Abstract
Blastocystis is a common enteric protist that colonizes humans and a wide range of animals. Although some studies have reported incidences of Blastocystis in humans and animals in China, there is no information available on the prevalence of Blastocystis in giant pandas, red pandas, or bird species. The aims of the present study were to determine the prevalence, subtype distribution, and genetic characterizations of Blastocystis in these animals in a captive situation in southwestern China, as well as assess the zoonotic potential of Blastocystis isolates. A total of 168 fecal specimens, including 81 from giant pandas, 23 from red pandas, 38 from black swans, 11 from ruddy shelducks, and 15 from green peafowl were collected at the Chengdu Research Base of Giant Panda Breeding in Sichuan province. The overall minimum prevalence of Blastocystis was 11.3% (19/168) based on PCR amplification of the barcode region of the SSU rRNA gene. The highest prevalence of Blastocystis was observed in ruddy shelduck (18.2%) and the lowest was found in green peafowl (6.7%). The prevalence of Blastocystis in giant pandas >5.5 years of age was higher than that in younger giant pandas. Two potentially zoonotic subtypes (ST1 and ST8) were identified, and ST1 (n = 12) was found to be more prevalent than ST8 (n = 7). To the best of our knowledge, this is the first report of the prevalence and subtypes of Blastocystis in giant pandas, red pandas, and bird species in China. The findings of this study will improve our understanding of the genetic diversity and public health potential of Blastocystis.
Keywords: Blastocystis, Pandas, Genetic diversity, Zoonotic potential, China
Graphical abstract
Highlights
-
•
This is the first report of Blastocystis in giant pandas, red pandas, and various bird species in China.
-
•
The overall minimum prevalence of Blastocystis was 11.3%.
-
•
Two zoonotic subtypes (ST1 and ST8) were identified.
-
•
This study will enrich the epidemiological data of Blastocystis infection in China.
1. Introduction
Blastocystis is one of the most common intestinal parasites that infect humans and animals worldwide (Mehlhorn et al., 2012). Generally, Blastocystis is commonly transmitted through cyst-contaminated water and food via the fecal-oral route, which is the primary mode of transmission (Yoshikawa et al., 2009). The pathogenicity of Blastocystis is not yet clear, there are studies associating it with symptoms of a variety of gastrointestinal disorders such as irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) (Boorom et al., 2008; Jimenez-Gonzalez et al., 2012; Dogruman-Al, 2009). However, the few microbiome studies revealed that Blastocystis is a common commensal in the human gut and it can increase the diversity of gut microbiota (Beghini F, 2017).
In addition to humans, this parasite is frequently found in a wide range of animal hosts, including non-human primates, and other mammals such as artiodactyls, perissodactyls, proboscideans, rodents, and marsupials as well as birds, reptiles, amphibians, fish, annelids, and insects (Wang et al., 2018).
Currently, based on sequence analysis of the small subunit ribosomal RNA gene of Blastocystis, at least 26 subtypes have been identified in humans and animals worldwide (Maloney et al., 2018; Li et al., 2018; Roberts et al., 2013). Subtypes 1–9 and ST12 have been found in humans (Zhang et al., 2017; Ramírez et al., 2016); some of which have also been observed in animals, such as ST3 in non-human primates, ST5 in cattle and pigs, ST7 in birds, and ST8 in non-human primates and birds, indicating zoonotic potential (Cian et al., 2017; Moosavi et al., 2012). Conversely, some subtypes such as ST10 and ST14 predominantly circulate in specific animal hosts and have never appeared in human infections (Stensvold and Clark, 2016), suggesting host specificity.
In China, giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) are classified as Category I and II protected species, respectively. Many factors have led to the endangered status of these animals, such as habitat destruction, low reproductive rates, accidental mortality, and infections from pathogens; the latter of which is considered to be the most serious threat. Since the first reported descriptions of Blastocystis infection in two chronic diarrhea cases of children in Guangdong province [12], an increasing number of cases have been reported in humans and various animals belonging to the orders Artiodactyla, Carnivora, Galliformes, Primates, Columbiformes, Anseriformes, Gruiformes, Rodentia, and Lagomorpha (Zanzani et al., 2016; Wang et al., 2018). However, no information is currently available regarding the genetic characteristics and subtype distribution of Blastocystis in giant pandas, red pandas, and various bird species in southwestern China. The objective of the present study was thus to determine the prevalence and subtype distribution of Blastocystis in these animals as well as to assess its zoonotic potential.
