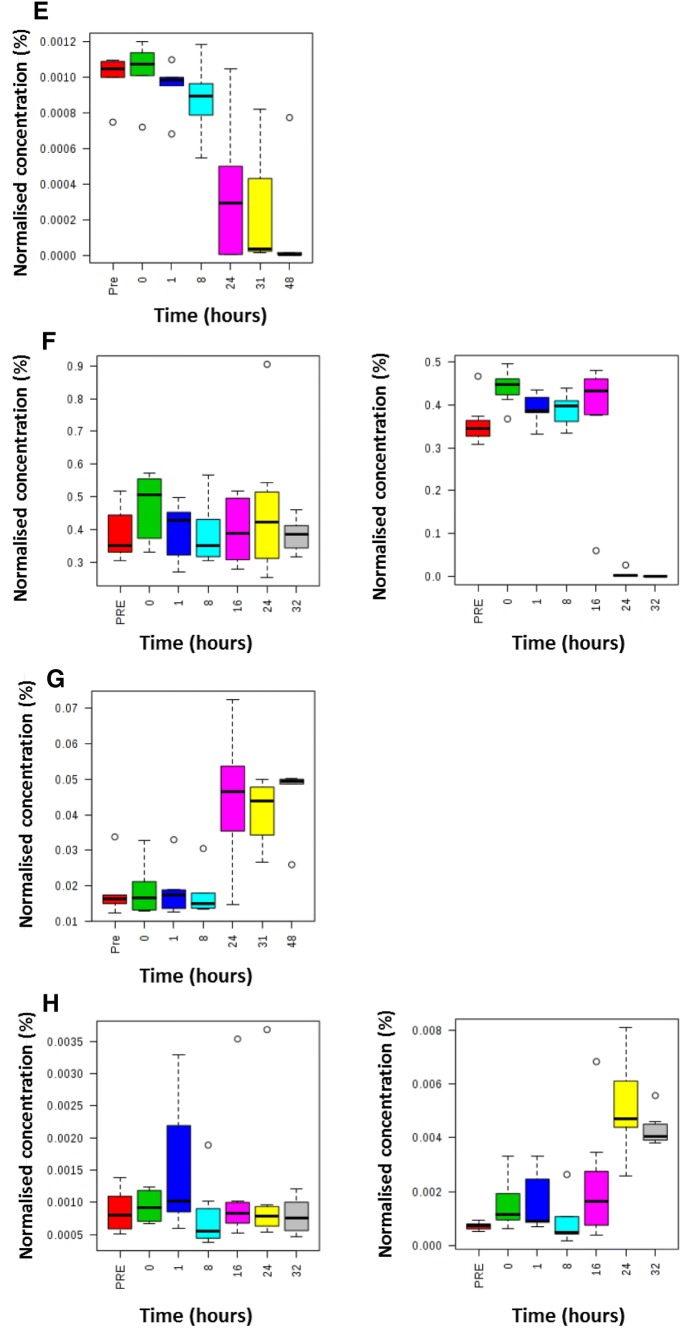
Metabolites were grouped into metabolic pathways/metabolite classes. It is important to note that multiple metabolites in each pathway/class were calculated to be statistically significant but not all metabolites in each pathway/class. Therefore these results indicate which metabolic pathways/metabolite classes warrant further investigation applying targeted analytical assays which measure all metabolites in these metabolic pathways/metabolite classes. Arginine and proline metabolism, bile acid metabolism, glycerophosphospholipid and triacylglyceride families were perturbed in both studies. The time-related changes in metabolite concentrations were investigated for these four metabolic pathways/metabolite classes. Triacylglycerides demonstrated the quickest response to exposure with increases or decreases in their concentrations from 8 h and later (Fig. 3A). Bile acids demonstrated large increases in their concentrations from 16 h and later (Fig. 3B). Arginine and ornithine demonstrated concentration changes from 24 h with arginine showing a decrease (Fig. 3C) and ornithine showing an increase in concentration (Fig. 3D).
