Abstract
Heart failure occurs in over 30% of the worldwide population and most commonly originates from cardiovascular diseases such as myocardial infarction. microRNAs (miRNAs) target and silence specific mRNAs, thereby regulating gene expression. Because the endogenous miR-155-5p has been ascribed to vasculoprotection, loading it onto positively charged, core-shell poly(isobutylcyanoacrylate) (PIBCA)-polysaccharide nanoparticles (NPs) was attempted. NPs showed a decrease (p < 0.0001) in surface electrical charge (ζ potential), with negligible changes in size or shape when loaded with the anionic miR-155-5p. Presence of miR-155-5p in loaded NPs was further quantified. Cytocompatibility up to 100 μg/mL of NPs for 2 days with human coronary artery endothelial cells (hCAECs) was documented. NPs were able to enter hCAECs and were localized in the endoplasmic reticulum (ER). Expression of miR-155-5p was increased within the cells by 75-fold after 4 hours of incubation (p < 0.05) and was still noticeable at day 2. Differences between loaded NP-cultured cells and free miRNA, at days 1 (p < 0.05) and 2 (p < 0.001) suggest the ability of prolonged load release in physiological conditions. Expression of miR-155-5p downstream target BACH1 was decreased in the cells by 4-fold after 1 day of incubation (p < 0.05). This study is a first proof of concept that miR-155-5p can be loaded onto NPs and remain intact and biologically active in endothelial cells (ECs). These nanosystems could potentially increase an endogenous cytoprotective response and decrease damage within infarcted hearts.
keywords: myocardial infarction, cytoprotection, microRNA, nanoparticle, endothelial cell
Introduction
Heart failure occurs in over 30% of the worldwide population1 and is a leading cause of death and disability in developing countries. It most commonly arises from coronary artery disease that causes severe impairment of the coronary artery blood supply.2, 3, 4, 5 The sudden complete occlusion of coronary vessels leads to myocardial infarction (MI), with the resulting hypoxia triggering a cascade of events that culminates in loss of viable cardiac tissue.3, 4, 6 If early reperfusion occurs,4, 7 damaged tissue is replaced with scar tissue.4 However, after 20–40 min of sustained severe ischemia, irreversible tissue damage typically develops,4 ultimately leading to heart failure.3, 5 The immediate clinical approach is to restore blood flow by thrombolytic treatments or coronary angioplasty,2, 5 although long-term complications and high death rates remain frequent,3, 8 particularly for the most severe cases.4 Consequently, developing new therapeutic strategies is a vital goal of both clinical and research fields.3, 9, 10
Nanoparticles (NPs) in suspension can be easily injected,11 and using an NP injection as a complement to reperfusion after MI would be feasible. With sizes above the renal clearance threshold (about 5 nm in diameter12), NPs can circulate within the body and deliver biologically active molecules into the target tissue.11 Interestingly, biological entities such as microRNAs (miRNAs) can act intracellularly or can be secreted by cells and contribute to intercellular or cell-tissue communication.13 miRNAs are evolutionarily conserved, ubiquitously expressed (albeit with a degree of tissue specificity), short (∼22 nucleotides), non-protein-coding RNA molecules involved in post-transcriptional gene regulation. At the target, miRNAs can generate strong, sustained, and comprehensive biological effects, through endogenous gene silencing,14 with very low concentrations (at least 0.5 nM)15. In addition, miRNAs affect multiple pathways and cellular processes rather than specific targets.16
miRNA-based therapies include the delivery of exogenous miRNA mimics, which are oligonucleotides with the same nucleotide sequence as an endogenous miRNA designed to mimic that miRNA.17 After the miRNAs are loaded onto the NPs, an intravenous route of administration18 would be the least invasive option. Different therapeutic candidate miRNAs have been described,3, 19, 20, 21, 22 and different NPs for miRNA delivery are now being explored as potential treatments for cardiovascular pathologies—namely, viral,23, 24, 25, 26, 27, 28, 29, 30 lipid,31, 32, 33, 34 and polymer-based35, 36, 37 platforms.
miR-155-5p mimics have been shown to silence the mRNA BACH1 in vitro in endothelial cells (ECs)38 and to display cytoprotective effects during inflammation through the induction of heme oxygenase 1 (a known antioxidant and cytoprotective enzyme). BACH1 has been confirmed as a direct target of miR-155-5p, in a study of the cell line HEK293.38
Knowing that an miRNA’s behavior is highly dependent on cell type,39 it is essential to pinpoint an adequate receptor to guide the nanosystem and avoid off-target effects.16 On one hand, P-selectin, an adhesion molecule expressed at the surface of activated ECs and platelets, is highly expressed at the infarct region during a MI.40 On the other hand, fucoidan, a naturally occurring sulfated polysaccharide developed in our laboratory,41 has high affinity to P-selectin.41, 42, 43, 44, 45 Coating NPs with fucoidan would confer on them the ability to target P-selectin-activated ECs,40, 45, 46 which are directly reachable through intravenous injection.
Hence, the strategy proposed here as a proof of concept involves the use of miRNAs adsorbed onto nanocarriers to evaluate their assembly and internalization by ECs, envisioning their use as a potential injectable cardio-protective therapy for MI. Work included miR-155-5p loading onto core-shell NPs.47, 48, 49 The NP shell included a cationic polysaccharide able to bind electrostatically to the miRNA. The NPs were characterized regarding their size and surface electrical charge (ζ potential), morphology, and miRNA presence. miRNA stability, release, and desorption were monitored. In vitro cell culture studies in hCAECs included cytocompatibility assays and cell internalization, in addition to mir-155-5p and BACH1 expression level analyses under pro-inflammatory conditions.
Results
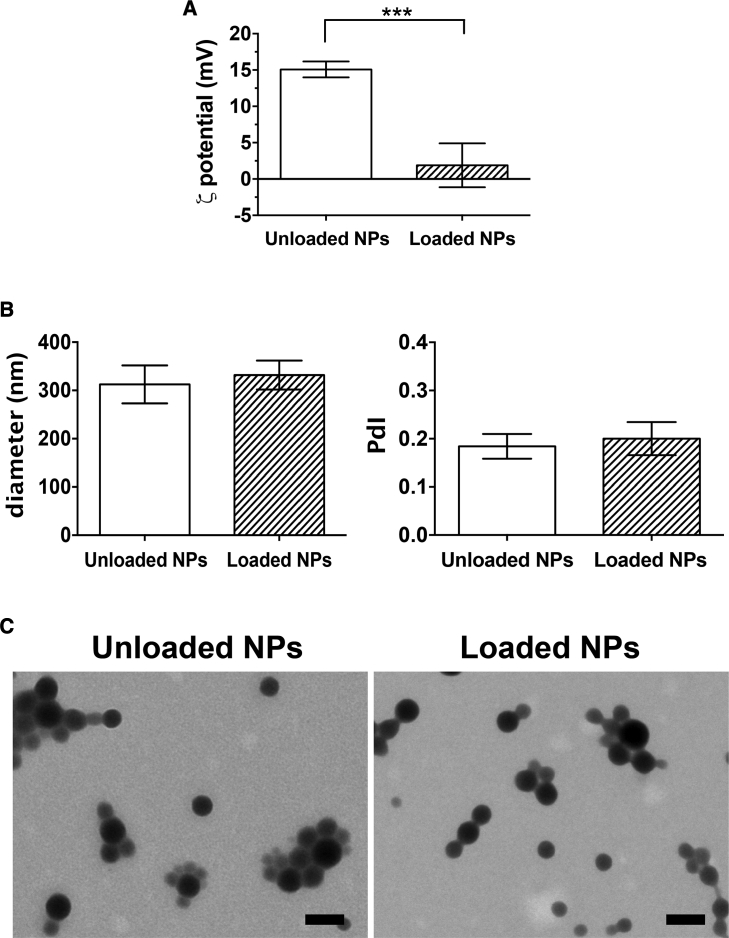
Unloaded NPs were first prepared through redox radical emulsion polymerization with a core of poly(isobutylcyanoacrylate) (PIBCA) and a shell containing fucoidan, diethylaminoethyl (DEAE)-dextran, and dextran. The nitrogen content of the DEAE-dextran of the unloaded NP shell was 0.44% (Table S1), indicating availability of 26 μg of nitrogen per NP dispersion of 6 mg. After miR-155-5p addition in sterile conditions onto unloaded NPs previously subjected to UV exposure and subsequent centrifugation to remove free or poorly adsorbed miRNA, the loaded NPs were obtained. The phosphorous content of the miRNA used for the miRNA loading was 5 μg per miRNA solution of 111 μg, using the optimized N/P (nitrogen to phosphorus) molar ratio of 10. Figure 1 highlights the NP design - namely the chemical bonds driving miRNA loading. As such, the ζ potential values of positively charged unloaded NPs decreased (p < 0.0001) from 15.1 ± 1.1 to 1.9 ± 3.0 mV, indicating that negatively charged molecules were added to the surface (Figure 2A). Both unloaded and loaded NPs were spherical in shape, with miRNA addition tending to increase NP diameter (d) (312.4 ± 39.3–331.9 ± 30.1 nm), while keeping a monodispersed size distribution (polydispersity index [PdI]) (0.18 ± 0.02–0.20 ± 0.03; Figure 2B). The NPs appeared on transmission electron microscopy (TEM) as electron-dense internal cores encircled by a less dense shell (Figure 2C), thereby substantiating the expected core-shell structure.50
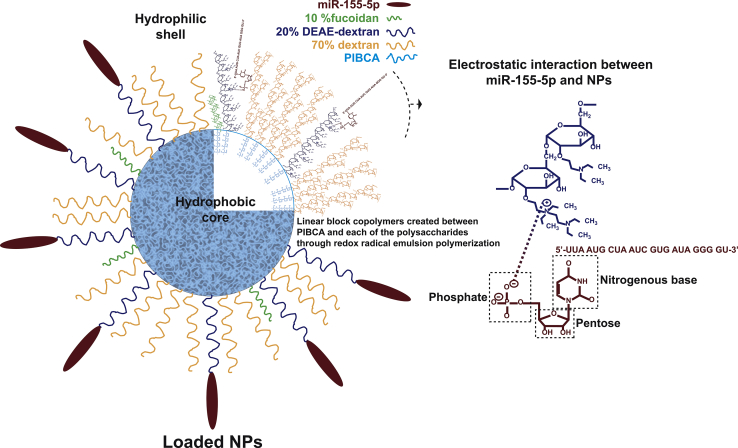
Figure 1.