2. Materials and methods
2.1. Ethics statement
This study complied with the guidelines of the Regulations for the Administration of Affairs Concerning Experimental Animals and was approved by the Animal Ethical Committee of Sichuan Agricultural University. No animals were harmed during the sampling process. Permission was obtained from the China Giant Panda Protection and Research Center for the collection of fecal specimens. All the procedures were conducted in accordance with the approved guidelines.
2.2. Sample collection
Between August 2017 and October 2018, a total of 168 fecal samples from 81 giant pandas, 23 red pandas, 38 black swans (Cygnus atratus), 11 ruddy shelducks (Tadorna ferruginea), and 15 green peafowl (Pavo muticus) were collected from the Chengdu Research Base of Giant Panda Breeding in Sichuan province, southwestern China. Both types of pandas reside in open enclosures at this facility. Panda fecal samples were collected with sterile gloves from the ground immediately after defecation. Samples were then transferred to sterile plastic containers marked with the age, gender, and sampling date. All the black swans and ruddy shelducks were raised in a small recreational park within the panda facility, and the green peafowls were free-ranging. Fecal samples were collected from bird cages with care to collect only the portion that did not have direct contact with the cage to avoid contamination. Samples were placed into sterile plastic containers labelled with the species of bird and sampling time. All fecal samples were stored in 2.5% potassium dichromate solution at 4 °C prior to analysis. All study animals were examined and appeared to be in good health, and no diarrhea was observed.
2.3. DNA extraction
Potassium dichromate was removed from the fecal specimens with distilled water by centrifugation at 1500 × g for 10 min. Genomic DNA was extracted from ∼200 mg of each fecal sample using a QIAamp DNA Stool Mini Kit (QIAgen, Hilden, Germany). Extracted DNA was stored at −20 °C until PCR analysis.
2.4. PCR amplification
All DNA preparations were screened for the presence of Blastocystis by PCR amplification of the barcode region (a fragment of ∼600 bp) of the SSU rRNA gene, and the primers and cycling parameters were used as previously described by Santín et al. (2011). PCR-positive DNA preparations were further analyzed to determine the subtypes of Blastocystis isolates by sequence analysis of the barcode region according to terminology for Blastocystis subtypes - a consensus (Stensvold et al., 2007). TaKaRa Taq DNA polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all of the PCR reactions. A negative control with no DNA added was included in all of the PCR tests. PCR products were subjected to electrophoresis in a 1.5% agarose gel and visualized by staining the gel with ethidium bromide.
2.5. Nucleotide sequencing and analysis
All positive PCR products were directly sequenced on an ABI PRISMTM 3730 DNA Analyzer (Applied Biosystems, USA), using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster, CA, USA). Nucleotide sequences obtained in the present study were subjected to BLAST searches (http://www.ncbi.nlm.nih.gov/blast/) and then aligned and analyzed with each other. Reference sequences were downloaded from the GenBank database using the program Clustal X 2.0 (http://www.clustal.org/) to determine the subtypes of Blastocystis isolates. The nucleotide sequences generated in present study have been deposited in GenBank under accession numbers MK742731–M K742737.
2.6. Phylogenetic analysis
A neighbor-joining tree was constructed to assess the genetic relationships among the Blastocystis subtypes obtained in the present study and those identified in previous studies, using the software Mega 6 (http://www.megasoftware.net/). Evolutionary distances were calculated using the Kimura two-parameter model. The reliability of the trees was assessed by bootstrap analysis with 1000 replicates.
2.7. Data analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). A chi-square test and 95% confidence intervals were used to compare the prevalence of Blastocystis between age groups, gender groups, and different species. Differences were considered statistically significant when P-values < 0.05.
3. Results
3.1. Prevalence of Blastocystis
PCR amplification of the barcode region of the SSU rRNA gene revealed that a total of 19 (11.3%) of the 168 samples had confirmed Blastocystis infections. The highest prevalence was observed in ruddy shelduck (18.2%, 2/11), followed by giant pandas (12.3%, 10/81). The prevalence of Blastocystis in black swans, red pandas, and green peafowl were 10.5%, 8.7%, and 6.7%, respectively (Table 1). There were no significant associations between prevalence and the different species (χ2 = 1.107, df = 4, P > 0.05). The prevalence of Blastocystis in giant pandas of different ages and genders is presented in Table 2; with the highest prevalence being observed in ages >5 years (14.8%, 8/54), followed by 8.7% (2/23) for ages 1.5–5.5 years, and 0% in ages < 1.5 years, however, the difference was not significant (χ2 = 1.654, df = 2, P > 0.05). Similarly, the difference was not significant between male (16.4%, 10/61) and female (16.9%, 14/83) giant pandas (χ2 = 0.014, df = 1, P > 0.05).
