Abstract
Functional modeling of normal breast epithelial hierarchy and stromal-epithelial cell interactions have been difficult due to inability to obtain sufficient stem-progenitor-mature epithelial and stromal cells. Recently reported epithelial reprogramming assay has partially overcome this limitation, but cross contamination of cells from the feeder layer is a concern. The purpose of this study was to develop a feeder-layer independent inexpensive method to propagate multiple cell types from limited tissue resources. Cells obtained after enzymatic digestion of tissues collected at surgery or by core-needle biopsies were plated on tissue culture dishes pre-coated with laminin-5-rich conditioned media from the rat bladder tumor cell line 804G and a defined growth media with inhibitors of ROCK, TGFβ, and BMP signaling. Cells were characterized by flow cytometry, mammosphere assay, 3D cultures, and xenograft studies. Cells from the healthy breasts included CD10+/EpCAM- basal/myoepithelial, CD49f+/EpCAM+ luminal progenitor, CD49f-/EpCAM+ mature luminal, CD73+/EpCAM+/CD90- rare endogenous pluripotent somatic stem, CD73+/CD90+/EpCAM-, Estrogen Receptor alpha (ERα)-expressing ALCAM (CD166)+/EpCAM+, and ALDFLUOR+ stem/luminal progenitor subpopulations. Epithelial cells were luminal (KRT19+), basal (KRT14+) or dual positive luminal/basal hybrid cells. While breast cells derived from BRCA1, BRCA2, and PALB2 mutation carriers did not display unique characteristics, cells from women with breast cancer protective alleles showed enhanced differentiation. Cells could also be propagated from primary tumors and metastasis of breast, ovarian, and pancreatic cancer-neuroendocrine subtype. Xenograft studies confirmed tumorigenic properties of tumor-derived cells.
Keywords: Primary epithelial cells, TGFβ and BMP inhibition, breast cancer, luminal progenitor, culture condition
Introduction
The breast tissue is composed of the epithelial and stromal cells cushioned by layers of adipose cells (1). The ability to culture and propagate various type of cells that form the breast hierarchy, which include the stem-progenitor-mature epithelial cells and stromal cells is of vital importance for all functional studies related to in vitro characterization of different cell types, epithelial-stromal interaction, elucidation of molecular mechanisms of normal cell differentiation, cancer initiation, and progression. Several protocols have been developed that support the propagation of breast epithelial cells, which in large part are biased towards outgrowth of basal cells [summarized in (2)]. Several of these culture protocols utilize irradiated mouse embryonic fibroblast or human foreskin fibroblasts as feeder cells to maintain the pluripotent state of stem cells (3,4). Normal breast stromal cells have morphological features and characteristics of fibroblasts. It is also difficult to distinguish mesenchymal and adipose stem cells from fibroblasts without profiling for cell surface markers. Therefore, an ideal system should allow growth of as many cell types of a tissue as possible and should utilize alternatives to feeder layers such as extracellular matrix proteins and small molecules that enable the maintenance of adult stem cells and their lineage commitment properties. In this respect, vitamin C and inhibitors of Rho associated coil-coil containing protein kinase (ROCK) have been shown to be effective in stem cell reprogramming and in preventing matrix detachment-induced apoptosis of stem cells, respectively (3,5).
A recent study reported that plating adult epithelial cells from lungs on tissue culture dishes pre-coated with laminin-5-rich conditioned media from the rat bladder cancer cell line 804G and in media containing dual inhibitors of SMAD/BMP pathways permits propagation of diverse epithelial cells (6). We had previously shown that maintaining primary breast epithelia cells in low glucose containing media and limiting the use of DMSO as a solvent for small molecules enable long-term culture of breast epithelial cells from a core needle biopsy on a feeder layer and these cultured cells maintain stem-progenitor-differentiated cell hierarchy (7). The goals of this study were to determine whether combining these two methods would permit growth of multiple cell types of the breast without the need for feeder layer and whether the technique can be extended to other cancer types as well as biopsies from metastatic sites. Since there is growing evidence for heterogeneity between primary tumor and metastasis within an individual patient, despite sharing large number of driver mutations (8), there is an urgent need to develop methods that enable characterization of metastatic cells in vitro for therapeutic decision making. We show that the method described here is effective in propagating primary epithelial cells that include but is not limited to 1) normal breast, 2) carriers of BRCA1, BRCA2, PALB2 mutations or breast cancer protective alleles, 3) primary breast cancer, 4) pleural effusions of breast cancer patients, 5) ascites fluids from ovarian cancer patients, and 6) liver metastasis of breast cancer and pancreatic cancer of neuroendocrine origin. The majority of cells cultured with this protocol maintained the epithelial phenotype and could be cultured for ~10 −12 passages and provided >5 million cells for cryopreservation, functional studies and/or to immortalize/transform in case of primary non-transformed cells. Non-epithelial cells could be easily separated from epithelial cells by flow cytometry and cultured for further evaluation of the microenvironment. In addition, tumor cells formed tumors in NSG mice thus allowing simultaneous characterization of patient-derived primary and metastatic tumor cells both in vitro and in vivo.
Materials and Methods
Creating and propagating primary cells
Fresh or cryopreserved, de-identified tissues samples were obtained from the Indiana University (IU) Simon Cancer Center Tissue Bank or Komen Tissue Bank (KTB) at IU School of Medicine, Indianapolis, after informed consent from the donors. All experiments were carried out in accordance with the approved guidelines of the Indiana University Institutional Review Board. The 804G cells were a generous gift from Dr. Rajagopal (Harvard Medical School). The 804G cells were grown in RPMI media (10–0400CV, Corning) with 10% FBS and 1% penicillin and streptomycin. Conditioned medium (CM) from the 804G cells was harvested once the cells were confluent. No more than 2–3 batches of CM were collected from one culture. The CM was filtered through 22-μm filters, divided into aliquots in 50 ml tubes and frozen at −20°C for future use. A day before the tissues were processed, 60 mm culture dishes were prepared by adding 5 ml of the thawed 804G CM followed by overnight incubation at 37°C. Just prior to use, the CM was aspirated and the dish was washed with PBS once. This CM is enriched for laminin-related components (9).
Table S1 provides details of tissues used in this study. Solid tissues dissociation protocol along with primary cell culture media has been described previously (7). The primary cell media was supplemented with 1 μM of BMP inhibitor DMH-1 (#4126, Tocris), 1 μM of TGFβ inhibitor A-83–01 (#2939, Tocris), 5 μM of ROCK inhibitor Y-27632, and 4 μl/ml of adenine (6 mg/ml stock, A9001, Sigma-Aldrich). Pleural effusion or ascites fluids were centrifuged at 1000 RPM for five minutes to collect cell pellet and the pellet was washed twice in PBS before platting on 804G CM coated plates. Medium was changed the next day to remove cellular debris and floating cells. The cells could be cultured for 10–12 passages using this method.