Schematics of Loaded NP Composition
Unloaded nanosystems prepared through redox radical emulsion polymerization included linear block copolymers composed of a hydrophobic PIBCA core and a hydrophilic shell containing fucoidan (10%), cationic DEAE-dextran (20%), and dextran (70%). miR-155-5p is bound to the NPs electrostatically by the cationic DEAE-dextran. The complete miRNA structure has been omitted for clarity.
Figure 2.
Surface Electrical Charge, Size, and Shape after Loading the miR-155-5p onto the Nanosystem
In ultrapure water, unloaded NPs were characterized after preparation and UV exposure before miRNA loading (white bars), and purified NPs were obtained by re-suspension after centrifugation (hatched bars). (A) ζ Potential (mV), (B) hydrodynamic diameter (nm), and polydispersity index (PdI) were determined by electrophoretic and dynamic light scattering (ELS and DLS, respectively). Results are shown as the mean ± SD of four independent experiments (n = 4). ***p < 0.0001, using the Wilcoxon matched-pairs signed rank test. (C) Representative transmission electron microscope (TEM) images of unloaded and loaded NPs. Scale bars represent 200 nm.
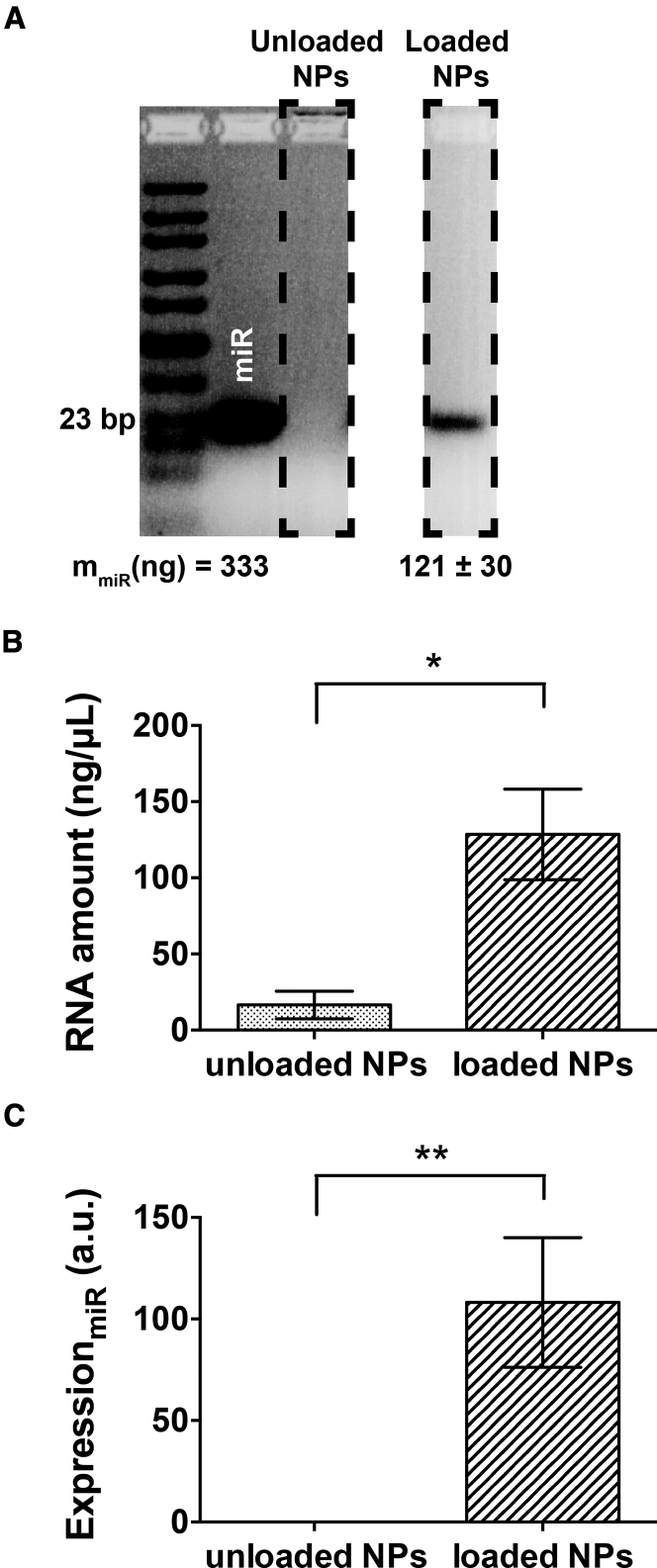
The presence of miR-155-5p was further evaluated by means of agarose gel electrophoresis (AGE), using highly concentrated agarose gels and a ladder sensitive to short nucleic acids (10–300 bp; lane 1), capable of detecting this miRNA (23 bp) within the gel. Figure 3A displays miR-155-5p (lane 2) at the same concentration as for the miRNA loading, plus unloaded and loaded NPs (lanes 3 and 4, respectively). An amount of 121 ± 30 ng of the added miRNA was kept by the NPs afterward, corresponding to approximately 36% of the initially added miR-155-5p. RNases were absent from both unloaded and loaded NPs after preparation (data not shown). The RNA quantification by UV spectrophotometry after NP disruption and RNA isolation, displayed a similar trend (p < 0.05) as in AGE results (Figure 3B). qPCR showed that loaded NPs (Figure 3C) had a higher RNA amount than did unloaded NPs (p < 0.001), showing that the miR-155-5p was present within the loaded NPs.
Figure 3.
Stability of Loaded Nanoparticles and miRNA Content
(A) Detection of nucleic acids within the loaded NPs (n = 4), as determined by 5% agarose gel electrophoresis (AGE). (B) RNA level (ng/μL) quantified by UV absorbance, and (C) miR-155-5p expression levels within unloaded and loaded NPs, evaluated through qPCR, with 2 ng/μL of each sample. Results are shown as the mean ± SD from four independent experiments. Statistical significance was determined with the Wilcoxon matched-pairs signed rank test; *p < 0.05, **p < 0.001.