Table 1.
Prevalence and subtype distribution of Blastocystis in giant pandas, red pandas, and birds.
| Hosts (Scientific name) | No. of examined | No. of positive | Prevalence (%) | 95% CI | Subtypes (n) |
|---|---|---|---|---|---|
| Mammals | |||||
| Giant panda (Ailuropoda melanoleuca) | 81 | 10 | 12.3 | 5.1–19.5 | ST1 (10) |
| Red pandas (Ailurus fulgens) | 23 | 2 | 8.7 | −2.8–20.2 | ST1 (2) |
| Subtotal | 104 | 12 | 11.5 | 5.3–17.7 | ST1 (12) |
| Birds | |||||
| Black Swan (Cygnus atratus) | 38 | 4 | 10.5 | 0.7–20.3 | ST8 (4) |
| Ruddy Shelduck (Tadorna ferruginea) | 11 | 2 | 18.2 | −4.6–40.9 | ST8 (2) |
| Green peafowl (Pavo muticus) | 15 | 1 | 6.7 | −5.9–19.3 | ST8 (1) |
| Subtotal | 64 | 7 | 10.9 | 3.3–18.6 | ST8 (7) |
| Total | 168 | 19 | 11.3 | 6.5–16.1 | ST1 (12), ST8 (7) |
Table 2.
Prevalence and subtype distribution of Blastocystis in giant pandas by age and gender.
| Factors | No. of examined | No. of positive (%) | 95% CI | Subtypes (n) |
|---|---|---|---|---|
| Age (years) | ||||
| <1.5 | 4 | 0 | ||
| 1.5–5.5 | 23 | 2 (8.7) | −2.8–20.2 | ST1 (2) |
| >5 | 54 | 8 (14.8) | 5.3–24.3 | ST1 (8) |
| Gender | ||||
| Male | 31 | 4 (12.9) | 1.1–24.7 | ST1 (4) |
| Female | 50 | 6 (12.0) | 3.0–21.0 | ST1 (6) |
3.2. Subtype distributions in giant pandas, red pandas, and various birds
Among the 19 Blastocystis isolates, two subtypes, ST1 and ST8, were identified as a result of sequence analysis of the barcode region of the SSU rRNA gene. ST1 was the predominant subtype (63.2%, 12/19) and was only found in giant and red pandas. ST8 was exclusively identified in black swans, ruddy shelducks, and green peafowl (Table 1).
3.3. Genetic characteristics of Blastocystis subtypes
A total of 7 representative sequences were obtained from 19 Blastocystis isolates in the present study. Among them, ST1 (n = 7) and ST8 (n = 7) have been described previously: ST1 was identical to that of Blastocystis in non-human primates found in the Philippines (KY929112), and ST8 showed 100% similarity to that of Blastocystis reported in a human in Spain (MG807921). The remaining 5 novel ST1 sequences matched 17 single nucleotide polymorphisms (SNPs) for KY929112. The genetic diversity in the barcode region of the SSU rRNA gene from the five ST1 novel isolates is presented in Table 3.
Table 3.
Nucleotide variations in the barcode region of the SSU rRNA gene of Blastocystis ST1 isolates.