Flow cytometry analysis
Adherent cells were collected by trypsinization, stained using antibodies CD49f-APC (FAB13501A), CD140b-FITC (FAB1263F) (R&D Systems), PROCR (CD201)-PE (130-105-256), EpCAM-PE (130-091-253), EpCAM-APC (130-091-254) (Miltenyi Biotech Inc.), CD271-APC (345108) (Biolegend), CD44-APC (559942), CD24-PE (555428), CD73-PE (561014), CD90-APC (559869), CD166-PE (559263), JAM-1-PE (552556), MUC-1-FITC (559774) (BD Pharmingen), CD10-PE (340920) (BD Biosciences), and CD117-FITC (11-1178-42) (eBioscience), and were acquired using a BD LSR II flow cytometer. Data were analyzed using CellQuest or FlowJo software. Forward and side scatter were used to ensure that only live cells were considered in the analysis. Gating was done using appropriate FITC (555573), PE (555749) and APC (555576) (BD Pharmingen) isotype control antibodies and only a representative isotype control for two fluorescent markers are shown.
Mammosphere formation assay
Mammosphere assay in 6-well plate at a density of 5000 cells/mL has been described previously (10). Phase contrast images were taken at day 5. Mammospheres were collected, washed, and trypsinized to obtain single-cell suspensions. Cells were stained with indicated antibodies and subjected to flow cytometry.
Determination of in vitro growth of primary cells
We used the KTB200, KTB201, and KTB202 primary cells to characterize the growth kinetics and in vitro survival of primary cells in culture. Growth kinetics was evaluated from successive cell counts. 2.5 × 105 cells per cell type were seeded on 60-mm dishes at day zero and incubated under standard conditions. Cells were harvested every two days by trypsinization, counted using a hemocytometer, and 2.5 × 105 cells were reseeded for the next culture.
Culturing and characterization of cells in 3D collagen or matrigel scaffolds
Floating collagen scaffold was prepared based on a protocol from Linnermann et al., with some modifications as detailed in Kumar et al. (2,11). Matrigel basement membrane (354234, Corning) was slowly thawed on ice. 8-chambered cover-glass system (155409, Lab-Tek II) was coated with 40 μl of matrigel per well and spread evenly to cover the bottom surface. The chamber was incubated for 30 minutes at 37 °C for the gel to solidify. 400 ul of overlay media comprising primary cell media as described above with 2% matrigel and 6000 cells per well were added. The medium was changed every 3–4 days and the cells were cultured to 10 to 12 days. Immunofluorescence staining of 3D structures has been described previously (2).
RNA isolation and quantitative RT-PCR
Total RNA was isolated using RNeasy Kit from Qiagen and 1 μg of RNA was used to synthesize cDNA with Bio-RAD iScript cDNA Synthesis Kit. Quantitative RT-PCR (qRT-PCR) was performed using Taqman universal PCR mix and predesigned Taqman assay primers with best coverage from Applied Biosystems. The following assays were used in our study: ACTB (Hs01060665_g1), ESR1 (Hs01046816_m1), GATA3 (Hs00231122_m1) FOXA1 (Hs04187555_m1) and FOXC1 (Hs00559473_s1). Applied Biosystems StepOneplus real time PCR system was used for PCR and fluorescence detection and the StepOne software was used for analysis.
Immunofluorescence staining of monolayer culture
Cells were cultured in glass bottomed microwell dish (P35G-0–14-C, MatTek Corporation) and stained with KRT14 (ab7800, Abcam), KRT19 (ab52625, Abcam), and KRT17 (ab53707, Abcam) primary antibody according to protocol described previously (2). Cells were stored in PBS in dark and images were taken within 48 hours of staining with Olympus FV1000 MPE inverted confocal microscope.
Xenograft studies
The Indiana University Animal Care and Use Committee approved the use of animals in this study and all procedures were performed as per NIH guidelines. Primary cells created from pleural effusions of breast cancer or TNBCs (5×105 cells) with 50% Basement Membrane Matrix (BME) Type 3 (100 μl volume) were implanted into the mammary fat pad of 5–6 week old female NSG (NOD/SCID/IL2Rgnull) mice. A 60-day slow release estradiol (0.72 mg) was implanted at the time of mammary fat pad injection, irrespective of the injected breast cancer subtype. Tumor growth was measured weekly and tumor volume was calculated as described previously (12). After 2–5 months, lungs and primary tumors were collected and processed for hematoxylin and eosin (H&E), ERα, PR, GATA3, and FOXA1 staining.
Immunohistochemistry
H&E, ERα, PR, GATA3, and FOXA1 immunostaining was performed at the CLIA certified Indiana University Health Pathology Laboratory and the whole slide digital imaging system of Aperio (ScanScope CS) was used for imaging. The following antibodies were used: ER (clone EP1, Dako IR 084), PR (#RB-9017-P, NeoMarkers), FOXA1 (Santa Cruz sc-6553), and GATA3 (Santa Cruz sc-268).
Statistical analysis
Statistical analyses were performed using GraphPad Prism software program (version 6.0). P values below 0.05 were considered statistically significant.
Results
We have developed a method for propagating the primary cells from normal, cancerous, and metastatic tissue samples with appropriate modifications to growth media. Cells propagated from more than 30 primary breast tissues (>6 healthy donors, two BRCA1-mutants, three BRCA2-mutants with one right and left breasts of the same donor, three PALB2 high-risk, two protective alleles, tumors with adjacent normal tissues from the same patients, several triple-negative breast cancers (TNBC), three breast cancer pleural effusion fluids, four breast cancer metastasis from different organs, one liver metastasis of pancreatic cancer-neuroendocrine subtype, and three ascites fluids of ovarian cancer patients) were analyzed (Table S1). These extensive analyses recognized enormous phenotypic heterogeneity in the normal breast and tumors.