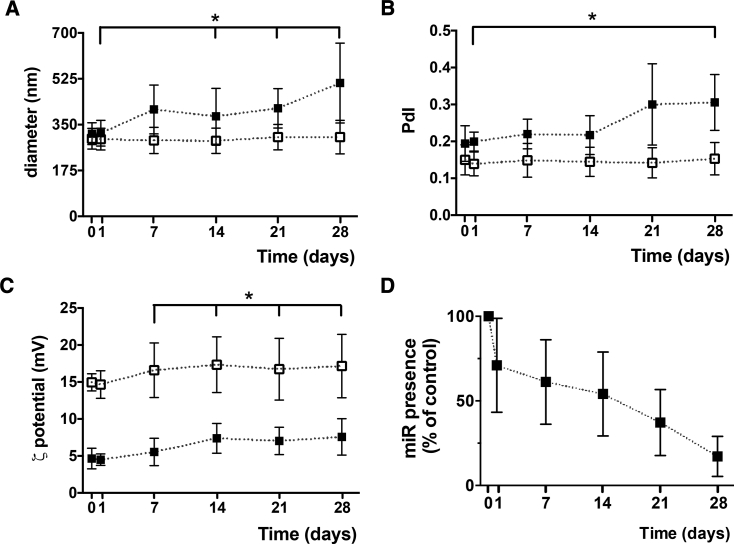
Loaded and unloaded NPs were highly stable at 4°C for 1 day without any measurable change (Figure 4). Then, a progressive trend was observed with diameters from 317.4 ± 49.1 nm (day 1) to 381.6 ± 106.6 nm (day 14, p < 0.05) and PdI from 0.20 ± 0.03 (day 1) to 0.22 ± 0.05 (day 14). The ζ potential from 5.5 ± 1.8 mV (day 7) to 7.4 ± 2.0 mV (day 14, p < 0.05) suggests that negatively charged molecules were removed from the surface. The miRNA content decreased with storage time (Figure 4C), with 61.2% ± 25.0% nucleic acids remaining at day 7.
Figure 4.
Stability of miR-155-5p-Loaded NPs after Preparation (Day 0) and after 1, 7, 14, 21, and 28 Days of Storage at 4°C
NP diameter (nm) (A) and PdI (B) were measured by DLS and ζ potential values (mV) by ELS (C). miRNA presence (% in relation to the control at day 0) within loaded NPs was observed through AGE (D). Outputs from unloaded NPs are also represented (white symbols). Results are shown as the mean ± SD from three to four independent experiments. Significant differences were detected for loaded NPs using the Wilcoxon matched-pairs signed rank test; *p < 0.05.
When subjected to strong orbital shaking at 37°C in PBS, loaded NPs released nucleic acids for 6 hours, with a half-life of 25 min after incubation (Figure 5, black symbols). After centrifugation and re-suspension in 1.5 M NaCl, an additional release of 13% was observed. As expected, we did not detect any miRNA on unloaded NPs (Figure 5, white symbols).
Figure 5.
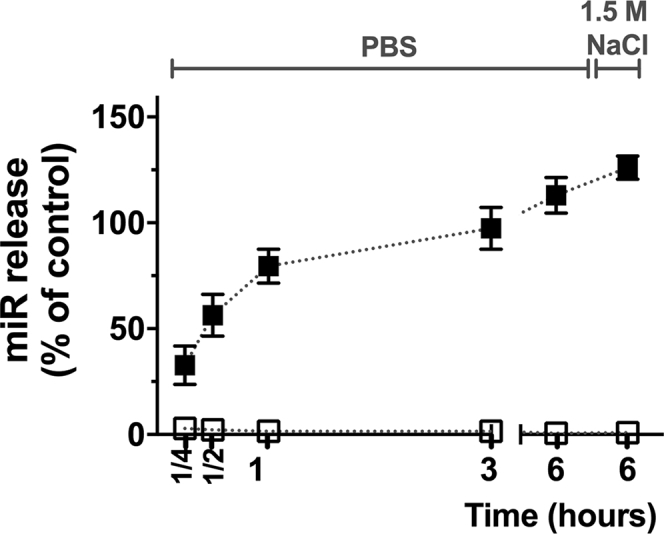
miRNA Release and Desorption from the NPs
NPs were re-suspended in PBS and incubated at 37°C with orbital shaking at 250 rpm, for 1/4, 1/2, 1, 3, and 6 hours, with aliquots removed at each time point. NPs at 6 hours were then centrifuged and re-suspended in 1.5 M NaCl to promote desorption of remaining miRNA. Results tracked by AGE are shown as the mean ± SD of three to four independent experiments. Loaded NPs, black symbols; unloaded NPs, white symbols.
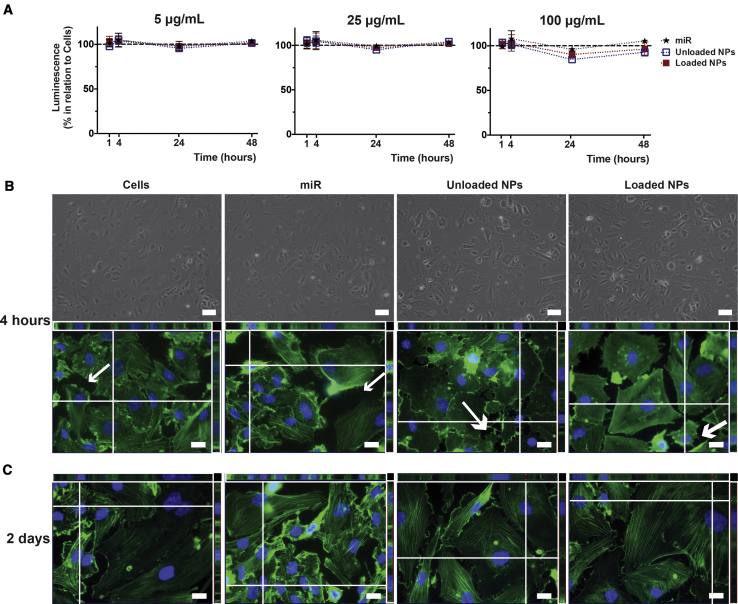
The biological response of unloaded and loaded NPs toward hCAECs was evaluated in vitro, after dynamic light scattering (DLS) measurements showed retention of the NP architecture and nanoscale size in the tested physiological conditions (data not shown), thereby showing NP stability in cell culture medium. The presence of metabolically active cells through ATP conversion during the 2-day period after NP addition, compared with the controls (cells alone, miR), demonstrated that unloaded and loaded NPs were not cytotoxic for hCAECs at the selected concentrations (5, 25, and 100 μg/mL) (Figure 6A). Light (Figure 6B) and fluorescent images of hCAECs were obtained (Figures 6B and 6C). miR-cultured cells tended to be of a slightly smaller size than the control cells, and cells incubated with unloaded NPs showed swollen features, particularly after 4 hours of incubation. miR-incubated cultures tended to have an increased number of cells, most evident after 2 days of incubation. Centrally located stress fibers (green fluorescent actin filaments) are clearly visualized in all culture conditions, but were less evident in unloaded NP-supplemented cultures, where fluorescence tended to concentrate at the cell margins. Unloaded NP-supplemented cellular membranes were also unique, as large protrusions (white arrow; Figure 6B) replaced the majority of the protruding thin filaments (thinner white arrows) that resemble the actin-rich filopodia and lamellipodia51, 52, 53 commonly observed in migratory cells, such as the hCAECs. All evidence showed that hCAECs incubated with loaded NPs were morphologically closer to the main control cells.
Figure 6.
Cytocompatibility of Unloaded and Loaded NPs toward hCAECs in Basal Static Conditions
(A) Luminescence measurements following ATP conversion after incubation for 1 and 4 hours and 1 and 2 days, with NPs at 5, 25, or 100 μg/mL. Results are shown as the mean ± SD from three independent experiments, with no significant differences detected using the Wilcoxon matched-pairs signed rank test. (B) hCAEC morphology at an early time point (4 h) of incubation with NPs at 100 μg/mL, visualized in light micrographs of cultures in 96-well plates (scale bars represent 100 μm) and fluorescence images of hCAECs in 8-well LabTek chamber slides emphasizing cell nuclei (Hoechst; nucleic acids; blue) and cytoplasm (phalloidin; actin fibers; green). Scale bars represent 20 μm for Z-stack-assembled (25–40 slices) images. (C) Fluorescence images taken after 2 days of incubation. Images are representative of three independent experiments.
