Abstract
Background
Gram-negative bacteria actively secrete outer membrane vesicles into the surrounding environment and these vesicles have been shown to play various physiological and protective roles such as carrying antibiotic-degrading enzymes and acting as decoys against host defences, therefore promoting the pathogenesis of the bacterium. It has been shown that avian pathogenic Escherichia coli species can increase vesicle biosynthesis through the acquisition of the hlyF gene but the effect this has on the cell by scavenging outer-membrane associated proteins (OmpA, OmpF) into the vesicles during vesicle release have not yet been investigated.
Results
Relative quantitative real-time PCR data obtained from hlyF expressing and non-expressing cells showed that during hlyF induction, ompF showed a nearly 2-fold down regulation relative to the non-expressing cells during the entire 24 hours, while ompA was expressed at the same level as the non-expressing cells during the first 8 hours of expression. At 24 hours post-hlyF expression, ompA was up-regulated 4-fold.
Conclusions
The regulatory effects of the newly described outer-membrane vesicle biosynthesis-related gene, hlyF, on E. coli has not previously been investigated. As hlyF-induced vesicles contain OmpA and OmpF scavenged from the bacterial outer-membrane, potential regulatory effects on the host was investigated. An increase in ompA expression and an insignificant decrease in ompF expression was observed during hlyF induction demonstrating that hlyF-related biosynthesis is not related to decreased ompA expression, which is one of the potential mechanisms discussed in literature for biosynthesis. Outer-membrane vesicle biosynthesis during hlyF over-expression could potentially be accomplished through a different mechanism(s).
Keywords: Microbiology, Escherichia coli, Gene expression, Gene regulation, Polymerase chain reaction, Proteins, hlyF, OMV, Biogenesis, Gene regulation, ompA, ompF, APEC, Haemolysin F
1. Background
Gram-negative bacteria actively secrete outer membrane vesicles (OMVs), which are nanosized lipid vesicles with periplasmic proteins and surface associated membrane proteins [1, 2]. The production of OMVs have been shown to play various physiological and protective roles and are secreted by bacteria throughout their lifecycles [3, 4]. Secreted OMVs accomplish these roles through interacting with the environment of the bacterium during survival situations such as the enzymatic neutralization of human serum [5], acting as decoys to be targeted by antibiotics and carrying antibiotic-degrading enzymes [6] and through the formation of biofilms [7], therefore promoting the pathogenesis of the bacterium.
It has been shown that some avian pathogenic Escherichia coli (APEC) species can increase OMV biosynthesis through the acquisition of the hlyF gene [8]. Hemolysin F (hlyF) as the name suggests, was alleged to be an avian haemolysin often associated with avian pathogenic Escherichia coli [9]. The erroneous naming of this haemolysin was due to the haemolytic activity gained by E. coli MG1655 cells after the introduction and subsequent over-expression of hlyF. This was later shown by Murase et al. [8] to have been due to the transport of a native haemolysin, cytolysin A which is natively produced by this strain, within E. coli MG1655 to the extracellular environment by the hlyF-induced OMVs. Literature indicates that hlyF is an important virulence factor and of pathological importance in screening of potential APEC isolates [10, 11].
Subsets of genes in various Gram-negative bacteria have been found to be associated with OMV biosynthesis and their functions on the cells have been investigated [12, 13, 14]. However, the association of hlyF over-expression with OMV biogenesis has newly been described and therefore the effects on the expressing cell is partial [8]. During OMV biosynthesis, outer-membrane proteins are incorporated into the OMVs, where proteins are scavenged from the cell-membrane, potentially leading to the loss of functions attributed by these cell wall proteins, unless there are regulatory changes involved in the expression levels of the genes to counteract the scavenging of these cell wall/membrane proteins. This study thus investigates the gene-regulation of two abundant outer-membrane proteins, namely outer-membrane proteins A and F, encoded by ompA and ompF associated with hlyF-induced OMVs. Outer-membrane protein A has been shown to be involved in stabilizing the structure of the peptidoglycan and the outer-membrane of E. coli [15], which can potentially be influenced by cell-membrane destabilization during OMV formation and release. The gene of Outer-membrane protein F has been shown to be regulated by the ompR gene product during osmotic pressures applied to E. coli cells [16], so during OMV production, speculatively, it is unlikely to be influenced by hlyF expression unless the formation of OMV influences the osmolarity of the cell, by some other unknown mechanism.