Normal breast contains multiple subpopulations of cells
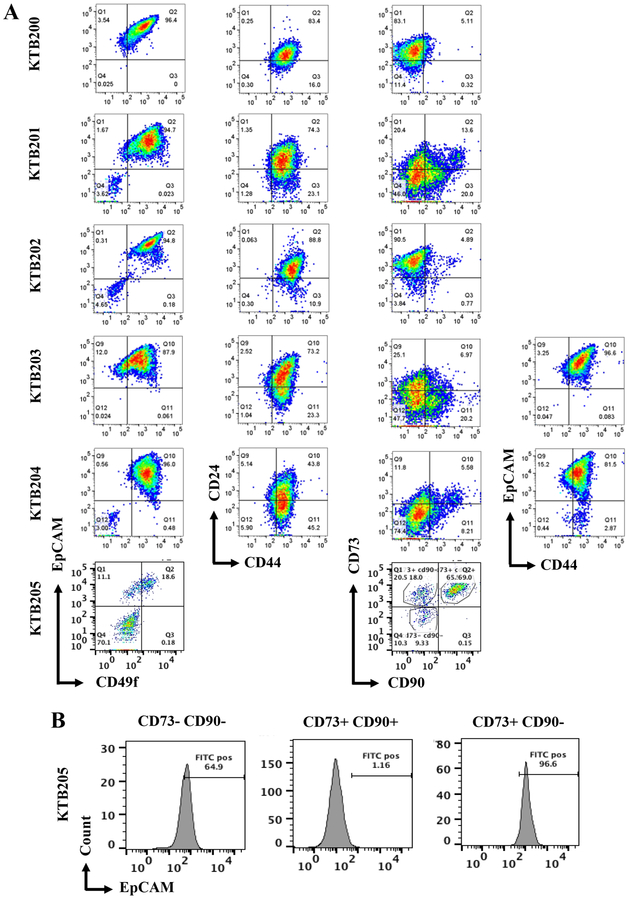
We subjected primary breast cells to phenotypic analyses using established and recently discovered markers that define the normal and cancer stem cells (CSCs), pluripotent somatic stem cells, progenitor, differentiated, luminal, basal/myoepithelial cells, and ERα-positive subpopulation of cells (2,13,14). Similar to results obtained with cells propagated using the previous epithelial reprogramming assay (7), this new culturing method enabled us to document enormous phenotypic heterogeneity in the normal breast. The expanded cell populations are described as CD49f+/EpCAM- stem/basal-enriched cells, CD49f+/EpCAM+ luminal progenitor cells, CD49f-/EpCAM+ mature luminal cells, CD44+/CD24- stem/basal cells, CD44+/CD24+ stem/luminal progenitor cells, CD44-/CD24+ differentiated cells, CD201+/EpCAM- multi-potent stem cells, CD271+ rare basal cells in luminal breast cancer (relevant in anti-estrogen resistance models), CD73+/EpCAM+/CD90- rare endogenous pluripotent somatic stem cells, CD73+/CD90+/EpCAM- potential mesenchymal stem cells, CD10+/EpCAM- basal/myoepithelial cells, ALCAM (CD166)+ cells that are enriched for ERα, and JAM-A+ cells, which are enriched for cancer stem cell phenotype (15–17). Flow cytometry characterization of breast epithelial cells from six healthy women (KTB200–205) showed remarkable variability in stem/basal, progenitor and mature/differentiated cell subpopulations (Figure 1A and Figure S1). All samples were from pre-menopausal women (20–55 age range, 23–41 BMI range, Table S1). Using a combination of CD44 and EpCAM, we were able to document the presence of luminal (CD44-/EpCAM+ or CD44low/EpCAM+) and basal (CD44+/EpCAMlow) cells (18). Proportion of cells that express the cell adhesion molecule MUC1 also varied between samples (Figure S1). MUC1 is expressed predominantly in CD24+ luminal cells and these cells often display higher self-renewal capacity and are metastatic (19,20). With respect to rare pluripotent somatic stem cells (13), our technique expanded this population, as a significant portion of CD73+/CD90- cells was EpCAM+ (Figure 1B). Cells propagated using this assay are free of fibroblasts as they lacked the expression of CD140b, which identify fibroblasts (21) (Figure S1). We also did not find cells expressing CD117, a putative marker of luminal progenitor cells (22) (Figure S1). These results suggest inter-individual heterogeneity among the normal breasts, particularly in number of breast epithelial cells expressing markers that identify cells as stem, basal or luminal cells, similar to our previous report (7). However, as we indicated in our previous report, because of limited sample size, the observed heterogeneity cannot be attributed to differences in age, BMI or time of sample collection (luteal or follicular phase).
Figure 1:
Breast epithelial cells from the normal breast contain multiple cell types and show inter-individual heterogeneity. A) Breast epithelial cells isolated from the normal breasts of five clinically healthy women show inter-individual differences in staining patterns with CD49f, CD44, CD24, CD90, CD73, and EpCAM antibodies. B) The culturing method enriches for rare endogenous pluripotent somatic stem cells based on CD73/CD90/EpCAM staining patterns. EpCAM expression was seen in greater than 96% of CD73+/CD90- cells in KTB205.
We performed serial dilution cell proliferation assay to determine the number of passages these cells can be maintained. Proliferation kinetics varied from samples to samples and declined by around passage 7 (Figure S1). However, by the time cells reached passage 7, couple of million cells could be easily obtained for immortalization and functional studies.
Breast epithelial cells from women carrying high-risk mutations:
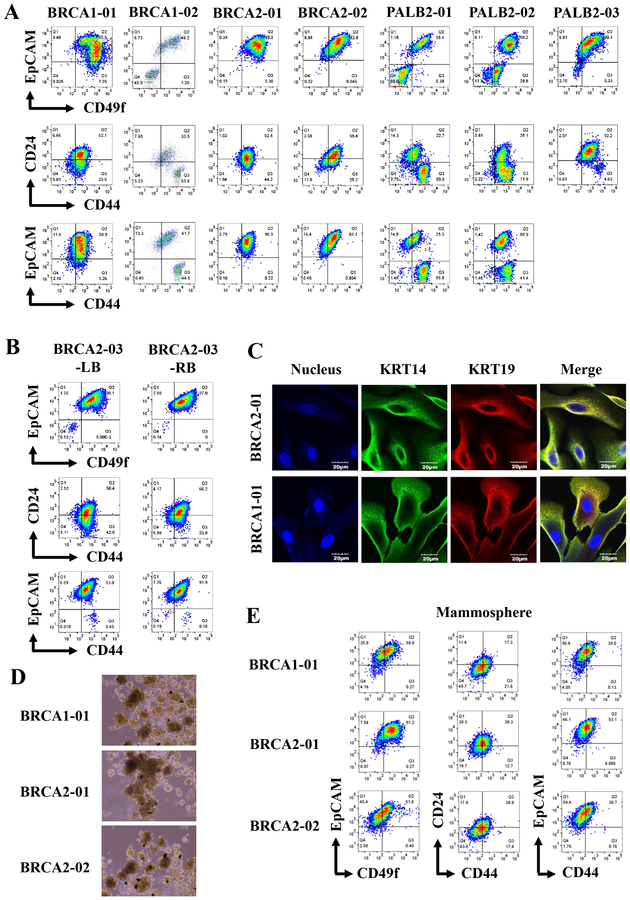
To evaluate the suitability of the assay for growing cells from high-risk women, we created primary cells from prophylactic mastectomy or biopsies of women carrying germline mutations in BRCA1 (two cases), BRCA2 (three cases, one with tissues from both breasts) or PALB2 (three cases). Staining with multiple antibody combinations did not show any patterns that are distinct from the cells cultured from healthy breasts (Figure 2A and Figure S2). It is also remarkable that inter-individual differences in staining patterns were noted among high-risk patients analyzed in the study (Figure S2). For example, CD271/EpCAM, CD10/EpCAM and MUC1/EpCAM staining patterns of two BRCA2 mutants and three PALB2 mutants were different (Figure S2). Thus, it is unlikely that the breasts of high-risk women contain phenotypically distinct population of cells compared to breasts of healthy women. In general, growing epithelial cells from women with PALB2 and CHEK2 mutations was more difficult than others because of outgrowth of cells with fibroblastic appearance or epithelial cells undergoing rapid epithelial to mesenchymal transition (EMT).