Fig. 3.
Box and whisker plots defining the time-related concentration for selected metabolites following administration of ricin i.v. Metabolites discovered as predictive of ricin exposure to rats using an untargeted metabolomics approach applying UHPLC-MS on blood samples. Samples were analysed at − 16 h (pre-exposure baseline control), 0 h (time of exposure), 1 h, 8 h, 24 h, 32 h and 48 h post exposure for the discovery study. Samples were analysed at − 16 h (pre-exposure baseline control), 0 h (time of exposure), 1 h, 8 h, 24 h and 32 h post exposure for the validation study. The seven box and whisker plots on the left are related to control animals (PBS exposed) and the seven box and whisker plots on the right are related to ricin exposed animals. Metabolites comprise TG[60:10] (panel A = discovery and panel B = validation), sulfolithocholylglycine (panel C = discovery and panel D = validation), arginine (panel E = discovery and panel F = validation) and ornithine (panel G = discovery and panel H = validation)
Multivariate AUROC analysis was performed for the validation study only because no control animals were included in the discovery study. Metabolites were assessed individually and in all possible combinations to identify the most powerful AUROC. An AUROC greater than 0.94 was observed at 8 h, 16 h and 24 h (Table 3). Caution should be taken relating to translation into human ricin intoxication because this is a well-controlled animal model study and human studies provide greater natural biological variability and potential confounders.
Table 3.
Area under the receiver operator curve for putative biomarker panels applying multiple metabolites in each biomarker panel at 8 h, 16 h and 24 h
| Time point (h) | AUROC | Metabolites included in biomarker panel |
|---|---|---|
| 8 | 0.94 | TG(54:6) and TG(60:10) |
| 16 | 0.98 | TG(54:6) and TG(60:10) and TG(60:9) and TG(58:4) and TG(53:8) |
| 24 | 1.00 | Ornithine and arginine and urocanic acid and/or isomer and cholesterol sulphate and sulfolithocholylglycine |
Discussion
Ricin toxin is a potential biothreat agent due to its ease of acquisition (castor bean plant) and lack of medical countermeasures. Early detection and diagnosis of ricin intoxication is essential in reducing morbidity and mortality associated with the disease. This is made more difficult when ricin toxin rapidly binds to cells and is quickly taken up into cells and trafficked to the Golgi apparatus (Jenner et al. 2018) meaning levels of detectable ricin in the blood are likely to be low.
In this study we confirm that when rats were exposed to ricin by the i.v. route standard ELISA techniques can only detect ricin in the blood at early time points post exposure (up to ~ 5 h post-exposure), the free toxin is quickly removed from the blood prior to overt signs of intoxication. Nonetheless, and for the first time, we have demonstrated that metabolic changes are detectable at 8 h post exposure. This is important because this is at a time when ricin is not detectable by ELISA in this model, but is prior to signs of intoxication which become apparent at ~ 24 h, thus it could provide a trigger for administration of treatment within a time frame that would ensure efficacy of treatment. Specific metabolic pathways/metabolite classes were perturbed and a putative ricin toxin diagnostic biomarker panel has been identified. A total of 37 metabolites were statistically significantly changed in both the discovery and validation studies with 32 of these metabolites demonstrating the same dynamic time profiles in both studies. The significantly altered metabolites were heavily associated with arginine and proline metabolism, bile acid metabolism and the triacylglyceride metabolite class.