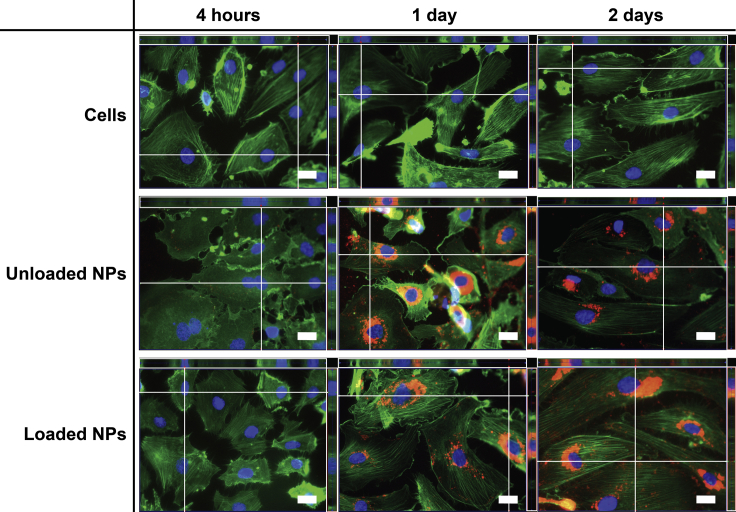
Fluorescence imaging was used to monitor internalization by hCAECs of NPs fluorescently labeled with Nile red (ftNPs) in pro-inflammatory conditions after stimulation with tumor necrosis factor-α (TNF-α). The images revealed that ftNPs (red) were strongly engulfed by hCAECs (Figure 7). No difference was perceived regarding uptake of ftNPs by loaded or unloaded NPs. The increased number of Nile red-labeled features were clearly visualized within the cells with increasing culture time. In NP-enriched cells, ftNPs underwent uptake with perinuclear localization.
Figure 7.
Fluorescence Microscopy of ftNPs (Nile Red) and Intracellular Distribution in hCAECs in TNF-α-Stimulated Cells after 4 hours and 1 and 2 Days of Incubation
Z-stack-assembled (25–40 slices) fluorescence images of hCAECs cultured with unloaded or loaded NPs, and stained to highlight the cell nucleus (Hoechst 33342; nucleic acids; blue) and cytoplasm (phalloidin; actin fibers; green) in monolayer cultured under static conditions. Scale bars represent 20 μm. Images are representative of three independent experiments. Color settings were optimized for loaded NPs at day 1 of incubation, and then used for all images, apart from the nuclear staining.
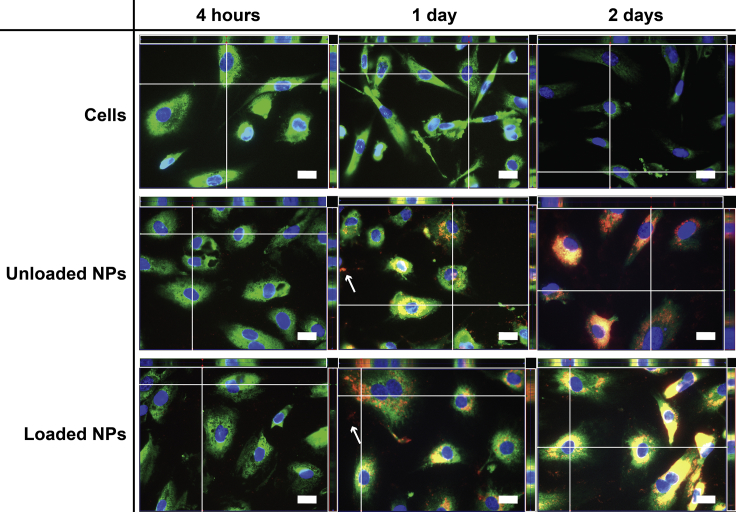
Since the ftNP juxtanuclear position suggested co-localization with the endoplasmic reticulum (ER), an additional ER staining with protein disulfide isomerase was performed. ftNPs were found within the ER of hCAECs (Figure 8), particularly after 2 days, with sparse red labeling detected outside the ERs (thin white arrows). No difference was noticed in the uptake of loaded or unloaded NPs cultured in pro-inflammatory conditions, other than the increase of red staining with time for unloaded and loaded NPs.
Figure 8.
Fluorescence Microscopy of ftNPs (Nile Red) and Intracellular Distribution in hCAECs in TNF-α-Stimulated Cells after 4 hours and 1 and 2 Days of Incubation
Z-stack-assembled (25–40 slices) fluorescence images of hCAECs cultured with unloaded or loaded NPs, and stained to highlight cell nucleus (Hoechst; nucleic acids; blue) and endoplasmic reticulum (protein disulfide isomerase; enzyme; green) in monolayer cultured under static conditions. Scale bars represent 20 μm. Images are representative of three independent experiments. Color settings were optimized for loaded NPs at day 1 of incubation, and then used for all images, apart from the nuclear staining.
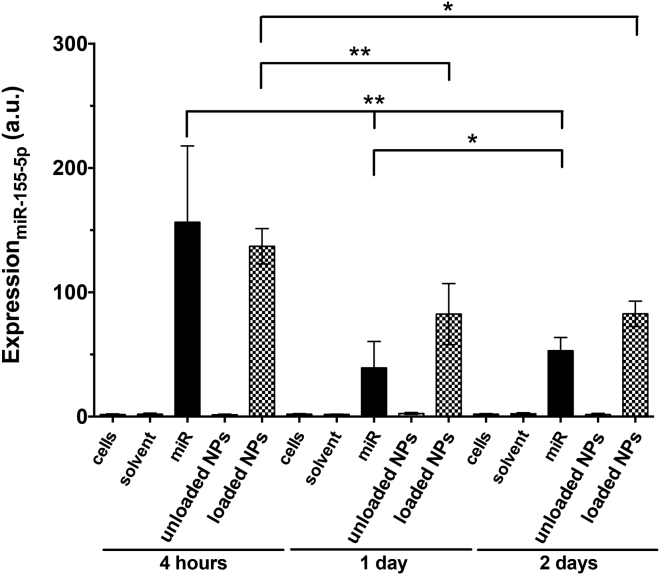
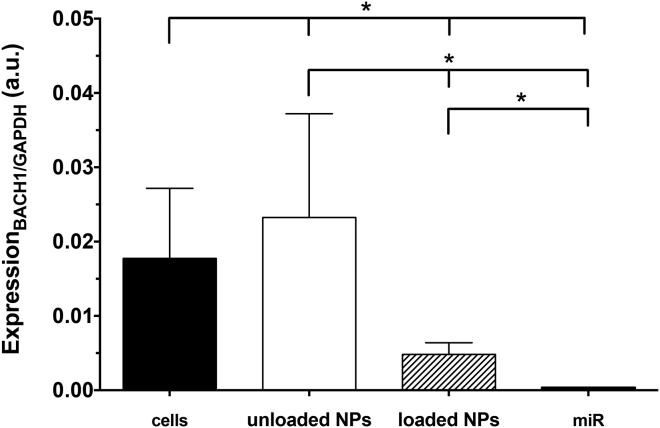
Finally, miR-155-5p and BACH1 levels within hCAECs were examined by qPCR with cells cultured in pro-inflammatory conditions (Figures 9 and 10). In the miR-negative control groups (such as unloaded NPs), miRNA levels were minimal (expression of 1.8 ± 0.5 a.u. after 4 h, with levels being maintained with culture time). Within the TNF-α-stimulated cells, miR-supplemented cultures revealed 85- or 75-fold upper levels, respectively, for the miRNA and loaded NPs, in comparison with the unloaded NPs (p < 0.05). miR-155-5p levels decreased from 156.7 ± 61.4 a.u. (4 h), to 39.5 ± 21.3 a.u. (1 day), to 53.3 ± 10.6 a.u. (2 days, p < 0.05), whereas loaded NP values decreased from 137.3 ± 14.2 a.u. (4 h), to 82.8 ± 24.6 a.u. (1 day), to 83.1 ± 10.2 a.u. (2 days, p < 0.05). However, the main noticeable differences were the increased miR-155-5p values for the loaded NP condition with respect to cells cultured with miRNA, suggesting a slower load release in these physiological conditions, in contrast to the possible degradation of exogenous, free miRNA in the physiological media at 1 (p < 0.05) and 2 (p < 0.001) days of incubation. On the other hand, loaded NP-enriched cells (48.16 × 10−4 ± 15.70 × 10−4 a.u.) led to a decrease (p < 0.05) of BACH1 levels after 1 day of incubation, when compared to the control (177.37 × 10−4 ± 94.29 × 10−4 a.u.). Unloaded NP-related cultures showed no indication of BACH1 reduction through the exogenous miR-155-5p, but free miRNA-supplemented cultures resulted in further BACH1 silencing (p < 0.05), as expected (3.52 × 10−4 ± 0.53 × 10−4 a.u.).