Figure 2:
Breast epithelial cells with luminal progenitor characteristics can be grown from breast cancer high-risk carriers. A) Breast epithelial cells from BRCA1, BRCA2, and PALB2 mutation carriers show variable subpopulation of cells based on CD49f/EpCAM, CD44/CD24 and CD44/EpCAM staining patterns. B) Breast epithelial cells propagated from randomly selected regions from two breasts of a BRCA2 carrier show similar phenotypes based on CD49f/EpCAM, CD44/CD24 and CD44/EpCAM staining patterns. C) BRCA1 and BRCA2 mutant breast epithelial cells are enriched for KRT14+/KRT19+ luminal progenitor cells as determined by immunofluorescence. D). BRCA1 and BRCA2 mutant cells form mammospheres. E) Cells in mammospheres show different degree of differentiation compared to cells grown in 2D culture. For example, CD49f-/EpCAM+ cells, which are considered differentiated luminal cells, increased under mammosphere condition compared to 2D culture as shown in A.
Cells with BRCA1 mutations, similar to cells from healthy women, formed acini on collagen gel and these acinis were composed of KRT5/6 and KRT17+ cells (Figure S3). We compared the staining pattern of cells from both the breasts of a BRCA2 carrier to ensure that observed staining pattern is not due to collection of biopsy from specific regions of the breast. Randomly collected breast tissues from both the breasts showed similar cell surface marker profiles (Figure 2B and Figure S3). Immunofluorescence with basal cell marker KRT14 and luminal cell marker KRT19 showed that the majority of cells from high-risk women are hybrid cells that expressed both KRT14 and KRT19 (Figure 2C), which is a suggested characteristic of cells with luminal progenitor phenotype (23).
We had previously demonstrated that immortalized cell lines derived from healthy breast self-renew as well as undergo differentiation when cultured under mammosphere growth conditions (2). To determine whether cells from high-risk women differ in their capacity to undergo self-renewal and differentiation, we subjected cells from BRCA1 and BRCA2 carriers to mammosphere assay followed by flow cytometry (Figure 2D and E). Phase contrast images of these mammospheres showed similarity between samples. However, as noted previously with cells from healthy breasts (2), inter-individual variability in differentiation capacity was noted with cells from high-risk women. For example, the proportion of cells with differentiated luminal phenotype (CD49f-/EpCAM+ or CD44-/EpCAM+ cells) increased under mammosphere condition compared to 2D growth condition but the degree of increase varied between samples (Figure 2A and E). Staining patterns of mammospheres with additional markers are shown in Figure S3.
Cells grown from women with protective alleles show enhanced differentiation capacity:
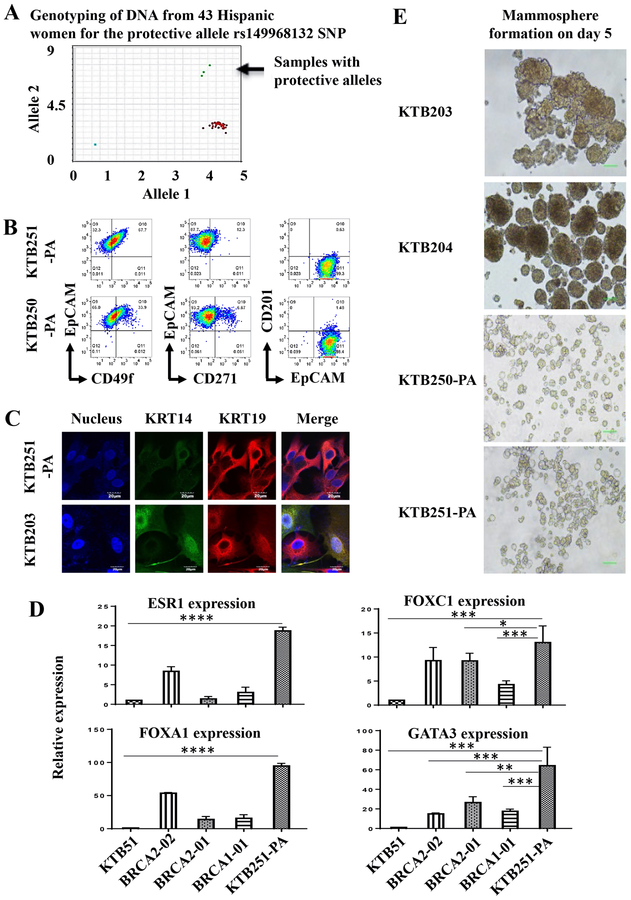
Although epidemiologic and genome-wide association studies have identified various risk, predisposition, and protective alleles including new 65 risk loci for breast cancer (24,25), functional analyses of these risk loci have been difficult because of limited cell resources. To determine whether culture system developed here is useful in this regard, we focused on a recently described breast cancer protective variant in Latinas on chromosome 6q25 located 5’ of Estrogen Receptor 1 (ESR1) gene, which is limited to Indigenous American Ancestry (26). We examined DNA from self-reported 43 Hispanic women from whom cryopreserved normal breast tissues were available at the Komen Tissue Bank (KTB). Among these women, three carried the protective allele (Figure 3A) but viable cells could be generated from two samples. These cells were less proliferative than cells generated from healthy or high-risk women and only limited number of experiments could be performed. Cell surface markers profiles showed clear enrichment of cells with differentiated/luminal characteristics (Figure 3B and Figure S4A and B). For example, unlike in cases of healthy or high-risk women, the majority of cells from women with protective alleles were CD49f-/EpCAM+, CD271-/EpCAM+ and CD201-/EpCAM+. In addition, these cells had higher proportion of cells that were KRT14-/KRT19+ compared to cells from non-carriers of protective alleles or high-risk individuals (Figure 3C), and were KRT17+ (Figure S4B). Furthermore, cells from one protective allele carrier, which we were able to grow for additional analyses, expressed higher levels of luminal cell markers ESR1, FOXA1, and GATA3 compared to cells from normal (KTB51) and high-risk individuals (Figure 3D). In mammosphere assays, cells with protective alleles generated very small spheres compared to cells generated from two Hispanic women not carrying the protective allele (Figure 3E). Thus, the assay established here can be used to characterize cells from women carrying distinct risk loci.
Figure 3:
Breast epithelial cells propagated from women with breast cancer protective alleles show differentiated characteristics. A) Distribution patterns of the breast cancer protective SNP rs140068132 in 43 self-reported Hispanic women. B) Breast epithelial cells from women with protective alleles show higher number of CD49f-/EpCAM+, CD271-/EpCAM+ and CD201-/EpCAM+ differentiated cells. C) Cells with protective alleles are predominantly KRT14-/KRT19+ differentiated cells compared to cells from another Hispanic women not carrying protective allele (KTB205). KTB205 contained both KRT14+/KRT19+ and KRT14-/KRT19+ cells. D) Cells with protective alleles express higher levels of luminal genes such as ESR1, FOXA1, and GATA3 compared to non-carriers (KTB51) as well as BRCA1 and BRCA2 mutant carriers. mRNA levels measured through qRT-PCR are shown. Differences in expression between non-carrier (KTB51) and protective allele carrier (KTB251-PA) with all genes are statistically significant (p<0.05, indicated by asterisks). Similarly, differences in expression between high-risk carriers and protective allele carrier are also significant. E). Breast epithelial cells from protective carriers generate small mammospheres compared to breast epithelial cells from Hispanic women without the protective allele.
Individualizing tumor characterization through propagation of tumor and adjacent normal from the same patient.