3.4. Phylogenetic analysis
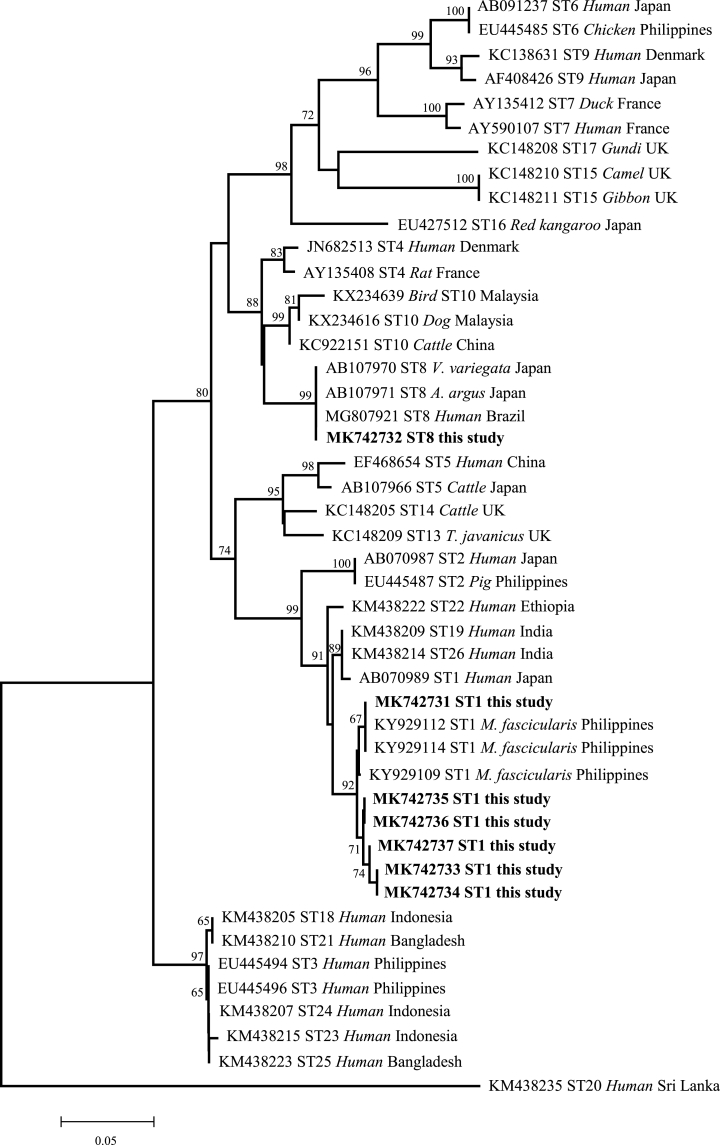
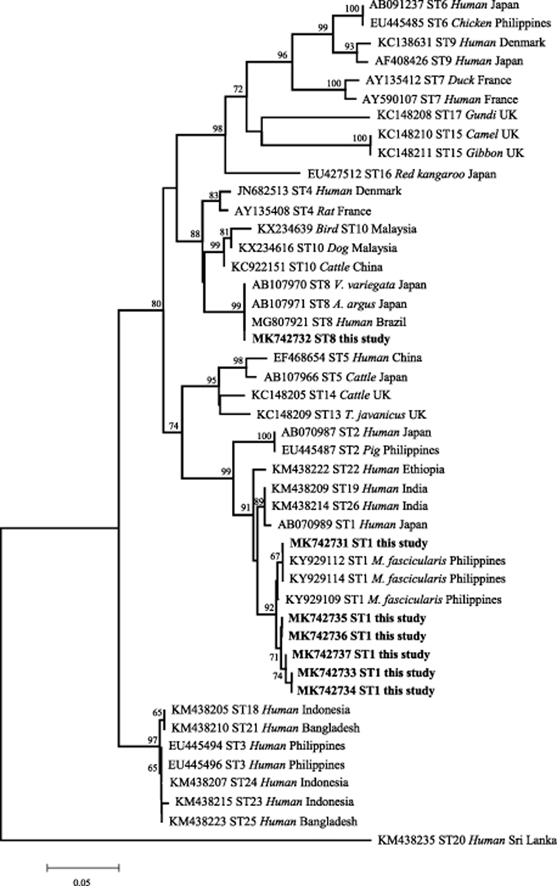
The sequences identified in this study were aligned to known sequence databases in GenBank, which revealed two known and five new partial SSU rRNA sequences. All five novel sequences were grouped with known isolates belonging to subtype ST1 during phylogenetic analysis using the neighbor-joining method (Fig. 1).
Fig. 1.
Phylogenetic relationships among nucleotide sequences of Blastocystis partial small subunit ribosomal RNA (SSU rRNA) genes (see Additional file 1: Table S1). The neighbor-joining method was used to construct the trees by the Kimura-2-parameter model. The number on the branches are percent bootstrapping values from 1000 replicates, with values of more than 50% shown in the tree. Each sequence is identified by its accession number, subtypes, host origin, and country. Blastocystis subtypes identified in the present study are indicated in bold-type.