Figure 9.
Expression of miR-155-5p within hCAECs after 4 hours and 1 and 2 Days, as Determined by qPCR for TNF-α-Stimulated Cells
Results are shown as the mean ± SD from four independent experiments. Significant differences were detected using the Wilcoxon matched-pairs signed rank test; *p < 0.05 and **p < 0.001. ns, nonsignificant.
Figure 10.
Expression of BACH1within hCAECs after 1 Day, as Determined by qPCR
Results are shown as means ± SD (n = 6). Significant differences were detected by the Wilcoxon matched-pairs signed rank test; *p < 0.05.
Discussion
This study evaluated the loading of miR-155-5p onto core-shell, polymer-based NPs through electrostatic interactions (Figure 1), their internalization by hCAECs, and the delivery of their miRNA cargo as a proof of concept of the feasibility of the use of miR-155-5p-loaded NPs for MI. Unloaded NPs were prepared through redox radical emulsion polymerization, containing a PIBCA core coated with a polysaccharide brush comprising 10% fucoidan for targeting, 20% DEAE-dextran for miRNA binding and promotion of cell internalization, and 70% dextran for surface stabilization. After miRNA loading under sterile conditions, unloaded and loaded NPs were analyzed through the combination of different techniques. Round-shaped NPs with a hydrodynamic size of approximately 300 nm were obtained (Figure 2), with a decrease of their ζ potential after miR-155-5p adsorption. Nucleic acids with the same size as our miRNA were detected in the loaded NPs through AGE runs (Figure 3). The RNA quantification, by UV spectrophotometry after NP disruption and RNA isolation, displayed a trend similar to that seen with AGE. miR-155-5p presence was further demonstrated by qPCR, with the additional information that the miRNA remains intact within loaded NPs, since the probe can detect only whole target sequences.
While loaded NP integrity is essential for the intended clinical application, miRNA release in physiological conditions is another essential step. Figures 4A–4C show that the main loaded NP features were highly stable up to 1 day of storage at 4°C in sterile conditions. A slight increase in NP diameter and PdI, particularly after days 1 and 14, respectively, was observed. The ζ potential showed a small increment in 28 days, which was accompanied by an apparent loss of miRNA of 38.8%, measured through AGE (Figure 4D). Since aliquots for DLS and ELS and AGE measurements were handled simultaneously, it is assumed that miRNA was released from the NP surface with some disruption of the ionic bond and/or media interaction with NP hydrophilic chain. Regardless, total miRNA release is achievable in physiological conditions, as illustrated in Figure 5 (see also Figure S1).
The previous cytocompatibility studies with human umbilical vein endothelial cells (HUVECs) performed by Matuszak and colleagues,54 with similar but not identical compositions of core-shell (PIBCA-polysaccharide) - 10% 7 kDa fucoidan, 10% 20 kDa DEAE-dextran, and 80% 70 kDa dextran - placed the upper limit of NP concentration in direct contact with the cells used at 100 μg/mL. No cytotoxicity could be detected in our conditions until 100 μg/mL, although there was some evidence of cytotoxicity with hCAECs incubated with ≥200 μg/mL NPs (data not shown). For 100 μg/mL of NPs, a loaded amount of 650 ng/mL miRNA was obtained, corresponding to 43.1 nM of loaded biomolecule, which is well above the efficacy threshold (0.5 nM) defined by the miRNA supplier.
Analyses of cell morphology were performed with hCAECs (Figure 6). miRNA alone in solution led to increased cell shrinkage and preservation of heightened stress fiber intensity with culture time51, 53; with unloaded NPs, cells displayed signs of oncosis and/or necrosis with substantial loss of typical topography with respect to visible actin fibers, swollen appearance, rounded periphery, and presence of large membrane protrusions suggesting membrane blebbing,51, 53, 55 even though short-length polymers (7 and 20 kDa) were used for NP fabrication.55 Regardless, cells exhibited healthier features at day 2, suggesting that these features were reversible after their eventual initial activation. Loaded NP-enriched cells showed the highest resemblance of all tested conditions to the control hCAECs cultured in basal conditions at all time points. ATP analyses showed no significant alterations in hCAEC metabolic activity. EC dysfunction and activation toward a pro-inflammatory phenotype is a key promoter of the cascade of detrimental signaling triggers to an infarcted region, which could further impair the tissue from recovering after an MI.3 Loaded NPs maintained hCAEC basal characteristics, so promising prospects can be perceived for future treatments based on such nano-objects.
The fluorescent molecule (Nile Red) added to the NP core during NP polymerization enabled their labeling, as previously shown.56 ftNP size decreased approximately 17 nm in comparison to the non-fluorescent counterpart, and ζ potential remained unchanged (data not shown). A main interest in working with NPs is to have them cross cellular membranes and enter living cells for cargo release after intracellular targeting.55 Some ftNPs were observed inside the cells after the 4 hour-period, but most were still outside (Figure S2). NP internalization was clearly observed after 1 day of incubation (result also noticed in basal conditions; data not shown), with even more NP accumulation within hCAEC borders after 2 days (Figure 7) and NP translocation to the perinuclearly located ER (Figure 8). The ER is the site where target mRNA binds to Argonaut-2 (Ago2) and miRNA, which precedes miRNA-mediated translation repression of target mRNA in mammalian cells,57 events importantly linked to increased biological efficacy.16, 57, 58 Possible ER transient disruption was also suggested by the cell morphology, and sparse red labeling distribution in several NPs all around cell nuclei55, 58 reinforce the latter hypothesis. Loaded NPs appeared to contain slightly more NPs encircling the hCAEC nucleus and within the ER, but internalization occurred similarly for either unloaded or loaded NPs. Endosomal entrapment is a usual endpoint of many loaded nanosystems guided toward natural cellular degradation processes through lysosomes.55, 59 No NPs have yet been reported to be capable of crossing EC ER membranes. Interestingly, our NPs clearly accumulated within hCAEC ER after 2 days.
Intracellular miR-155-5p level was studied (Figure 9) to evaluate if the load had been delivered within the cells. The massive presence of loaded ftNPs within the ER of the hCAECs reinforces the potential that the miRNA detected within the cells has been delivered to the required cell organelle for the miRNA to proceed. No information can be given, however, regarding intracellular location of the miRNA through qPCR. Even if it cannot be discounted that both exogenous and endogenous miRNAs were detected through this technique, it is clear that 4 h of incubation was sufficient to yield a 75-fold increase in miRNA within the loaded NP-supplemented TNF-α-hCAECs when compared to unloaded NPs. From 4 hours to 2 days, the miR-155-5p may have been consumed or degraded through the classic routes.38 On the other hand, cultures with miR-155-5p alone also produced 85-fold increased miRNA expression levels. miRNAs are expected to be degraded in serum and have poor cellular uptake.12, 60 The results showed that miRNA expression was consistently lower for free miRNA than for miRNA complexed to the NPs. The intracellular level of BACH1 was also studied to assess delivered miRNA’s ability to target this gene and lead to its reduction within hCAECs. Loaded NP-enriched cultures resulted in a 4-fold reduction of BACH1 within hCAECs in comparison to the control with cells alone. When incubated with NPs, BACH1 levels within hCAECs were five times lower when miR-155-5p was loaded. Free miR-supplemented cultures also induced a decrease in BACH1 expression levels when compared to the control or loaded NPs (50- or 14-fold, respectively). The results therefore indicate that the hCAECs’ contact with miR-155-5p is followed by endogenous BACH1 silencing, thus showing in vitro functional validation of miR-155-5p delivery through reduction of this downstream target.