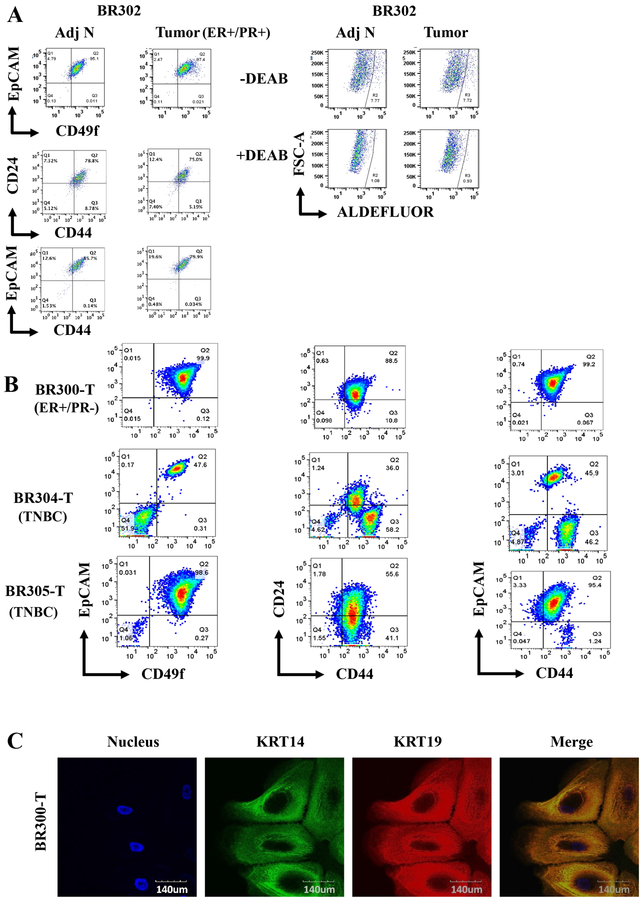
Since gene expression in normal tissues shows inter-individual variations due to single nucleotide polymorphism in the gene regulatory regions (27), we had recently proposed that normal cells from the same patient need to be used to determine cancer specific gene expression changes (28). Although the use of cells from normal tissues adjacent to tumor is far from perfect because of cancer-induced field defects on adjoining cells (29,30), it is still better to use those cells or from the unaffected contralateral breast than normal cells from unrelated individuals as controls to identify cancer-specific gene expression changes. Towards achieving this goal, we established culture conditions to grow tumor and tumor adjacent normal from the same patient. Representative flow cytometry profiles with various markers are shown in Figure 4A and Figure S4C and D. Both tumor and the adjacent normal showed similar marker profiles except that tumors had a minor CD49fhigh/EpCAM+ population. Cells derived from not all tumors that were characterized showed similar profiles as samples from triple negative breast cancers (TNBCs) showed a distinct profile than the sample described above (Figure 4B and Figure S4D). Tumor cells were KRT14+/KRT19+ suggesting their luminal progenitor characteristics, similar to flow cytometry stain pattern (Figure 4C). We have created a bank of tumor cells from at least 15 patients with few cases of paired tumor and tumor adjacent normal, which could be utilized for functional studies.
Figure 4:
Characteristics of cells propagated from tumors and tumor adjacent normal cells. A) ER+/PR+ tumor cells and tumor adjacent normal cells of the same patient show limited heterogeneity. B) Distinct profiles of tumor cells propagated from an ER+/PR- tumor and two TNBCs. C) KRT14 and KRT19 staining patterns of ER+/PR- tumor cells. Based on merged staining pattern, KRT19 staining appears dominant over KRT14.
Tumor cells form ductal and acinar-like structures on collagen gels:
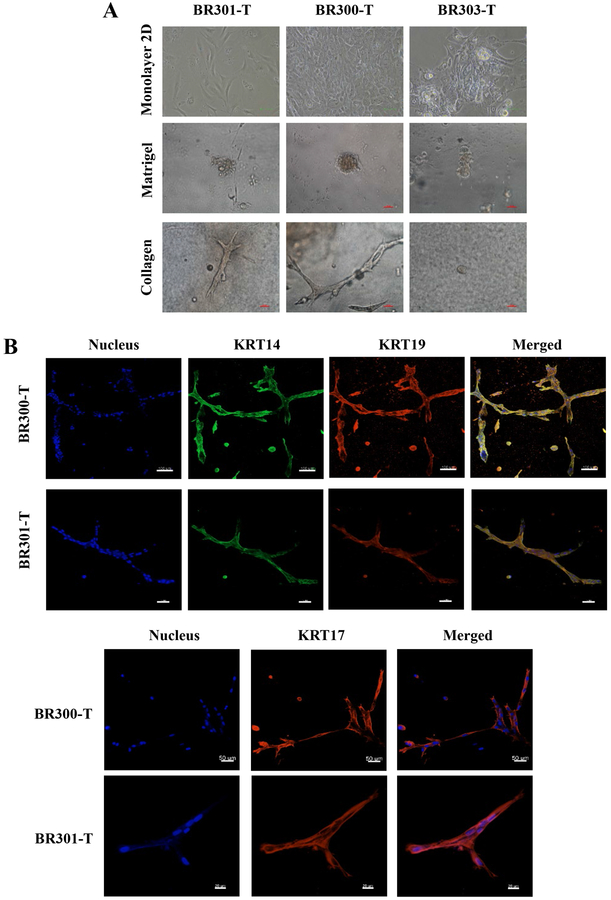
Since organoid culture system is gaining traction for various types of studies including screening for anti-cancer therapies (31), we determined whether tumor cells form organoid-like structures in 3D. On matrigel, tumor cells formed spheres, whereas on collagen gels, cells formed elongated structures that resembled breast ducts (Figure 5A). Immunofluorescence of collagen gel structures with KRT14, KRT17 and KRT19 showed these ducts are KRT17+ and are lined by KRT14+/KRT19+ double positive cells (Figure 5B). Most breast cancers are believed to originate from luminal progenitor cells, which are typically KRT14+/KRT19+ (23).
Figure 5:
Tumor cells form acinar structures on matrigel and ductal structures on collagen gel. A) Phase contrast images of tumor cells on monolayer, matrigel, and collagen gel. B) KRT14, KRT17, and KRT19 staining patterns of ductal structures on collagen gel.