4. Discussion
To the best of our knowledge, the present study is the first report to provide information on Blastocystis in giant pandas, red pandas, and various bird species within Sichuan province, southwestern China. The prevalence of Blastocystis varies in mammals and birds, ranging from 0.3 to 100% (Table 4). In the present study, the prevalence of Blastocystis ranged from 6.7% to 18.2%, which is similar to that in recent studies on reindeer (6.7%), pigs (8.8%), cattle (9.5%), domestic chicken (13.0%), red crowned crane (14.0%) in China (Wang et al., 2018; Li et al., 2018).
Table 4.
Subtypes and prevalence of Blastocystis detected in various animals in the China.
| Hosts | No. of Examined | No. of positive | Prevalence (%) | Subtypes (n) | References | |
|---|---|---|---|---|---|---|
| Mammals | ||||||
| Pig | 560 | 419 | 74.8% | ST5 (397), ST1 (15), ST3 (6), ST10 (1) | Song et al. (2017) | |
| Pig | 16 | 16 | 100% | ST5 (16) | Yan et al. (2007) | |
| Pig | 68 | 6 | 8.8% | ST5 (6) | Wang et al. (2018) | |
| Cattle | 147 | 14 | 9.5% | ST10 (10), ST3 (2), ST14 (2) | Wang et al. (2018) | |
| Dairy cattle | 526 | 54 | 10.3% | ST10 (41), ST14 (10), ST4 (2), ST5 (1) | Song et al. (2017) | |
| Sheep | 109 | 6 | 5.5% | ST10 (3), ST1 (1), ST5 (1), ST14 (1) | Wang et al. (2018) | |
| Sheep | 832 | 50 | 6.0% | ST10 (25), ST14 (10), ST5 (8), Novel 1 (3), Novel 2 (1), Novel 3 (1), Novel 4 (2) | Li et al. (2018) | |
| Goat | 781 | 2 | 0.3% | ST1 (2) | Li et al. (2018) | |
| Reindeer | 104 | 7 | 6.7% | ST13 (4), ST10 (3) | Wang et al. (2018) | |
| Sika deer | 82 | 12 | 14.6% | ST10 (10), ST14 (2) | Wang et al. (2018) | |
| Racoon dog | 40 | 3 | 7.5% | ST3 (3) | Wang et al. (2018) | |
| Domestic dog | 136 | 4 | 2.9% | ST1 (3), ST4 (1) | Wang et al. (2018) | |
| Arctic fox | 213 | 4 | 1.9% | ST1 (2), ST4 (1), ST7 (1) | Wang et al. (2018) | |
| Brown rat | 108 | 4 | 3.7% | ST4 (4) | Wang et al. (2018) | |
| New Zealand white rabbit | 215 | 7 | 3.2% | ST4 (7) | Wang et al. (2018) | |
| Cynomolgus macaques | 97 | 85 | 87.6% | ST1 (4), ST2 (14), ST7 (2), ST2+ST1 (14), ST2+ST3 (5), ST2+ST7 (5), ST3+ST1 (3), ST5+ST2 (1), ST7+ST1 (7), ST7+ST3 (1), ST1+ST2+ST3 (10), ST1+ST2+ST7 (5), ST2+ST3+ST7 (3), ST1+ST3+ST7 (1), ST1+ST2+ST3+ST7 (10) | Zanzani et al. (2016) | |
| Birds | ||||||
| Domestic chicken | 46 | 6 | 13.0% | ST6 (3), ST7 (3) | Wang et al. (2018) | |
| Pigeon | 47 | 1 | 2.1% | ST6 (1) | Wang et al. (2018) | |
| Red crowned crane | 43 | 6 | 14.0% | ST6 (4), ST7 (2) | Wang et al. (2018) | |
| Ostrich | 9 | 3 | 33.3% | ST5 (1), ST10 (1), ST20 (1) | Zhao et al. (2017) | |
However, the prevalence of Blastocystis identified in this study was much lower compared with those in other studies conducted on pigs (74.8% and 100%), and cynomolgus macaques (87.6%) in China (Zanzani et al., 2016; Yan et al., 2007; Song et al., 2017a, Song et al., 2017b). The differences in the prevalence of Blastocystis may result from variations of animal ages, sample sizes, immune status of the animals.