The endothelial-enriched miR-155-5p is one of the TNF-α-inducible miRNAs predicted to bind to BACH1 mRNA,38 an important pleiotropic regulator of cell homeostasis known for its vasculoprotective effects through restrain of endothelial cell dysfunction, inflammation, and hypertension.38, 61, 62 miRNA-based therapies offer an approach that can reach levels of intra- and intercellular communication so that potentiated effects can be achieved with very low doses of the bioactive compound.16 The use of nanoscale injectable systems that are minimally invasive, able to protect and carry loads, avoid immediate renal clearance, interact with surface receptors, pass through biological barriers, be internalized into target cells, deliver loads, and influence cellular processes11, 44 is an interesting strategy for reducing the damaging effects of a MI.18, 44 Targeting this pathway is a promising in vivo strategy, because mice lacking BACH1 subjected to an experimental MI were shown to have an infarct size 48.4% smaller than that in wild-type mice.63, 64, 65 Although efficient EC targeting systems are still lacking,33, 66 miRNA carriers are now being used. Altogether, we believe that the findings in this study represent proof of concept that miR-155-5p can be successfully loaded onto polymer-based NPs and can be internalized into hCAECs, aiming at modulating hCAEC response toward their normal state through BACH1 silencing. Delivering this miRNA to the ECs of the heart during reperfusion is hence expected to reduce their oxidative stress and, as a consequence, their barrier function and their anti-inflammatory and anti-thrombotic functions, as well as their survival.67, 68, 69, 70 The ultimate goal is not to regenerate the heart and obtain native healthy tissue in the long-term, but instead to decrease the overall effects of the injury, so that a decrease in the chances for heart failure can be attained.
Conclusions
Herein, we have evaluated for the first time miRNA-loaded polymer-based NPs containing a core of poly(cyanoacrylate) and a shell with dextran, DEAE-dextran, and fucoidan targeting P-selectin on activated hCAECs. miRNA loading was successful and reproducible, as was load release in physiological conditions. Loaded NPs were cytocompatible for these cells, which presented typical hCAEC morphology. Cells cultured with the loaded NPs increased their content of miR-155-5p and decreased their content of BACH1. NPs were located within the ERs, thereby positioning them to bind with target mRNA and fulfill its function. Hence, this study is the first proof of concept that miR-155-5p can be loaded onto core-shell, poly(cyanoacrylate)-polysaccharide NPs and remain intact and biologically active, with the NPs located at the target site for mRNA degradation. Given BACH1 silencing within hCAECs, a potential decrease in the overall detrimental effects within infarcted hearts through intravenous injection is envisioned, with results opening new perspectives for therapeutic approaches for cardiovascular diseases.
Materials and Methods
Materials
Pharmaceutical grade fucoidan (7 kDa; Algues & Mer), DEAE-dextran (20 kDa; TdB Consultancy), dextran (20 kDa, Pharmacosmos) and isobutylcyanoacrylate (IBCA) monomers (AFINITICA) were used without further chemical modification. Polysaccharides were vacuum dried at 60°C overnight before use. Chemically modified, double-stranded RNA oligonucleotides from QIAGEN were used to mimic the natural human mature microRNA miR-155-5p (miRNA sequence: 5′-UUA AUG CUA AUC GUG AUA GGG GU-3′). The biomolecule was received lyophilized, and then it was reconstituted, aliquoted, and stored at −20°C, according to the manufacturer’s recommendations. The molecular units of the aforementioned materials are represented in Figure S3.
Preparation of the Unloaded and miR-Loaded NPs
Unloaded NPs were prepared as previously described,47, 48, 49 by redox radical emulsion polymerization, to obtain core-shell NPs, with the core composed of the synthetic and hydrophobic cyanoacrylate PIBCA and the shell constituted of natural and hydrophilic polysaccharides. The latter were added with a proportion of 10% fucoidan, 20% DEAE-dextran, and 70% dextran, at a total concentration of 17.2 mg/mL in a nitric acid solution (0.2 M HNO3), and then kept at 40°C under nitrogen (N2) bubbling and magnetic stirring until solubilization. Ammonium nitrate cerium (IV) ions ((NH4)2[Ce(NO3)6]; Merck), previously solubilized at 0.08 M in 0.2 M HNO3, were added to the mixture to initiate the formation of radicals, followed by addition of IBCA monomers (11.4 mg/mL) to form the NP’s core. Ethanol was added 2 min later,50, 56 to synthesize non-fluorescent NPs with properties similar to those of the fluorescent ones. Reaction continued at 40°C under magnetic stirring for 50 min, with N2 bubbling present for the first 10 min after (NH4)2[Ce(NO3)6] and IBCA addition. A sodium citrate tribasic dehydrate solution (1.02 M HOC(COONa)(CH2COONa)2.2H2O; Sigma-Aldrich) was added to the NP dispersion at room temperature to inhibit the reaction, the pH was adjusted to 6.5, and the final dispersion was dialyzed against distilled water (molecular weight cutoff [MWCO], 100 kDa; Spectrum Labs). Finally, before miRNA loading, three additional steps were performed: (1) freeze drying of unloaded NPs, to assess their mass concentration; (2) determination of their N content by elemental analysis, using a combustion method and detection through katharometry (CHNS Vario EL III Elemental Analyzer, Elementar); and (3) exposure of the NPs through UV.
miR-loaded NPs were prepared under sterile conditions, by dropwise addition of a miR-155-5p solution into a stirring NP suspension (4 mg/mL), at an N/P molar ratio of 10 at room temperature. Solutions and suspensions were prepared in UltraPure DNase/RNase-Free Distilled Water (Invitrogen), henceforth named ultrapure water. Then, the NPs were allowed to stabilize 20 min at 4°C, centrifuged (4°C, 50,000 g, 45 min) to remove free or poorly adsorbed miRNA, and after resuspension in ultrapure water, the loaded NPs were obtained. NPs were then maintained at 4°C until further use (maximum 24 hours). NP preparation is summarized in Figure S4. See also Figure 1.
ftNPs were used for the internalization studies. For that, NP preparation was subjected to the following modifications: Nile Red, a hydrophobic fluorescent dye (Sigma-Aldrich), was dissolved in ethanol at 0.1 mg/mL and added to the mixture 2 min after the IBCA (similarly as aforementioned) to label the hydrophobic PIBCA core.50, 56
Characterization of the Unloaded and Loaded NPs
The size (diameter), PdI, and surface electrical charge (ζ potential) of the NPs (Figures 2A and 2B) were determined using a Zetasizer Nano ZS (Malvern Panalytical) equipped with a 4 mW HeNe laser beam with a wavelength of 633 nm and a scattering angle of 173° (for size measurements) and 13° (for ζ potential measurements). Size and ζ potential values were automatically calculated through DTS Nano v.6.30 software, using the Stokes-Einstein equation and the Henry equation with the Smoluchowski approximation, respectively. The dispersion solution used was the solvent in which the NPs were dispersed for the DLS, i.e., ultrapure water, and 1 mM KCl for the ELS. Disposable solvent-resistant microcuvettes or folded capillary cells (DLS; Sigma) with gold-plated electrodes (ELS; Malvern) were used. Data were acquired at 25°C, in triplicate.
The morphology of the NPs was observed by a transmission electron microscope (TEM, 80 kV; Tecnai 12; FEI) with a 1 K × 1 K KeenView camera (OSiS). Samples were prepared by placing 3 μL of the NPs suspension onto a 400 μL mesh copper grid coated with carbon (Electron Microscopy Support). One minute after deposition, the grid was tapped with a filter paper. TEM images were obtained at different magnifications (Figure 2C).
Presence of miR-155-5p within Loaded NPs
The presence of the miR-155-5p was monitored by AGE and qPCR (Figures 3A and 3B, respectively). Five percent agarose gels, prepared in TBE (Tris-Borate-EDTA; Thermo Fisher Scientific) buffer and stained with ethidium bromide, were used to allow detection of nucleic acids within a 10–100 bp range, as well as the GeneRuler Ultra Low Range DNA Ladder (10–300 bp; Thermo Scientific). Samples included unloaded and loaded NPs, plus additional controls from different steps of the miRNA loading: miRNA alone (at the concentration used for the loading), loaded NPs right after miRNA addition, and supernatant from the ultracentrifugation. Samples were run by electrophoresis (50 V, 120 min) and detected through a UV transilluminator smart imaging system (Vilber Lourmat).