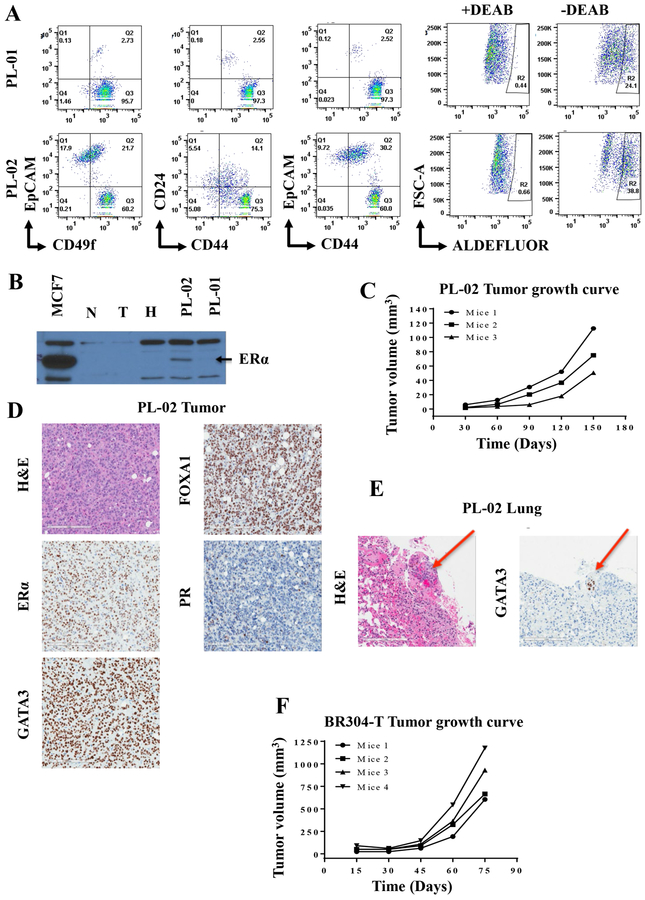
Breast cancer cells propagated from pleural effusions develop estrogen receptor alpha (ERα)-positive tumors in NSG mice.
Although it has proven extremely difficult to cultivate breast epithelial cells that express ERα, there have been few recent advances in culture methods that enabled propagation of ERα-positive cells (2,14). Similarly, patient-derived tumor xenograft (PDX) models heavily favor ERα-negative breast cancers relative to ERα+ PDXs (32). In addition, very rarely, tumor cells and PDXs have been created from the same tumors. Since we had >90% success rate in generating cells from tumors, we next investigated whether cells derived from breast tumors or pleural effusions can create tumors in NSG mice. Detailed characteristics of tumor cells derived from pleural effusions developed from two ERα+ tumors are described in Figure 6A and Figure S5. Flow cytometry patterns of cells grown from two pleural effusions were different with pleural effusion #2 containing higher proportion of EpCAM+ cells. Cells from both pleural effusions contained significant levels of ALDEFLUOR+ cancer stem cells (33). Western blotting showed expression of ERα in effusion #2, although very low levels of ERα were detectable in the effusion #1 (Figure 6B). Cells derived from effusion #2 generated tumors in the mammary fat pad of NSG mice by two months post-implantation (Figure 6C), which expressed all three markers of luminal breast cancer- ERα, FOXA1, and GATA3 but lacked progesterone receptor (PR), indicating luminal B characteristics (Figure 6D). A lung metastatic nodule, which expressed GATA3, was also detected (Figure 6E). We have generated tumors from primary TNBC cells, which could be re-implanted into additional animals similar to PDX models (Figure 6F). Whole genome sequencing of these TNBC cells showed mutation patterns observed in breast tumor samples (data not shown). Thus, the method described here provides resources of patient tumor-derived primary cells and tumors from these cells for functional studies.
Figure 6:
Tumorigenic properties of tumor cells. A) Flow cytometry staining patterns of cells propagated from pleural effusions of patients who had ER+ breast cancer. B) ERα expression pattern in MCF7 cells (positive control), and cells from tumor adjacent-normal (N), tumor (T), KTB normal (H) and two pleural effusions. C) Growth characteristics of tumors developed in the mammary fat pad of NSG mice injected with cells from pleural effusion 2 (PL-02). D) H&E staining and luminal marker expression patterns in PL-02. E) PL-02 -derived tumor metastasized to lungs (red arrow), which expressed GATA3. F) Growth characteristics of tumors derived from TNBC cells.
Propagation of cells derived from liver metastasis and from other cancers
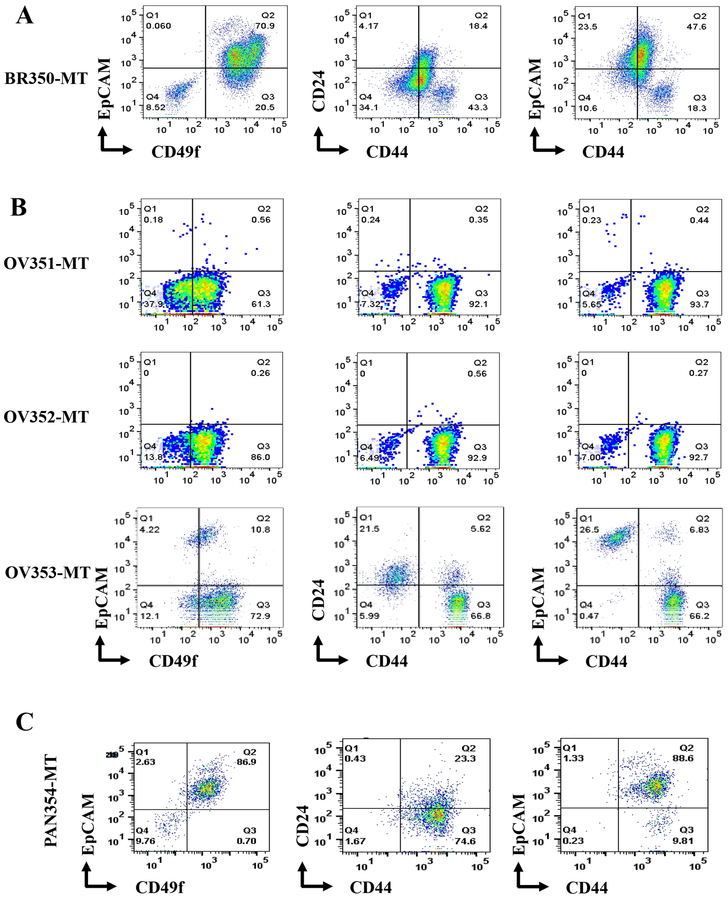
Metastasis is the main cause of cancer death and several studies have shown distinct differences between primary tumors and metastasis (34,35). We had previously shown that epithelial reprogramming assay could be used to grow and perform genomics of liver metastasis of various cancers (36). However, because mouse embryonic feeder layer cells were needed to grow cancer cells, sorting of cancer cells by flow cytometry was required before sequencing, and bioinformatics analyses of genomic data required additional steps to ensure that sequence reads are not from residual contaminating mouse DNA. To determine whether metastatic cells grown under reprogramming growth conditions can adapt to the current method, we transferred cells from a liver metastasis of a breast cancer grown initially under reprogramming assay to the new method. Indeed, cells readily adapted to the new method and a significant number of cells could be obtained for phenotypic characterization (Figure 7A and Figure S5). The majority of cells were CD49f+/EpCAM+, Jam-A+/EpCAM+ and CD271+/EpCAM+.
Figure 7:
Characterization of cells propagated from sites of metastasis of different cancers. A) Flow cytometry profiles of liver metastasis of a breast cancer. B) Flow cytometry profiles of cells propagated from ascites fluids of three patients with ovarian cancer. C) Flow cytometry profiles of liver metastasis of a pancreatic cancer (neuroendocrine).