In the present study, ST1 (63.2%, 12/19) was more prevalent than ST8 (36.8%, 7/19), and was exclusively reported in mammals. Previous studies have demonstrated the presence of ST1 in cancer patients with diarrhea in China (Zhang et al., 2017), humans with GI symptoms in Thailand (Sarinee et al., 2013), and humans with irritable bowel syndrome in Iran (Khademvatan et al., 2017). Moreover, ST1 has been identified in goats, sheep, domestic dogs, arctic foxes, and cynomolgus macaques from China (Song et al., 2017a, Song et al., 2017b; Zanzani et al., 2016; Wang et al., 2018). ST1 was found in both human and mammalian hosts, indicating that this subtype has the potential for zoonotic transmission. However, due to the unavailability of data regarding the subtyping of human-derived Blastocystis isolates from China and investigated areas, the actual pollution/contamination sources and transmission routes of mammals with Blastocystis infection were not elucidated in the present study.
Currently, a total of nine subtypes (ST1, ST2, ST4-8, ST10 and ST20) have been identified in birds (Cian et al., 2017; Zhao et al., 2017; Yoshikawa et al., 2003) (Table 5). Although ST6 and ST7 are the most common subtypes in birds, we only identified ST8 in the present study, which is consistent with previous report in pheasants (Abe et al., 2003). In addition to birds, ST8 has been identified in humans in Colombia (Ramírez et al., 2014), symptomatic patients in Italy (Dionigia et al., 2011), and Brazil (Barbosa et al., 2018), as well as in stored water reserves in the Peninsular Malaysia (Noradilah et al., 2016). Furthermore, ST8 has been reported more commonly in primate handlers, suggesting a zoonotic spread from primates to their handlers (Stensvold et al., 2009). However, due to people who have close contact with birds for occupational or recreational reasons in this province have not reported cases of this ST, more intensive research should be performed to clarify the potential zoonotic transmission routes and other sources of Blastocystis in the investigated area.
Table 5.
Subtypes and prevalence of Blastocystis detected from birds in the world.
| Host | Number of samples | Subtypes (n) | References |
|---|---|---|---|
| Chicken | 5 | ST1 (2), ST2 (1), ST4 (1), ST5 (1) | Yoshikawa et al. (2003) |
| 6 | ST6 (3), ST7 (3) | Wang et al. (2018) | |
| Pigeon | 1 | ST6 (1) | Wang et al. (2018) |
| Red crowned crane | 6 | ST6 (4), ST7 (2) | Wang et al. (2018) |
| Ostrich | 3 | ST5 (1), ST10 (1), ST20 (1) | Zhao et al. (2017) |
| Peasant | 5 | ST4 (3), unknown (2) | Yoshikawa et al. (2003) |
| 4 | ST6 (1), ST8 (1), unknown (2) | Abe et al. (2003) | |
| Quail | 9 | ST2 (5), ST4 (4) | Yoshikawa et al. (2003) |
| Ostrich | 3 | ST5 (1), ST10 (1), ST20* | Zhao et al. (2017) |
| Turkey | 1 | ST7 (1) | Hess et al. (2006) |
| Partridge | 1 | ST7 (1) | Abe et al. (2003) |
| Guineafowl | 1 | ST6 (1) | Abe et al. (2003) |
| Passer domesticus | 17 | ST6 (17) | Ramírez et al. (2014) |
| Thraupis episcopus | 6 | ST6 (6) | Ramírez et al. (2014) |
| Oryzoborus maximiliani | 8 | ST6 (8) | Ramírez et al. (2014) |
| Sicalis flaveola | 5 | ST6 (5) | Ramírez et al. (2014) |
| Petrochelidon pyrrhonota | 11 | ST6 (11) | Ramírez et al. (2014) |
5. Conclusions
The present study is the first report on the prevalence, subtype distribution, and genetic characteristics of Blastocystis in giant pandas, red pandas, and bird species in southwestern China. Phylogenetic relationship analysis revealed five novel nucleotide sequences of Blastocystis isolated from the studied animals. The overall minimum prevalence of Blastocystis was 11.3%, and two potentially zoonotic subtypes (ST1 and ST8) were identified. As visitors and keepers frequently come in contact with the animals and birds in the investigated area, proper advice should be given to these susceptible human populations to reduce zoonotic transmission at this panda breeding facility. Furthermore, future studies should aim to identify high-resolution molecular markers to better understand the dynamics of Blastocystis transmission.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
The study was financially supported by the Chengdu Giant Panda Breeding Research Foundation (CPF2017-12, CPF2015-09, CPF2015-07). The funders contributed to the study design and data collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.06.007.
Contributor Information
Chan-Juan Yue, Email: Chanjuan_Yue@163.com.