2. Methods
2.1. Production of Haemolysin F pET22b(+) constructs in E. coli BL21 (DE3) cells
The hlyF gene was amplified from the APEC strain 1080, which had previously tested positive for the presence of hlyF [17]. The translated sequence of the hlyF gene of this strain shares 99,7 % amino acid homology to the reference sequence in the NCBI database (Accession number NC_011964.1). The hlyF gene was amplified using the primers hlyF-pet22-FP 5′-GGGAATTCATGGATCCTCGTCTTGATG-3′ and hlyF-pet22-RP 5′-GGCTCGAGTTATTTAAAATCAACTTCCATTTGTTG-3’ (Integrated DNA Technologies). The forward and reverse primers were designed with incorporated EcoRI and XhoI restriction recognition sites (underlined) respectively, for cloning into pET22b(+) (Novagen).
Standard molecular biology techniques were carried out as described by literature [18], and enzymes were applied according to the manufacturers' specifications. Amplicons from PCR were sub-cloned into the pGEM®-T Easy Vector System (Promega), using E. coli 10-β cells (New England Biolabs). The sequences of the cloned inserts were determined using Sanger sequencing with the SP6-promotor and T7-terminator primers (Integrated DNA Technologies). Consensus sequences were subjected to BLAST searches against the NCBI sequence database [19] for the confirmation of the sequences.
The hlyF gene was transferred from the pGEM®-T Easy vector into the pET22b(+) vector using the EcoRI and XhoI restriction endonucleases (Thermo Fisher Scientific). The digested DNA fragments were purified from 2 % LM-sieve agarose gels (Lonza) using the Wizard® SV Gel and PCR Clean-Up System (Promega). The digested gene inserts were ligated into pET22b(+), and the recombinant plasmids were used to transform E. coli BL21 (DE3) (New England Biolabs).
The plasmids were extracted using the Qiagen QIAprep Spin Miniprep kit (Whitehead Scientific) after a 24-hour incubation period in Luria-Bertani (LB) broth (10 g/L tryptone, 10 g/L NaCl, 5 g/L yeast extract) at 37 °C supplemented with 100 ug/mL ampicillin. The presence of the inserted hlyF region was confirmed by PCR, and the sequences were obtained using Sanger sequencing with the T7-promotor and T7-terminator primers (Integrated DNA Technologies), which amplifies the cloning site of pET22b(+). As a control, an empty pET22b(+) was cloned into E. coli BL21 (DE3) as described above.
2.2. Transmission electron microscopy (TEM) analysis of OMV production
The transformed cells described above were inoculated into two LB media tubes containing 100 ug/mL ampicillin. One of the tubes received IPTG to a final concentration of 1 mM to induce hlyF expression while another tube served as control. Both tubes were incubated at 37 °C for 16 hours. Cell pellets were harvested through centrifugation and mixed with bacteriological agar. Small cubes were cut (1 mm3), fixed with osmium tetroxide and glutaraldehyde and dehydrated sequentially with increasing acetone concentrations. The dehydrated cubes were impregnated into epoxy and subjected to an ultramicrotome for sectioning to roughly 50 nm slices. The sections were placed on a copper grid and stained with uranyl acetate and lead citrate followed by transmission electron microscopy (TEM) analysis on a Philips CM100 TEM (Philips Electron Optics).
2.3. RNA extraction and quality analysis
Total RNA was extracted and purified using the Qiagen RNeasy Mini Kit (Whitehead Scientific) following the manufacturer's specifications and an on-column DNase (RNase-free DNase set by Qiagen) was performed to remove any contaminating DNA from the extracted RNA. Concentrations of the extracted total RNA was measured using a NanoDrop ND-1000 spectrophotometer. RNA quality was determined by a 2100 Agilent Bioanalyzer using the Agilent RNA 6000 Pico Kit according to the manufacturer's specifications.