As for qPCR, miR-155-5p was detected using the Hs_miR-155_2 miScript Primer Assay (5′UUAAUGCUAAUCGUGAUAGGGGU-3′; QIAGEN), a primer able to detect specifically the human mature miRNA miR-155-5p, following the supplier’s recommendations. For that, NPs were centrifuged (0.7 mL at 4°C, 20,000 g, 30 min), QIAzol lysis reagent (QIAGEN) was added (0.7 mL) to each sample, and the samples were vortexed until homogenization. RNA was isolated using the miRNeasy Mini Kit (QIAGEN). The quality and quantity of total RNA were determined by UV spectrophotometry. The miScript II RT Kit (QIAGEN) with a final volume of 20 μL, containing 90 ng RNA, 4 μL 5× miScript Hiflex buffer, 2 μL 10× miScript Nucleics Mix, and 2 μL miScript Reverse Transcriptase mix were used for cDNA synthesis. The final concentration of cDNA was 4.5 ng/μL. The qPCR analysis was performed using the miScript SYBR Green PCR kit (QIAGEN) for miR-155-5p with StepOnePlus Real-Time PCR System (Applied Biosystems), according to the manufacturer’s recommendations. The amplification was done with a final volume of 25 μL, containing 2× QuantiTect SYBR Green PCR Master Mix, 10× miScript Universal Primer, 10× miScript Primer Assay, and 2.5 μL of each sample at 2 ng/μL. For each DNA amplification, a standard range was also generated from cDNA of a miR-155-5p solution, to determine the efficiency of the primer, as well as the negative control (without the reverse transcriptase). The qPCR conditions included 1 cycle for enzyme activation (95°C for 15 min), followed by 40 cycles of denaturation, annealing, and extension (94°C for 15 s, 55°C for 30 s, and 70°C for 34 s). By means of a standard curve, the expression of the gene encoding the miRNA was calculated using the standard curve and a geometric mean.
Loaded NP Stability, Release, and Desorption
Loaded NP stability, while stored at 4°C, was evaluated by DLS (Figures 4A and 4B), ELS (Figure 4C), and AGE (Figure 4D), right after miRNA loading (day 0) and 1, 7, 14, 21, and 28 days after miRNA inclusion. Aliquots for each time point were prepared under sterile and RNase-free conditions at day 0. Furthermore, to assess if all miRNA could be released and desorbed from the NPs under physiological and sterile conditions, loaded NPs were resuspended in PBS at 4°C (instead of the ultrapure water) after centrifugation, transferred into a clear, flat-bottomed, ultra-low attachment plate (Corning) and subjected to strong orbital shaking at 250 rpm at 37°C. At each time point (15 and 30 min and 1, 3, and 6 h), the plate was removed from the orbital shaker and incubator, and one-tenth of the volume of each well was removed and placed at −20°C and replaced by PBS. At the last time point, the remaining content of each well was centrifuged (4°C, 20,000 g, 30 min). Samples were resuspended in 1.5 M NaCl, supernatants collected, and a final run of re-suspension and centrifugation in 1.5 M NaCl was performed to obtain the final NPs. Samples were analyzed by AGE (Figure 5).
In Vitro Cell Culture Studies
hCAECs (PromoCell) were expanded in monolayer culture in cell culture medium (EC basal medium MV 2 plus a supplemental pack containing 5% fetal calf serum, 5 ng/mL recombinant human epidermal growth factor, 10 ng/mL recombinant human basic fibroblast growth factor, 20 ng/mL recombinant human Long R3 insulin-like growth factor, 0.5 ng/mL recombinant human vascular endothelial growth factor 165, 1 μg/mL ascorbic acid, and 0.2 μg/mL hydrocortisone (PromoCell). Additional supplements included 1% penicillin-streptavidin (Life Technologies), 0.2% Primocin (InvivoGen), and 0.02% Plasmocin (InvivoGen), in a humidified atmosphere with 5% CO2 at 37°C. At a subconfluent-to-confluent stage, the cells were detached with 0.05% trypsin-EDTA (Gibco) and re-seeded until the required number of cells for the subsequent experiments was obtained. Flasks, plates, and chamber slides were always pre-coated with 50 μg/mL fibronectin (PromoCell). Cells were used at passage numbers 6–7 (P6–P7) for all in vitro cell culture studies.
Cytocompatibility Studies
Unloaded and loaded NP cytocompatibility toward hCAECs was assessed by the CellTiter-Glo luminescent cell viability assay (Promega), which measures ATP presence by luminescence (integration time: 1,000 ms), with the result being directly proportional to the number of viable cells within the wells (Figure 6A). Bright-field images (EVOS XL Core Cell Imaging System; Thermo Fisher Scientific, Figure 6B) and fluorescence images (Axiovert Observer Z1 inverted microscope; Zeiss; Figures 6B and 6C) were taken during culture to monitor hCAEC morphology in the presence of unloaded and loaded NPs.
hCAECs were seeded at 6.25 × 104 cells/cm2 onto Nunc white, opaque, flat-bottomed plates (Fisher Scientific) so that ATP conversion could be measured. After 24 hours and then twice per week, the medium was changed until the cells reached confluence. At this point, the medium was changed again to include the following conditions: unloaded or loaded NPs at 5, 25, and 100 μg/mL; miRNA solutions at 0.03, 0.16, and 0.65 μg/mL; with the NP dispersions and miRNA solutions within the solvent ultrapure water at one-tenth of the well volume. A blank control comprising only medium was also included, together with solvent alone at the aforementioned volume proportion and controls of all tested conditions without cells. A standard curve built from ATP sodium salt (Sigma) was prepared in the plate at each time point, according to the supplier’s recommendations. Measurements were performed 1 and 4 hours and 1 and 2 days after incubation with all conditioned media.
Bright-field images were acquired at magnification 10×, after 4 hours of incubation in the tested conditions: conditioned medium with unloaded and loaded ftNPs at 100 μg/mL, miRNA solution at 0.65 μg/mL, solvent, and blank control with medium. Cells (P6–P7) were previously seeded at 6.25 × 104 cells/cm2 onto Corning clear, flat-bottomed, 96-well plates. For acquiring fluorescence images, cells were seeded onto eight-well Nunc Lab-Tek chamber slides (Thermo Scientific) and then incubated during 4 h with each imaging condition (cells, solvent, miR, unloaded ftNPs, and loaded ftNPs). Next, the wells were fixed and cell nuclei and cytoplasm were stained. For this, cells were fixed in paraformaldehyde (PFA; 3.7%) for 15 min, washed in PBS, and stored at 4°C. Subsequently, a blocking step was introduced to reduce nonspecific background staining with 1 hour of incubation in PBS containing 1% BSA (Bio-techne). Cells were then incubated overnight in the dark at 4°C in Alexa Fluor 488-phalloidin (dilution 1:40 in blocking solution; Life Technologies). Cell nuclei were counterstained by immersion in 10 mg/mL Hoechst 33342 trihydrochloride trihydrate (dilution, 1:1,000 in PBS; Life Technologies) for 10 min at room temperature, in dark conditions. After final washes in PBS, the samples were mounted with ProLong diamond anti-fade mounting medium (Life Technologies) and sealed. Images were captured with a 40× oil objective, as Z-stack-assembled (25–40 slices, 7.44–12.09 μm) images. Color settings were optimized for each condition and time point.
Cell Internalization
To evaluate the capacity of hCAECs to internalize ftNPs, unloaded and loaded NPs were incubated with these cells under pro-inflammatory conditions (method adapted from Ollivier et al.71). Cell internalization was then visualized through fluorescence microscopy, as detailed in the previous section (Figures 7 and 8).
hCAECs were seeded at 6.25 × 104 cells/cm2 onto the eight-well chamber slides described in the previous section. Once confluent, the medium was changed to include cells stimulated overnight with TNF-α-supplemented medium (10 ng/mL, TNF-α; Millipore). In the following day, medium was changed again to include unloaded and loaded NPs at 100 μg/mL, miRNA solution at 0.65 μg/mL, solvent, and control with medium (detailed in the previous section). At 4 h and 1 and 2 days of incubation, the cells were fixed and stained according to two protocols in parallel, with the first protocol staining cell nuclei and cytoplasm (detailed in the previous section), and the second protocol staining cell nuclei and ER, where SelectFX Alexa Fluor 488 Endoplasmic Reticulum Labeling Kit (Life Technologies) was used, according to the supplier’s ER staining protocol. Images were captured with a 40× oil objective, as z stack-assembled (25–40 slices; 7.44–12.09 μm) images. Color settings were optimized for loaded NPs at day 1 of incubation and then used for all images, apart from the nuclear staining.