Guang-Neng Peng, Email: pgn.sicau@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abe N., Wu Z., Yoshikawa H. Molecular characterization of Blastocystis isolates from birds by PCR with diagnostic primers and restriction fragment length polymorphism analysis of the small subunit ribosomal RNA gene. Parasitol. Res. 2003;89:393–396. doi: 10.1007/s00436-002-0782-5. [DOI] [PubMed] [Google Scholar]
- Barbosa C.V., Barreto M.M., Andrade R.J., Sodré F., D'Avila-Levy C.M., Peralta J.M., Igreja R.P., de Macedo H.W., Hlc S. Intestinal parasite infections in a rural community of Rio de Janeiro (Brazil): prevalence and genetic diversity of Blastocystis subtypes. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini F., Pasolli E., Truong T.D., Putignani L., Cacciò S.M., Segata N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11:2848–2863. doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorom K.F., Smith H., Nimri L., Viscogliosi E., Spanakos G., Parkar U., Li L.H., Zhou X.N., Ok Ü.Z., Leelayoova S. Oh my aching gut: irritable bowel syndrome, Blastocystis, and asymptomatic infection. Parasites Vectors. 2008;1 doi: 10.1186/1756-3305-1-40. 40-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian A., El S.D., Osman M., Moriniere R., Gantois N., Benamrouz-Vanneste S., Delgado-Viscogliosi P., Guyot K., Li L.L., Monchy S. Molecular epidemiology of Blastocystis sp. in various animal groups from two French zoos and evaluation of potential zoonotic risk. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionigia M., Giovanna S., Philippe P., Hicham E.A., Magali C., Laurence D., Eduardo D.C., Frederic D., Pier L.F., David D.C. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol. Res. 2011;109:613–619. doi: 10.1007/s00436-011-2294-7. [DOI] [PubMed] [Google Scholar]
- Dogruman-Al F., Kustimur S., Yoshikawa H., Tuncer C., Simsek Z., Tanyuksel M. Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem. Inst. Oswaldo Cruz. 2009;104:724–727. doi: 10.1590/s0074-02762009000500011. [DOI] [PubMed] [Google Scholar]
- Hess M., Kolbe T., Grabensteiner E., Prosl H. Clonal cultures of Histomonas meleagridis, Tetratrichomonas gallinarum and a Blastocystis sp. established through micromanipulation. Parasitology. 2006;133:547–554. doi: 10.1017/S0031182006000758. [DOI] [PubMed] [Google Scholar]
- Jimenez-Gonzalez D.E., Martinez-Flores W.A., Reyes-Gordillo J., Ramirez-Miranda M.E., Arroyo-Escalante S., Romero-Valdovinos M., Stark D., Souza-Saldivar V., Martinez-Hernandez F., Flisser A. Blastocystis infection is associated with irritable bowel syndrome in a Mexican patient population. Parasitol. Res. 2012;110:1269–1275. doi: 10.1007/s00436-011-2626-7. [DOI] [PubMed] [Google Scholar]
- Khademvatan S., Masjedizadeh R., Rahim F., Mahbodfar H., Salehi R., Yousefi-Razin E., Foroutan M. Blastocystis and irritable bowel syndrome: frequency and subtypes from Iranian patients. Parasitol. Int. 2017;66:142–145. doi: 10.1016/j.parint.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Li W.C., Wang K., Gu Y. Occurrence of Blastocystis sp. and Pentatrichomonas hominis in sheep and goats in China. Parasites Vectors. 2018;11:93. doi: 10.1186/s13071-018-2671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney J.G., Lombard J.E., Urie N.J., Shivley C.B., Santin M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol. Res. 2018;118:575–582. doi: 10.1007/s00436-018-6149-3. [DOI] [PubMed] [Google Scholar]
- Mehlhorn H., Tan K.S.W., Yoshikawa H. 2012. Blastocystis: pathogen or passenger? [Google Scholar]
- Moosavi A., Haghighi A., E Nazemalhosseini M., Zayeri F., Alebouyeh M., Khazan H., Kazemi B., Zali M.R. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol. Res. 2012;111:2311–2315. doi: 10.1007/s00436-012-3085-5. [DOI] [PubMed] [Google Scholar]