2.4. cDNA synthesis using random oligos
cDNA was synthesized from the extracted RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as per the manufacturer's instructions.
2.5. Primer design for qPCR assays
Primer pairs as listed in Table 1 were designed for regions within the genes of interest and analysed with the Oligo Analyzer® v3.1 tool (Integrated DNA Technologies) to yield primer pairs with melting temperatures close to 58 °C to allow for optimal annealing of the primer pairs during the same cycling conditions and with thermodynamic attributes hindering the formation of hairpins and dimers.
Table 1.
Primer sequences and target regions are tabulated for the six primer pairs used in this study. The source of each primer pair is indicated in the last column.
| Target gene | Primer name | Primer sequence (5′ – 3′) | Category | Melting Temperature (°C) | Reference |
|---|---|---|---|---|---|
| hlyF | hlyF-pet22-FP | GGGAATTCATGGATCCTCGTCTTGATG | Cloning | 59,1 | This study |
| hlyF-pet22-RP | GGCTCGAGTTATTTAAAATCAACTTCCATTTGTTG | 58,6 | |||
| cysG | cysG-qPCR-FP | TTGTCGGCGGTGGTGATGTC | Reference | 60,3 | Zhou et al., [20] |
| cysG-qPCR-RP | ATGCGGTGAACTGTGGAATAAACG | 57,9 | |||
| hcaT | hcaT-qPCR-FP | GCTGCTCGGCTTTCTCATCC | Reference | 58,7 | Zhou et al., [20] |
| hcaT-qPCR-RP | CCAACCACGCTGACCAACC | 59,3 | |||
| idnT | idnT-qPCR-FP | CTGTTTAGCGAAGAGGAGATGC | Reference | 55,5 | Zhou et al., [20] |
| idnT-qPCR-RP | ACAAACGGCGGCGATAGC | 58,9 | |||
| ompA | ompA-qPCR-FP | TGGACCAACAACATCGGTGAC | Target | 57,6 | This study |
| ompA-qPCR-RP | CAACTACTGGAGCTGCTTCGC | 58,3 | |||
| ompF | ompF-qPCR-FP | TGCTTATGGTGCCGCTGAC | Target | 58,2 | This study |
| ompF-qPCR-RP | CGTAGTTCGACTGCCAGGTAG | 57,7 |
2.6. Relative quantitative real-time PCR (qPCR)
The assays were performed in biological duplicates and technical triplicates. The qPCR was performed as per the manufacturer's instructions using the LightCycler® FastStart DNA MasterPLUS SYBR Green I kit (Roche) on a LightCycler® 2.0 (Roche) capillary instrument. Each experimental run was performed with the primer pairs for the selected reference gene, ompA and ompF target genes, and no-template controls (NTC) for each primer pair. The following temperature cycling conditions were used for all reactions: An initial denaturation step at 95 °C for 10 minutes, followed by amplification, which included 95 °C for 10 seconds, 55 °C for 10 seconds and 72 °C for 7 seconds. Melting curve analysis was performed at 65 °C for 1 minute followed by 0.1 °C/s increases up to 95 °C. All the data was analysed on the LightCycler® 2.0 software version 4.1 (Roche).
2.7. Standard curves
Standard curves for all the qPCR primer pairs were performed in triplicate by creating two-fold dilution series of cDNA with five data points. The efficiency values for each primer pair were calculated on the LightCycler® 2.0 software version 4.1.
2.8. Reference gene selection
Zhou et al. [20] conducted a study during which microarray data was obtained for 63 E. coli strain BL21 (DE3) transformants during the over-expression of 63 distinct recombinant proteins [20]. This data was used to select for constitutively expressed genes during various conditions to select for candidate reference genes for qPCR. Three genes were identified as reliable and novel reference genes, namely cysG, idnT and hcaT, as ideal genes to monitor during transcription analysis during recombinant protein expression studies in E. coli. These three genes respectively encode for a siroheme synthase, L-idonate and D-gluconate transporter and a putative 3-phenylpropionic transporter and were tested in this study.