Arginine and proline are amino acids and are therefore essential for a number of key functions within the body such as detoxification, embryonic survival and organ homeostasis (Albaugh et al. 2017; Wu et al. 2009). The degradation of arginine produces proline, but also ornithine and creatine, both of these latter metabolites were also identified as significantly altered metabolites following ricin exposure. However due to the key nature of these amino acids, changes in the arginine and proline metabolic pathway and associated metabolites are seen in variety of diseases and injuries including sickle cell disease (Schnog et al. 2004), chronic kidney disease (Baylis 2006) and cancers (Ma et al. 1999). A total of seven metabolites that were significantly altered following ricin exposure were associated with bile acid metabolism. Bile acids are produced in the liver, are released as glycine or taurine conjugates and are essential for the emulsification of lipids in the intestines, breakdown of cholesterol and nutrient absorption. Bile acids also have the ability to signal via interaction with receptors (Makishima et al. 1999) and control inflammation (Guo et al. 2016). Defects in bile acid metabolism, not surprisingly, lead to a variety of liver associated diseases, but also obesity and diabetes (Chavez-Talavera et al. 2017). The observed increase in bile acids could be indicative of toxicity effects on the liver or indicative of gut cell membrane changes leading to their increased permeability and further studies are required to investigate this. Triacylglycerides are the main constituents of fat deposits in adipocytes and other organs. They can be broken down by lipases and bile into smaller fatty acids to aid absorption. High levels of triacylglycerides are associated with metabolic disease, heart disease and stroke (Tao et al. 2016). As all of the metabolic pathways altered can be prognostic for human diseases, the diagnostic potential for these metabolites are therefore limited. However, the rapid changes over hours provides an opportunity to collect and analyse multiple samples over 8 h and measure the change in concentration which will be quicker than with the chronic diseases listed above. When combined with known health status and risk of ricin exposure, greater successes of ricin intoxication diagnosis can be obtained.
The fact that arginine, bile acid and triacylglyceride metabolism are altered following exposure to ricin is not that surprising, especially when you consider the pathogenesis of the intoxication, where in rodent models there is severe weight loss and organ dysfunction following the apoptosis of cells. However, what is interesting is that the changes in these pathways and the corresponding metabolites are time specific and do not always follow the same pattern. The triacylglyceride metabolites are shown to be important at 8 h post exposure with two metabolites increasing and two metabolites decreasing. This is prior to any visually scored signs of intoxication being observed and was prior to weight loss in these animals. However, animal temperatures increased within a few hours of exposure to ricin and therefore increases in lipolysis to release fatty acids for fatty acid beta-oxidation and ATP generation is the most probable mechanism connecting temperature increase and triacylglyceride changes. The bile acid metabolites, along with ceramides, show significant concentration changes at 16 h post ricin exposure, but all increase compared to controls. Arginine and proline metabolism is only perturbed at 24 h post exposure, with ornithine levels increasing and arginine levels decreasing. However at this point the rats were showing signs associated to ricin intoxication.
Despite the disturbances being in common metabolite pathways it is the association of particular metabolic pathways with specific time points that has the potential to generate biomarkers that could be used in a pre-symptomatic or post-symptomatic setting. It is worth noting that the AUROC increases with time, but even at 8 h the combination of TG(54:6) and TG(60:10) gives a AUROC value of 0.94. When 5 metabolites are combined (ornithine, arginine, urocanoic acid/isomer, cholesterol sulfate and sulfolithocholylglycine) at 24 h post exposure the AUROC value is 1.0. This would indicate that it is possible to diagnose ricin exposure via the i.v route with 100% confidence compared to unchallenged controls from 8 h onwards. However, significant toxic effects can occur early following exposure and a biomarker which is applicable from 1 h following exposure would be ideal to allow efficient management and treatment. It these scenarios a combination approach of using ELISA and metabolomics maybe applicable.
We are aware that there are some limitations to this study. I.V is not the most likely route of ricin exposure in the human population, but was chosen to ensure ricin was definitely in the system and would cause intoxication. Other routes including inhalation and ingestion are considered to be more likely, especially in a potential bioterrorism scenario (Schep et al. 2009). However for these proof-of-principle studies it was important to reduce variability in the animal model therefore the I.V route was selected. The relevance of all animal model data and human diseases is regularly discussed and can never be completely transferable. However, the metabolites identified are detected in human blood samples and due to the large fold changes seen in this rat study, there is a high likelihood that these changes would be seen in humans also. We are, in the case of ricin and other biological threat agents, very reliant on animal data, as conducting clinical trials is not possible and there tends to be few natural cases.