We extended this method to propagate cells from other cancer types. Cells from ascites fluid of ovarian cancer patients (OV351-MT to OV353-MT) and liver metastasis of pancreatic cancer-neuroendocrine subtype were generated and characterized with various cell surface markers. Cells from ascites fluid of ovarian cancer patients displayed the phenotypic heterogeneity and exhibited predominant CD49f+/EpCAM-, CD44+/CD24-, CD44+/EpCAM-, CD73+/CD90+, CD201+/EpCAM-, CD166+/EpCAM, and CD10+/EpCAM- phenotypes (Figure 7B and Figure S6). However, it is likely that few of the cell types that do not express EpCAM are non-cancer stromal cells. Nonetheless, the method permitted propagation of EpCAM+ cells, which can be sorted by flow cytometry for functional studies.
The cells from liver metastasis of pancreatic cancer-neuroendocrine subtype contained predominantly CD49f+/EpCAM+, CD44+/CD24low, CD44+/EpCAM+, CD73+/CD90-, CD201low/EpCAM+, CD271-/EpCAM+, CD10-/EpCAM+, CD166+/EpCAM+, CD117-/EpCAM+, MUC-1-/EpCAM+, JAM-1+/EpCAM+, and CD140b-/EpCAM+ subpopulations (Figure 7C and Figure S6). A fraction of these cells were CXCR4-positive, which is expressed only in metastatic cancer stem cells of pancreatic cancer and is required for metastasis (37). Taken together, detailed characterization of various cells from different sources documents that our propagating method is suitable for growing primary cells, establishing heterogeneity of cancer cells in tumors, and provides a replenishable source of live primary cells for functional studies.
Discussion
Functional modeling of breast epithelial hierarchy and stromal-epithelial cell interactions have been difficult due to inability to obtain sufficient stem-progenitor-mature epithelial cells and stromal cells. Establishment of such techniques for modeling of the normal breast in vitro is critical if progress made in identifying various risk and protective alleles of cancers to have an impact in clinic. Similarly, further advancement is needed to translate knowledge gained through single cell sequencing technologies, which are currently descriptive cataloguing of different cell types in primary and metastatic tumors without allowing functional evaluation of different cell types (35). In this study, we report in vitro propagation method for primary cells from different sources including breast, ovary, and metastasis. These cells are suitable for “omics” studies as TNBC cells from one of the patients have already been sequenced and found to have mutations enriched in breast cancers and additional samples are currently being sequenced (data not shown). Most importantly, sequence information can be obtained when primary tissue is very limited. We expect these tumor-derived cells to be also compatible for proteomics and metabolomics studies with and without prior exposure to chemotherapeutics. Other advantages of the method are that it is relatively inexpensive, adaptable in any labs and cancer cells can be grown from cryopreserved tissues. We have found that cryopreserving tissues with ROCK inhibitor yields higher number of live cells (7). Although continuous passage leads to drift in proportion of different cell types, the original cell types remain after passage. Samples with disproportionately higher number of fibroblast-like cells compared to epithelial-like cells within a week of initiating culture, which can be identified through phase contrast microscopy, show a higher tendency of drift to non-epithelial (EpCAM-) cells with continuous passage. Differential trypsinization (two minutes at room temperature) or flow cytometry sorting can be used to enrich epithelial cells in culture. The only caveat of this modification is that cancer cells that have undergone EMT would get excluded from functional studies.
Single cell studies are gaining popularity for better understanding of heterogeneity of cell types in an adult organ. Fifteen and three epithelial cell types have been described in the mouse mammary gland and the human breast, respectively (38,39). Since the assay described here allows propagation of cells from a core biopsy and cells form distinct structures on a 3D matrix, functional studies of cells from different clusters can now be performed with and without addition of stromal cells. Additional advantage of this system is the availability of cells that carry breast cancer risk alleles. Depending on the type of risk alleles, these cells can be easily immortalized with human telomerase. While we could easily immortalize cells with BRCA1or BRCA2 mutations, generating cell lines from PALB2 and CHEK2 carriers as well as those with protective alleles has so far proved difficult. Phenotypic analyses using ~10 cell surface marker combinations did not reveal unique features of cells obtained from high risk individuals, which is bit surprising considering a previous study, which demonstrated enrichment of cells with luminal progenitor properties in the breast of BRCA1 mutant carriers (17). However, we were able to document enhanced differentiation properties of cells obtained from women who carry breast cancer protective alleles, which clearly indicates the role of proper differentiation in protecting against breast cancer.
We had previously demonstrated that short-term culturing followed by sequencing of cultured tumor cells could detect functionally important mutations that are otherwise undetectable upon direct sequencing of tumors (36). Culturing for short duration as such did not introduce mutations. Culturing method used at that time required propagation of tumor cells on a mouse fibroblast feeder layer and purifying tumor cells by flow cytometry before sequencing. The new method described here eliminates those requirements. Despite TNBCs displaying higher rate of mutations than other breast cancer subtypes (40) and data from many samples already available, very few are detected across multiple TNBCs and very rarely the impact of these mutations using the same tumor material could be studied. In general, cancers believed to contain 2–8 “driver” mutations and 30–60 protein-coding changes in passenger genes that alter cellular functions (41). We hope that the assay described here would not only enhance detection of additional mutations but also evaluation of presumed driver and passenger mutations in future. Furthermore, new mutations detected from these cell lines can be incorporated for screening of distant metastasis using ctDNA technology (42). In this respect, tumor cells generated from our culturing method form unique structures under 3D growth conditions and develop tumors in NSG mice, similar to a direct PDX model. Availability of millions of cancer cells from patients would permit further refinement of these models including characterizing growth properties of specific subclones with or without co-culture with stromal cells, in vitro high throughput drug screening on an individual patient basis and further validation of the effective drugs in xenograft models. Sufficient cells can be obtained within a month of initiating culture for rapid in vitro screening for drug sensitivity. In this respect, a recent study that utilized single cell genomics and TNBC samples from patients receiving neoadjuvant therapy concluded that drug resistant genotypes are pre-existing in tumors and adaptively selected during neoadjuvant therapy (43). Relatively inexpensive way to generate cells from TNBCs described here would enable identification and characterization of such pre-existing drug resistant clones. While breast cancer research community has placed significant emphasis on finding cure for TNBCs, similar degree of attention is also needed for ERα-positive breast cancer because of higher number of patients with this disease type and its late recurrence (44). Recent studies have identified set of genes that predict early and late recurrence (45). However, developing in vitro models with patient-derived cells to functionally characterize genes in the signature has proven to be a challenge. Our assay has overcome this challenge as the cultured cells derived from recurrent tumors maintain ERα and one of our future goals is to establish cultures from metastasis that carry mutations in ERα, which are frequently observed in metastasis but not in primary tumors (46). We do acknowledge that, similar to PDX model (47), clonal selection of different tumor cells during culturing is one of the limitations of this assay. Nonetheless, simplicity of the assay allows propagation of cells from multiple cancers and serves as a resource for validation of pan-cancer genomic signatures including DNA methylation and chromatin accessibility identified through bioinformatics analyses (48–50).
Supplementary Material
Implications:
Our method expands the scope of individualized studies of patient-derived cells and provides resources to model epithelial-stromal interactions under normal and pathological conditions.