The triplicate Cq values obtained for the three reference genes' (idnT, cysG and hcaT) standard curves were analysed on the RefFinder tool to select a stable reference gene for this study [21].
2.9. Experimental setup
Gene expression profiles for the three genes (cysG, ompA, ompF) were carried out on the cDNA synthesised from the total RNA extracted from lactose-induced transformed E. coli cells either with or without (as control) hlyF inserted into in the pET22b(+) plasmid at three different time points post-induction – 4, 8 and 24 hours. This process was repeated to serve as a biological replicate. The data obtained were used to calculate the relative expression ratios during this 24 hours of gene expression.
2.10. Calculation of relative expression ratios
The relative expression ratios of the ompA and ompF genes were calculated relative to the reference gene during both hlyF expressing and non-expressing cells by using the Pfaffl equation (Equation 1) [22]. The Pfaffl equation incorporates qPCR Cq values from two different conditions, in this case hlyF expressing and non-expressing cells, of the target two genes and reference gene to indicate any potential effects of the different conditions on the target genes' expression.
Equation 1: The Pfaffl equation adapted from Pfaffl [22]. Ratio is the relative expression ratio of the target gene of interest (ompA or ompF) to that of the reference gene (cysG). Etarget is the PCR efficiency value of the target gene (ompA or ompF) obtained from its standard curve. ΔCqtarget is the difference between the Cq values of the target gene (ompA or ompF) obtained in the control sample and the experimental sample. Ereference is the PCR efficiency value of the reference gene (cysG) PCR obtained from its standard curve. ΔCqreference is the difference between the Cq values of the reference gene (cysG) obtained in the control sample and the experimental sample.
| (1) |
3. Results
Cloning and transformation into E. coli 10-β and BL21 (DE3) was successful with both the empty pET22b(+) vector and the pET22b(+) containing the PCR-amplified hlyF insert. Sequence data obtained from the pET22b(+) containing the insert confirmed the correct orientation of the gene and no mutations which could result in the protein not being transcribed and translated correctly and 99.5 % homology was obtained with the hlyF reference sequence (Accession number NC_011964.1).
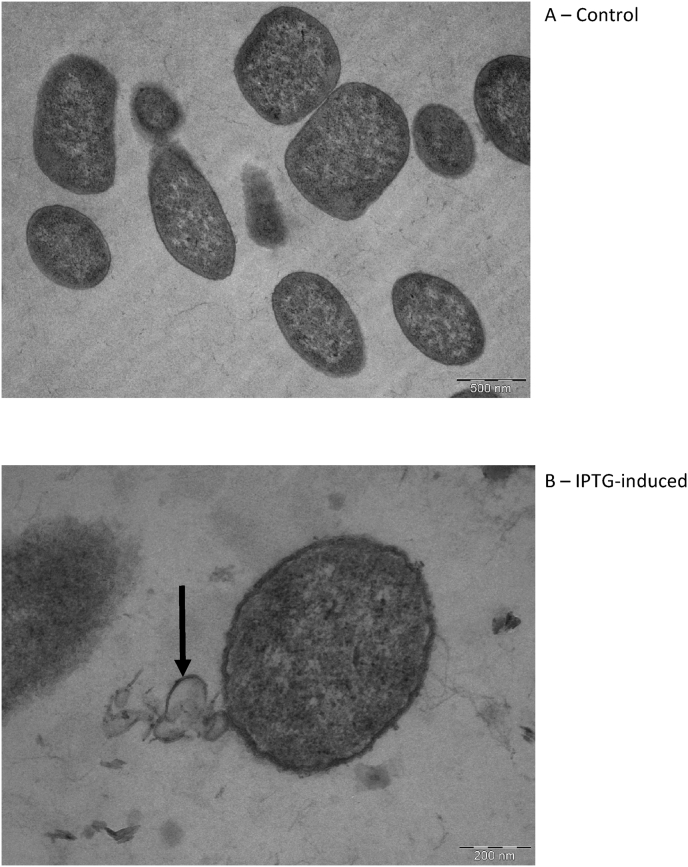
The effect of HlyF protein production was tested by preparing the IPTG-induced and uninduced transformant for TEM to observe any OMV-like structures. Transmission electron microscope micrographs indicated various electron-transparent extracellular membranous vesicles in the sample treated with 1 mM IPTG while the untreated cells show minimal extracellular material near the bacterial cells (Fig. 1).