Additional studies must be conducted to further support these exciting findings, such as investigating different routes of exposure, lower sub-lethal challenge doses, and comparing the data to related toxins. It will only then be possible to state that a highly-specific diagnostic biomarker for ricin has been discovered. Nonetheless the data presented here clearly demonstrates that metabolomics is a useful tool to identify putative biomarkers of exposure and to reveal new insights into the biochemical changes during the rodent response to ricin intoxication.
Author Contributions
RVD, SAG, FMS, CD, VP, ACG, RAL and WBD conceived of and designed the study. CLW, ADS, RJMW, FMS, CD, VP, WBD performed the research. RVD, RJMW, MRV, ACG, RAL, WBD analysed the data. RVD, SAG, ACG, CLW, ADS, RJMW, WBD wrote the paper.
Compliance with ethical standards
Conflict of interest
This work was funded by the UK Ministry of Defence. The authors would like to thank the Medical Research Council for funding the construction of Phenome Centre Birmingham (MR/M009157/1). All authors declare that they have no conflict of interest.
Ethical approval
All animal studies were conducted under an approved Home Office licence. DSTL hold regular Animal Welfare and Ethical Review Body (AWERB) meetings for all organizational HO licences. All individual studies conducted for the generation of the data presented in this manuscript underwent further internal and independent ethical review before commencement.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Riccardo V. D’Elia, Email: RVDELIA@mail.dstl.gov.uk
Warwick B. Dunn, Phone: +44 (0)121 4145458, Email: w.dunn@bham.ac.uk
References
- Albaugh VL, Stewart MK, Barbul A. Chapter 27—cellular and physiological effects of arginine in seniors. In: Watson RR, editor. Nutrition and functional foods for healthy aging. Cambridge: Academic Press; 2017. pp. 317–336. [Google Scholar]
- Baylis C. Arginine, arginine analogs and nitric oxide production in chronic kidney disease. Nature Clinical Practice Nephrology. 2006;2:209–220. doi: 10.1038/ncpneph0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, et al. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics. 2011;27:1108–1112. doi: 10.1093/bioinformatics/btr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Talavera O, Tailleux A, Lefebvre P, Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152:1679–1694.e3. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- Chen HY, Tran H, Foo LY, Sew TW, Loke WK. Development and validation of an ELISA kit for the detection of ricin toxins from biological specimens and environmental samples. Analytical and Bioanalytical Chemistry. 2014;406:5157–5169. doi: 10.1007/s00216-014-7934-1. [DOI] [PubMed] [Google Scholar]
- Crompton R, Gall D. Georgi Markov–death in a pellet. Medico-Legal Journal. 1980;48:51–62. doi: 10.1177/002581728004800203. [DOI] [PubMed] [Google Scholar]
- D’Elia RV, Harrison K, Oyston PC, Lukaszewski RA, Clark GC. Targeting the “cytokine storm” for therapeutic benefit. Clinical and Vaccine Immunology. 2013;20:319–327. doi: 10.1128/cvi.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertzbaugh MT, Rossi CA, Paddle BM, Hale M, Poretski M, Alderton MR. Monoclonal antibodies to ricin: in vitro inhibition of toxicity and utility as diagnostic reagents. Hybridoma (Larchmt) 2005;24:236–243. doi: 10.1089/hyb.2005.24.236. [DOI] [PubMed] [Google Scholar]
- Diakite ML, et al. Point-of-care diagnostics for ricin exposure. Lab on a Chip. 2015;15:2308–2317. doi: 10.1039/c5lc00178a. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society Reviews. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- Franz DR, Jaax NK. Ricin toxin. In: Sidell FR, Takafuji ET, Franz DR, editors. Textbook of Military Medicine. Washington DC: Borden Institute, Walter Reed Army Military Centre; 1997. [Google Scholar]