Acknowledgements:
We thank Dr. Rajagopal from Harvard Medical School of 804G cell line. We thank the members of IU Simon Cancer Center tissue procurement facility, flow cytometry core, clinical trial office, IU School of Medicine immunohistochemistry core, Komen Tissue Bank for their service and countless number of women who donated their breast tissue for research purpose as well as volunteers who facilitated tissue collection. Department of Defense DOD-W81XWH-15-1-0707, Susan G Komen for the Cure SAC110025, 100 Voices of Hope and R03CA195250–01A1 (to HN) funded this study. Susan G. Komen for the Cure, Breast Cancer Research Foundation and Vera Bradley Foundation for Breast Cancer Research and the Catherine Peachy Foundation provide funding support to the Komen Normal Tissue Bank.
List of abbreviations:
- ALCAM
activated leukocyte cell adhesion molecule
- BME
basement membrane extract
- BMP
bone morphogenic extract
- BRCA
breast and ovarian cancer susceptibility protein 1
- CHEK2
checkpoint kinase 2
- CLIA
clinical laboratory improvement amendments
- CM
conditioned medium
- ctDNA
circulating tumor DNA
- C-x-C
chemokine receptor type 4
- DMSO
dimethylsulfoxide
- EMT
epithelial to mesenchymal transition
- ER
estrogen receptor
- FBS
fetal bovine serum
- KRT
keratin
- KTB
Komen tissue bank
- MUC1
mucin 1
- NSG
NOD/SCID/IL2Rgnull
- PALB
partner and localizer of BRCA
- PBS
phosphate buffered saline
- PDX
patient derived xenograft
- PR
progesterone receptor
- ROCK
Rho-associated kinase
- TGFβ
transforming growth factor
- TNBC
triple negative breast cancer
Footnotes
The authors declare no potential conflicts of interest
References:
- 1.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 2014;28(11):1143–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar B, Prasad MS, Bhat-Nakshatri P, Anjanappa M, Kalra M, Marino N, et al. Normal breast-derived epithelial cells with luminal and intrinsic subtype-enriched gene expression document inter-individual differences in their differentiation cascade. Cancer Res 2018;78(17):5107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 2012;180(2):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, Qiu Y, Zeng X, Ding Y, Zeng J, Lu K, et al. Effect of a feeder layer composed of mouse embryonic and human foreskin fibroblasts on the proliferation of human embryonic stem cells. Exp Ther Med 2016;11(6):2321–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimmino L, Neel BG, Aifantis I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol 2018;28(9):698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, et al. Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 2016;19(2):217–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakshatri H, Anjanappa M, Bhat-Nakshatri P. Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Scientific reports 2015;5:13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 2018;361(6406):1033–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langhofer M, Hopkinson SB, Jones JC. The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. J Cell Sci 1993;105 (Pt 3):753–64. [DOI] [PubMed] [Google Scholar]
- 10.Bhat-Nakshatri P, Goswami CP, Badve S, Sledge GW Jr., Nakshatri H. Identification of FDA-approved drugs targeting breast cancer stem cells along with biomarkers of sensitivity. Scientific reports 2013;3:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linnemann JR, Miura H, Meixner LK, Irmler M, Kloos UJ, Hirschi B, et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development 2015;142(18):3239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Kishimoto H, Chua HL, Badve S, Miller KD, Bigsby RM, et al. Interleukin-1 alpha promotes tumor growth and cachexia in MCF-7 xenograft model of breast cancer. Am J Pathol 2003;163(6):2531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Gascard P, Dumont N, Zhao J, Pan D, Petrie S, et al. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc Natl Acad Sci U S A 2013;110(12):4598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridriksdottir AJ, Kim J, Villadsen R, Klitgaard MC, Hopkinson BM, Petersen OW, et al. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nature communications 2015;6:8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lathia JD, Li M, Sinyuk M, Alvarado AG, Flavahan WA, Stoltz K, et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell reports 2014;6(1):117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CH, Hale SJ, Cox CV, Blair A, Kronsteiner B, Grabowska R, et al. Junctional Adhesion Molecule-A Is Highly Expressed on Human Hematopoietic Repopulating Cells and Associates with the Key Hematopoietic Chemokine Receptor CXCR4. Stem Cells 2016;34(6):1664–78. [DOI] [PubMed] [Google Scholar]
- 17.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 2009;15(8):907–13. [DOI] [PubMed] [Google Scholar]
- 18.Skibinski A, Breindel JL, Prat A, Galvan P, Smith E, Rolfs A, et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell reports 2014;6(6):1059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res 2010;16(3):876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget 2014;5(9):2622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewitt KJ, Shamis Y, Knight E, Smith A, Maione A, Alt-Holland A, et al. PDGFRbeta expression and function in fibroblasts derived from pluripotent cells is linked to DNA demethylation. J Cell Sci 2012;125(Pt 9):2276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Villadsen R. Expression of Luminal Progenitor Marker CD117 in the Human Breast Gland. J Histochem Cytochem 2018:22155418788845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol 2007;177(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman N Realizing the promise of cancer predisposition genes. Nature 2014;505(7483):302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005;436(7051):720–4. [DOI] [PubMed] [Google Scholar]
- 26.Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nature communications 2014;5:5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501(7468):506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anjanappa M, Cardoso A, Cheng L, Mohamad S, Gunawan A, Rice S, et al. Individualized Breast Cancer Characterization through Single-Cell Analysis of Tumor and Adjacent Normal Cells. Cancer Res 2017;77(10):2759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teschendorff AE, Gao Y, Jones A, Ruebner M, Beckmann MW, Wachter DL, et al. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nature communications 2016;7:10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakshatri H, Kumar B, Burney HN, Cox ML, Jacobsen M, Sandusky GE, et al. Genetic ancestry-dependent differences in breast cancer-induced field defects in the tumor-adjacent normal breast. Clin Cancer Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takebe T, Wells JM, Helmrath MA, Zorn AM. Organoid Center Strategies for Accelerating Clinical Translation. Cell Stem Cell 2018;22(6):806–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittle JR, Lewis MT, Lindeman GJ, Visvader JE. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res 2015;17:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007;1(5):555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter KW, Amin R, Deasy S, Ha NH, Wakefield L. Genetic insights into the morass of metastatic heterogeneity. Nat Rev Cancer 2018;18(4):211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol 2018;20(12):1349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anjanappa M, Hao Y, Simpson ER, Bhat-Nakshatri P, Nelson JB, Tersey SA, et al. A system for detecting high impact-low frequency mutations in primary tumors and metastases. Oncogene 2018:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1(3):313–23. [DOI] [PubMed] [Google Scholar]
- 38.Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nature communications 2017;8(1):2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nature communications 2018;9(1):2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017;31(2):172–79. [DOI] [PubMed] [Google Scholar]
- 43.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018;173(4):879–93 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 2017;377(19):1836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buus R, Yeo B, Brentnall AR, Klintman M, Cheang MCU, Khabra K, et al. Novel 18-gene signature for predicting relapse in ER-positive, HER2-negative breast cancer. Breast Cancer Res 2018;20(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer discovery 2017;7(3):277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015;518(7539):422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell reports 2018;25(4):1066–80 e8. [DOI] [PubMed] [Google Scholar]
- 49.Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362(6413). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhawan A, Scott JG, Harris AL, Buffa FM. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nature communications 2018;9(1):5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.