Fig. 1.
TEM view of a section of control cells (A) showing no visible OMVs in the vicinity of the cells. Cells over-expressing hlyF under IPTG induction (B) with a visible OMV being released from the cell as indicated by the arrow.
The standard curves generated during qPCR for each gene produced efficiency values between 1.83 and 2.0 for the primer pairs. The Cq values from the standard curves for the potential reference genes, which were subjected to the RefFinder tool indicated that idnT and cysG were the most stable of the three reference genes and cysG was ultimately selected as the reference gene of choice for this study due to the lower expression levels of idnT (Supplementary Data 1).
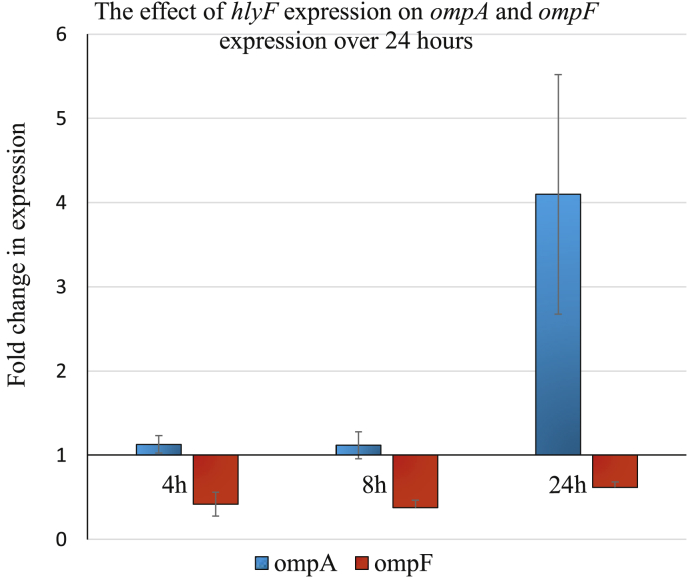
The Cq values obtained from the qPCR of the target genes ompA and ompF with cysG as the reference gene were analysed using the Pfaffl equation and the data plotted in Fig. 2 (Supplementary Data 2). Outer-membrane protein F showed a nearly 2-fold down regulation (0.616-fold change as shown in Table 2) during the 24 hours under hlyF expressing conditions while ompA was expressed at the same level as the non-expressing cells during the first 8 hours of expression. At 24 hours post-hlyF expression however, ompA was up-regulated 4-fold (4.099-fold change in Table 2).
Fig. 2.
Bar graph with standard deviations indicated by error bars, showing the fold change in gene expression levels relative to the reference gene during hlyF expressing and non-expressing conditions, according to the Pfaffl equation. An overall down regulation for ompF can be observed for the three time intervals, while ompA stays constant until a period between 8 and 24 hours is reached, leading to strong upregulation of the gene.
Table 2.
Data of the relative expression fold changes for each gene at the different time intervals are indicated for each biological replicate. Each value was calculated from the mean of triplicate Cq values used in the Pfaffl equation. The mean fold change is indicated in the last three columns.
| Gene | Biological Replicate 1 |
Biological Replicate 2 |
Mean Relative Expression Fold1 Change |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Relative Expression at 4h | Relative Expression at 8h | Relative Expression at 24h | Relative Expression at 4h | Relative Expression at 8h | Relative Expression at 24h | 4h | 8h | 24h | |
| ompA | 1.022 | 0.967 | 2.676 | 1.230 | 1.278 | 5.521 | 1.126 | 1.123 | 4.099 |
| ompF | 0.277 | 0.465 | 0.550 | 0.558 | 0.287 | 0.681 | 0.418 | 0.376 | 0.616 |
Fold change values <1 indicate a decrease in expression, = 1 no change in expression and >1 indicates increase in expression levels relative to the control cells lacking hlyF.