- Gal Y, Mazor O, Falach R, Sapoznikov A, Kronman C, Sabo T. Treatments for pulmonary ricin intoxication: current aspects and future prospects. Toxins (Basel) 2017;9:311. doi: 10.3390/toxins9100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GD. Understanding ricin from a defensive viewpoint. Toxins. 2011;3:1373–1392. doi: 10.3390/toxins3111373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths GD, Phillips GJ, Holley J. Inhalation toxicology of ricin preparations: animal models, prophylactic and therapeutic approaches to protection. Inhalation Toxicology. 2007;19:873–887. doi: 10.1080/08958370701432124. [DOI] [PubMed] [Google Scholar]
- Guo Z, et al. Proteomic study of differential protein expression in mouse lung tissues after aerosolized ricin poisoning. International Journal of Molecular Sciences. 2014;15:7281–7292. doi: 10.3390/ijms15057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:944. doi: 10.1016/j.immuni.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Hamm J, et al. Alternative approaches for identifying acute systemic toxicity: Moving from research to regulatory testing. Toxicology in Vitro: An International Journal Published in Association with BIBRA. 2017;41:245–259. doi: 10.1016/j.tiv.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner D, Chong D, Walker N, Green AC. An imaging flow cytometry method to assess ricin trafficking in A549 human lung epithelial cells. Methods. 2018;134–135:41–49. doi: 10.1016/j.ymeth.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AS, Morse S, Lillibridge S. Public-health preparedness for biological terrorism in the USA. Lancet. 2000;356:1179–1182. doi: 10.1016/s0140-6736(00)02769-0. [DOI] [PubMed] [Google Scholar]
- Kim JS, Anderson GP, Erickson JS, Golden JP, Nasir M, Ligler FS. Multiplexed detection of bacteria and toxins using a microflow cytometer. Analytical Chemistry. 2009;81:5426–5432. doi: 10.1021/ac9005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuca K, Pohanka M. Chemical warfare agents. EXS. 2010;100:543–558. [PubMed] [Google Scholar]
- Leith AG, Griffiths GD, Green MA. Quantification of ricin toxin using a highly sensitive avidin/biotin enzyme-linked immunosorbent assay. Journal of the Forensic Science Society. 1988;28:227–236. doi: 10.1016/S0015-7368(88)72840-6. [DOI] [PubMed] [Google Scholar]
- Lord JM, Roberts LM, Robertus JD. Ricin—structure, mode of action, and some current applications. FASEB Journal. 1994;8:201–208. doi: 10.1096/fasebj.8.2.8119491. [DOI] [PubMed] [Google Scholar]
- Lukaszewski RA, et al. Presymptomatic prediction of sepsis in intensive care unit patients. Clinical and Vaccine Immunology. 2008;15:1089–1094. doi: 10.1128/cvi.00486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Williamson KE, O’Rourke D, Rowlands BJ. The effects of l-arginine on crypt cell hyperproliferation in colorectal cancer. Journal of Surgical Research. 1999;81:181–188. doi: 10.1006/jsre.1998.5512. [DOI] [PubMed] [Google Scholar]
- Makishima M, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- May KL, Yan Q, Tumer NE. Targeting ricin to the ribosome. Toxicon: Official Journal of the International Society on Toxinology. 2013;69:143–151. doi: 10.1016/j.toxicon.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men J, et al. Detection of residual toxin in tissues of ricin-poisoned mice by sandwich enzyme-linked immunosorbent assay and immunoprecipitation. Analytical Biochemistry. 2010;401:211–216. doi: 10.1016/j.ab.2010.02.033. [DOI] [PubMed] [Google Scholar]
- Morton DB, et al. Removal of blood from laboratory mammals and birds. Laboratory Animals. 1993;27:1–22. doi: 10.1258/002367793780745633. [DOI] [PubMed] [Google Scholar]
- Moshiri M, Hamid F, Etemad L. Ricin toxicity: Clinical and molecular aspects. Reports of Biochemistry & Molecular Biology. 2016;4:60–65. [PMC free article] [PubMed] [Google Scholar]