- Noradilah S.A., Lee I.L., Anuar T.S., Salleh F.M., Manap S.N.A.A., Mohtar N.S.H.M., Azrul S.M., Wan O.A., Moktar N. Occurrence of Blastocystis sp. in water catchments at Malay villages and Aboriginal settlement during wet and dry seasons in Peninsular Malaysia. PeerJ. 2016;4 doi: 10.7717/peerj.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez L.V., Bautista D.C., Corredor A.F., Flórez A.C., Stensvold C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014;22:223–228. doi: 10.1016/j.meegid.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez A., Hernández C., Flórez C., Bernal M.C., Giraldo J.C. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Roberts T., Stark D., Harkness J., Ellis J. Subtype distribution of Blastocystis isolates from a variety of animals from new south Wales, Australia. Vet. Parasitol. 2013;196:85–89. doi: 10.1016/j.vetpar.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Santín M., Gómez-Muñoz M.T., Solano-Aguilar G., Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011;109:205–212. doi: 10.1007/s00436-010-2244-9. [DOI] [PubMed] [Google Scholar]
- Sarinee J., Porntip P., Kookwan S., Somchai P., Arunnee S., Chotechana W., Wachanan W., Hisao Y. Subtype identification of Blastocystis spp. isolated from patients in a major hospital in northeastern Thailand. Parasitol. Res. 2013;112:1781–1786. doi: 10.1007/s00436-012-3218-x. [DOI] [PubMed] [Google Scholar]
- Song J.K., Yin Y.L., Yuan Y.J., Tang H., Ren G.J., Zhang H.J., Li Z.X., Zhang Y.M., Zhao G.H. First genotyping of Blastocystis sp. in dairy, meat, and cashmere goats in northwestern China. Acta Trop. 2017;176:277–282. doi: 10.1016/j.actatropica.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Song J.K., Hu R.S., Fan X.C., Wang S.S., Zhang H.J., Zhao G.H. Molecular characterization of Blastocystis from pigs in Shaanxi province of China. Acta Trop. 2017;173:130–135. doi: 10.1016/j.actatropica.2017.06.014. [DOI] [PubMed] [Google Scholar]
- Stensvold C., Alfellani M.L.,S., Prip K., Victory E., Maddox C., Nielsen H., Clark C. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Clark C.G. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Suresh G.K., Tan K.S.W., Thompson R.C.A., Traub R.J., Viscogliosi E., Yoshikawa H., Clark C.G. Terminology for Blastocystis subtypes - a consensus. Trends Parasitol. 2007;23:93–96. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Wang J., Gong B., Yang F., Zhang W., Zheng Y., Liu A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China's Heilongjiang Province. Infect. Genet. Evol. 2018;57:171–176. doi: 10.1016/j.meegid.2017.11.026. [DOI] [PubMed] [Google Scholar]
- Yan Y., Su S., Ye J., Lai X., Lai R., Liao H., Chen G., Zhang R., Hou Z., Luo X. Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol. Res. 2007;101:1527–1532. doi: 10.1007/s00436-007-0672-y. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Wu Z., Pandey K., Pandey B.D., Sherchand J.B., Yanagi T., Kanbara H. Molecular characterization of Blastocystis isolates from children and rhesus monkeys in Kathmandu, Nepal. Vet. Parasitol. 2009;160:295–300. doi: 10.1016/j.vetpar.2008.11.029. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Abe N., Wu Z. Genomic polymorphism among Blastocystis isolates and development of PCR-based identification of zoonotic isolates. J. Eukaryot. Microbiol. 2003;50:710–711. doi: 10.1111/j.1550-7408.2003.tb00698.x. [DOI] [PubMed] [Google Scholar]
- Zanzani S.A., Gazzonis A.L., Epis S., Manfredi M.T. Study of the gastrointestinal parasitic fauna of captive non-human primates (Macaca fascicularis) Parasitol. Res. 2016;115:307–312. doi: 10.1007/s00436-015-4748-9. [DOI] [PubMed] [Google Scholar]
- Zhang W., Ren G., Zhao W., Yang Z., Shen Y., Sun Y., Liu A., Cao J. Genotyping of Enterocytozoon bieneusi and subtyping of Blastocystis in cancer patients: relationship to diarrhea and assessment of zoonotic transmission. Front. Microbiol. 2017;8:1835. doi: 10.3389/fmicb.2017.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.H., Hu X.F., Liu T.L., Hu R.S., Yu Z.Q., Yang W.B. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol. Res. 2017;116:2327–2333. doi: 10.1007/s00436-017-5506-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.