4. Discussion
Outer membrane vesicles are extracellular lipid structures produced by Gram negative bacteria and have been linked to virulence [8], antimicrobial resistance [3, 23] and resistance to bacteriophages [3]. The recently observed effect of outer-membrane vesicle formation during hlyF overexpression and the potential benefits a cell gains from the biogenesis of OMVs, as discussed, gives a potential explanation as to why this gene has been highly associated with APEC and is considered one of the five minimal predictors of virulence in poultry [10].
The OMVs produced during hlyF overexpression, similarly reported as with other E. coli OMV biosynthesis processes, contain various proteins from the host cell found in the periplasmic space and the outer membrane of the cell wall [1, 2]. Outer-membrane protein A is required by the cell to maintain membrane-peptidoglycan stability [24, 25]. The OmpA protein was observed in OMVs formed during hlyF overexpression and is expected to have been scavenged from the membrane of expressing cells. Therefore, the cell membrane stability would be compromised unless the cell compensates for the loss by increased expression of ompA or by some other unknown mechanism. Outer-membrane protein F, which is also present in hlyF-induced OMVs, is involved during changing osmotic pressure conditions of cells [16]. As they are also scavenged from the host cell during OMV formation, this could lead to the cell being susceptible to changes in osmotic pressure unless there is a mechanism in place to counter this. Therefore, scavenging of proteins such as OmpA and OmpF from the cell wall can compromise the cell integrity and ability to survive osmotic stresses. However, gene expression regulation could potentially be playing a role in replenishing the cell wall with these proteins. In contrast, it has been shown that low levels of expression of ompA can also lead to increased OMV production [26] – which is expected as a lack of OmpA will lead to a destabilized cell wall due to decreased peptidoglycan-outer-membrane interactions, however this was not the case as supported by the qPCR data obtained in this study nor was any cell wall destabilisation observed during TEM. There is the additional possibility that hlyF over-expression leads to decreased ompA expression, which leads to OMV formation due to this effect. As OmpF is only situated in the outer membrane and does not directly interact with the peptidoglycan, it is doubtful that a change in ompF expression will have a similar role.
In this study, qPCR results show that OMV biogenesis during hlyF overexpression is not related to a decrease in ompA expression and is therefore speculated to be accomplished through a different mechanism. We hypothesise that the eventual increase in ompA expression observed between 8 and 24 hours was possibly due to some as yet uncharacterised mechanism in the cells to stabilize the cell wall to prevent extensive cellular leakage and cell lysis.
The expected decrease in OmpF protein abundance on the outer cell membrane, due to the scavenging process of OMV biosynthesis, would likely leave the cells more susceptible to osmotic stresses. An insignificant down regulation of a less than 1 log fold-change as shown in Fig. 2 in ompF expression levels was observed during the three intervals tested during hlyF overexpression in comparison to the control. Therefore, the data does not support the possibility of a compensatory mechanism to counteract the scavenging of this protein.
5. Conclusions
The regulatory effects of the newly described OMV biosynthesis-related gene hlyF on E. coli has not been fully investigated. As hlyF-induced OMVs contain OmpA and OmpF scavenged from the host cell, potential regulatory effects on the host was investigated. An increase in ompA expression and an insignificant decrease in ompF expression was observed. It can be speculated that the increase in ompA expression is to counteract the scavenging of OmpA from the cell, which is necessary to maintain the outer-membrane-peptidoglycan interaction to stabilise the cell wall. From the results, it can also be deduced that OMV biosynthesis is not related to decreased ompA expression during hlyF overexpression, which is one of the potential mechanisms as shown in literature for OMV biosynthesis [27]. Outer-membrane vesicle biosynthesis during hlyF overexpression is thus potentially accomplished through a different mechanism(s).
Declarations
Author contribution statement
Wouter van der Westhuizen: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Chrispian Theron: Analyzed and interpreted the data; Wrote the paper.