- Olsnes S. The history of ricin, abrin and related toxins. Toxicon. 2004;44:361–370. doi: 10.1016/j.toxicon.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Patel VR, Dumancas GG, Kasi Viswanath LC, Maples R, Subong BJJ. Castor oil: Properties, uses, and optimization of processing parameters in commercial production. Lipid Insights. 2016;9:1–12. doi: 10.4137/LPI.S40233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patocka J, Streda L. Protein biotoxins of military significance. Acta Medica (Hradec Kralove) 2006;49:3–11. doi: 10.14712/18059694.2017.102. [DOI] [PubMed] [Google Scholar]
- Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli MA, Rivera VR, Hewetson JF, Merrill GA. Detection of ricin by colorimetric and chemiluminescence ELISA. Toxicon. 1994;32:1371–1377. doi: 10.1016/0041-0101(94)90409-X. [DOI] [PubMed] [Google Scholar]
- Pratt TS, Pincus SH, Hale ML, Moreira AL, Roy CJ, Tchou-Wong KM. Oropharyngeal aspiration of ricin as a lung challenge model for evaluation of the therapeutic index of antibodies against ricin A-chain for post-exposure treatment. Experimental Lung Research. 2007;33:459–481. doi: 10.1080/01902140701731805. [DOI] [PubMed] [Google Scholar]
- Prince H. John Eveln’s Elysium Britannicum and European gardening. Ecumene. 2000;7:482–483. doi: 10.1177/096746080000700410. [DOI] [Google Scholar]
- Ristanovic E. Bioterrorism—risk and threat: The misuse of science. In: Dishovsky C, Pivovarov A, editors. Counteraction to chemical and biological terrorism in east European countries. NATO science for peace and security series A—chemistry and biology. Dordrecht: Springer; 2009. pp. 121–125. [Google Scholar]
- Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerging Infectious Diseases. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CJ, Song K, Sivasubramani SK, Gardner DJ, Pincus SH. Animal models of ricin toxicosis. Current Topics in Microbiology and Immunology. 2012;357:243–257. doi: 10.1007/82_2011_173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep LJ, Temple WA, Butt GA, Beasley MD. Ricin as a weapon of mass terror—separating fact from fiction. Environment International. 2009;35:1267–1271. doi: 10.1016/j.envint.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Schier JG, et al. Public health investigation after the discovery of ricin in a South Carolina postal facility. American Journal of Public Health. 2007;97(Suppl 1):S152–S157. doi: 10.2105/ajph.2006.099903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnog JJ, et al. Evidence for a metabolic shift of arginine metabolism in sickle cell disease. Annals of Hematology. 2004;83:371–375. doi: 10.1007/s00277-004-0856-9. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LX, et al. Longitudinal associations between triglycerides and metabolic syndrome components in a Beijing adult population, 2007–2012. International Journal of Medical Sciences. 2016;13:445–450. doi: 10.7150/ijms.14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slyke G, et al. Humanized monoclonal antibody that passively protects mice against systemic and intranasal ricin toxin challenge. Clinical and Vaccine Immunology. 2016;23:795–799. doi: 10.1128/cvi.00088-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield SJC, et al. Production, characterisation and testing of an ovine antitoxin against ricin; efficacy, potency and mechanisms of action. Toxins (Basel) 2017;9:329. doi: 10.3390/toxins9100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs S, et al. Ricinus communis intoxications in human and veterinary medicine—a summary of real cases. Toxins. 2011;3:1332–1372. doi: 10.3390/toxins3101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, et al. Arginine metabolism and nutrition in growth, health and disease. Amino Acids. 2009;37:153–168. doi: 10.1007/s00726-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]