Charlotte Boucher: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Robert Bragg: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Kulp A., Kuehn M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge T.J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning A.J., Kuehn M.J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni H.M., Jagannadham M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology. 2014;160:2109–2121. doi: 10.1099/mic.0.079400-0. [DOI] [PubMed] [Google Scholar]
- 5.Grenier D., Bélanger M. Protective effect of Porphyromonas gingivalis outer membrane vesicles against bactericidal activity of human serum. Infect. Immun. 1991;59:3004–3008. doi: 10.1128/iai.59.9.3004-3008.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwechheimer C., Kuehn M.J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nature Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonezawa H., Osaki T., Kurata S., Fukuda M., Kawakami H., Ochiai K. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009;9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murase K., Martin P., Porcheron G., Houle S., Helloin E., Pénary M. HlyF produced by extraintestinal pathogenic Escherichia coli is a virulence factor that regulates outer membrane vesicle biogenesis. J. Infect. Dis. 2016;213:856–865. doi: 10.1093/infdis/jiv506. [DOI] [PubMed] [Google Scholar]
- 9.Morales C., Lee M.D., Hofacre C., Maurer J.J. Detection of a novel virulence gene and a Salmonella virulence homologue among Escherichia coli isolated from broiler chickens. Foodb. Pathog. Dis. 2004;1:160–165. doi: 10.1089/fpd.2004.1.160. [DOI] [PubMed] [Google Scholar]
- 10.Johnson T.J., Wannemuehler Y., Doetkott C., Johnson S.J., Rosenberger S.C., Nolan L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008;46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Oliveira A.L., Rocha D.A., Finkler F., De Moraes B.L., Barbieri N.L., Pavanelo D.B. Prevalence of ColV plasmid-linked genes and in vivo pathogenicity of avian strains of Escherichia coli. Foodb. Pathog. Dis. 2015;12 doi: 10.1089/fpd.2014.1934. 679:85. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa R., Takaya A., Ohya M., Mizunoe Y., Takade A., Yoshida S.-I. Biogenesis of Salmonella enterica serovar typhimurium membrane vesicles provoked by induction of PagC. J. Bacteriol. 2010;192:5645–5656. doi: 10.1128/JB.00590-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodero M.D., Pilonieta M.C., Munson G.P. Repression of the inner membrane lipoprotein NlpA by rns in enterotoxigenic Escherichia coli. J. Bacteriol. 2007;189:1627–1632. doi: 10.1128/JB.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashburn-Warren L.M., Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 2002;292:396–401. doi: 10.1006/bbrc.2002.6657. [DOI] [PubMed] [Google Scholar]
- 16.Kawaji H., Mizuno T., Mizushima S. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 1979;140:843–847. doi: 10.1128/jb.140.3.843-847.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Westhuizen W.A., Bragg R.R. Multiplex polymerase chain reaction for screening avian pathogenic Escherichia coli for virulence genes. Avian Pathol. 2012;41:33–40. doi: 10.1080/03079457.2011.631982. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Russell R.W. third ed. Cold spring harbor laboratory press; 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 19.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhou K., Zhou L., Lim Q.', Zou R., Stephanopoulos G., Too H.P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie F., Xiao P., Chen D., Xu L., Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciofu O., Beveridge T.J., Kadurugamuwa J., Walther-Rasmussen J., Høiby N. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 24.Sonntag I., Schwarz H., Hirota Y., Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clavel T., Germon P., Vianney A., Portalier R., Lazzaroni J.C. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol. Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 26.Deatherage B.L., Lara J.C., Bersbaken T., Barrett S.L.R., Lara S., Cookson B.T. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 2009;72:1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valeru S.P., Shanan S., Alossimi H., Saeed A., Sandström G., Abd H. Lack of outer membrane protein a enhances the release of outer membrane vesicles and survival of Vibrio cholerae and suppresses viability of Acanthamoeba castellanii. Internet J. Microbiol. 2014;2014:610190. doi: 10.1155/2014/610190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.