Detection of miR-155-5p and BACH1 within hCAECs
In order to verify miR-155-5p and BACH1 presence within hCAECs, loaded NPs were incubated with these cells, as described in section 2.4. The intracellular level of the genes was then evaluated by qPCR (Figures 9 and 10).
hCAECs were seeded at 3.8 × 106 cells/cm2 in clear, flat-bottomed, six-well plates (Corning), as described above. Once confluent, the cells were cultured under basal and pro-inflammatory conditions, and comprising the aforementioned conditioned media. At 4 hours and 1 and 2 days of incubation, the wells were washed with PBS, lysed with the QIAzol lysis reagent, and processed for qPCR, as described for digested NPs, with a final cDNA concentration of 250 ng/μL. Relative gene expression was determined for BACH1 after 1 day of incubation, normalized by GAPDH. Specific primers (Eurogentec) were used to quantify the gene expression of BACH1 (forward primer sequence: 5′-TCGCAAGAGAAAACTTGACTG-3′; reverse primer sequence: 5′-CCCAGAGTTGACAAAATGTGA-3′) and GAPDH (forward primer sequence: 5′-GTCGCCAGCCGAGCCACATC; reverse primer sequence: 5′-CCAGGCGCCCAATACGACCA-3′). For both genes, the amplification was done with a final volume of 25 μL, containing Absolute Blue qPCR SYBR Green ROX Mix (Thermo Fisher Scientific), forward primer, reverse primer, RNase-free water, and 10 μL of each sample at 0.8 ng/μL for BACH1 or 0.2 ng/μL for GAPDH. The qPCR conditions included annealing temperatures of 62°C or 69°C, respectively.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism (version: 5.0a). The parametric distribution of the data was first evaluated by the D’Agostino-Pearson omnibus normality test. Because the data followed a non-parametric distribution, statistical analysis was performed with the Wilcoxon matched-pairs signed rank test to compare each paired group. A confidence interval of at least 95% was chosen to define statistical significance (*p < 0.05, **p < 0.001, ***p < 0.0001).
Author Contributions
J.C.A. designed the study, performed the experiments, collected and interpreted the data, and wrote the draft version of the manuscript. L.B. optimized qPCR protocols, provided genetics expertise, and assisted in study design and data interpretation. F.C.M. aided in miRNA release experiments and data collection, M.J. advised on nanoparticle preparation. M.-S.G., M.A., and C.B. provided genetics expertise. F.C. advised on nanoparticle characterization. G.C. aided with the immunostaining of nanoparticles and collaborated in the design of the whole study, interpretation of the results (particularly at the cell-material interface), and correction of the manuscript, together with A.N. and V.O. A.N. was also instrumental in cell internalization studies and V.O. guided the agarose gel electrophoresis, the cells used and the in vitro cell culture model. D.L. provided the funding, supervised the study, discussed the results and corrected the manuscript. C.C. supervised the study, discussed the results, and corrected the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was financed by European Union project NanoAthero FP7-NMP-2012-LARGE-6-309820 and ANR-13-LAB1-0005-01 “FucoChem.” The authors would like to acknowledge F.C.M.’s PhD fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) from Brazil (Process 201245/2017-5). Thanks also to B. Li, M.-P. Jacob-Lenet, C. Choqueux, R. Aid-Launais, G. Even, M. Morvan, F. Andreata, S. Loyau, L. Venisse, A. Loste, J. Aerts, and A. Grodet for technical assistance and guidance during the experiments. The authors would also like to acknowledge Marie-Françoise Bricot from the Institut de Chimie des Substances Naturelles of the Université Paris-Saclay for microanalysis of the raw polymers and NPs. We also thank the ImagoSeine core facility of the Institut Jacques Monod, member of IBiSA and France-BioImaging (ANR-10-INBS-04) infrastructures, particularly Dr. Rémi le Borgne for sample preparation and assistance with TEM imaging.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.016.
Supplemental Information
References
- 1.Godier-Furnemont A., Vunjak-Novakovic G. Cardiac Muscle Tissue Engineering. In: Ratner B.D., Hoffman A.S., Schoen F.J., Lemons J.E., editors. Biomaterials Science: An Introduction to Materials in Medicine. Third Edition. Academic Press; 2002. pp. 1262–1276. [Google Scholar]
- 2.Chauvierre C., Letourneur D. The European project NanoAthero to fight cardiovascular diseases using nanotechnologies. Nanomedicine (Lond.) 2015;10:3391–3400. doi: 10.2217/nnm.15.170. [DOI] [PubMed] [Google Scholar]
- 3.Fiedler J., Thum T. MicroRNAs in myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2013;33:201–205. doi: 10.1161/ATVBAHA.112.300137. [DOI] [PubMed] [Google Scholar]
- 4.Frangogiannis N.G. The immune system and cardiac repair. Pharmacol. Res. 2008;58:88–111. doi: 10.1016/j.phrs.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sala V., Bergerone S., Gatti S., Gallo S., Ponzetto A., Ponzetto C., Crepaldi T. MicroRNAs in myocardial ischemia: identifying new targets and tools for treating heart disease. New frontiers for miR-medicine. Cell. Mol. Life Sci. 2014;71:1439–1452. doi: 10.1007/s00018-013-1504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost R.J.A., van Rooij E. miRNAs as therapeutic targets in ischemic heart disease. J. Cardiovasc. Transl. Res. 2010;3:280–289. doi: 10.1007/s12265-010-9173-y. [DOI] [PubMed] [Google Scholar]
- 7.Boersma E., Mercado N., Poldermans D., Gardien M., Vos J., Simoons M.L. Acute myocardial infarction. Lancet. 2003;361:847–858. doi: 10.1016/S0140-6736(03)12712-2. [DOI] [PubMed] [Google Scholar]
- 8.Thum T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol. Med. 2012;4:3–14. doi: 10.1002/emmm.201100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farías J.G., Molina V.M., Carrasco R.A., Zepeda A.B., Figueroa E., Letelier P., Castillo R.L. Antioxidant Therapeutic Strategies for Cardiovascular Conditions Associated with Oxidative Stress. Nutrients. 2017;9:E966. doi: 10.3390/nu9090966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spath N.B., Mills N.L., Cruden N.L. Novel cardioprotective and regenerative therapies in acute myocardial infarction: a review of recent and ongoing clinical trials. Future Cardiol. 2016;12:655–672. doi: 10.2217/fca-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antunes J.C., Pereira C.L., Teixeira G.Q., Silva R.V., Caldeira J., Grad S., Gonçalves R.M., Barbosa M.A. Poly(γ-glutamic acid) and poly(γ-glutamic acid)-based nanocomplexes enhance type II collagen production in intervertebral disc. J. Mater. Sci. Mater. Med. 2017;28:6. doi: 10.1007/s10856-016-5787-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y.-J., Larsson M., Huang W.-T., Chiou S.-H., Nicholls S.J., Chao J.-I., Liu D.-M. The use of polymer-based nanoparticles and nanostructured materials in treatment and diagnosis of cardiovascular diseases: Recent advances and emerging designs. Prog. Polym. Sci. 2016;57:153–178. [Google Scholar]
- 13.Cavarretta E., Frati G. MicroRNAs in Coronary Heart Disease: Ready to Enter the Clinical Arena? BioMed Res. Int. 2016;2016:2150763. doi: 10.1155/2016/2150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devaux Y., Stammet P., Friberg H., Hassager C., Kuiper M.A., Wise M.P., Nielsen N., Biomarker subcommittee of TTM trial (Target Temperature Management After Cardiac Arrest, NCT01020916) MicroRNAs: new biomarkers and therapeutic targets after cardiac arrest? Crit. Care. 2015;19:54. doi: 10.1186/s13054-015-0767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiagen . 2018. Guidelines for miRNA mimic and miRNA inhibitor experiments.https://www.qiagen.com/fr/resources/resourcedetail?id=3e1477ad-74a2-4ee6-9c31-54b1997f2941&lang=en [Google Scholar]
- 16.van Rooij E., Marshall W.S., Olson E.N. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ. Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Small E.M., Olson E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen M.M., Gianneschi N.C., Christman K.L. Developing injectable nanomaterials to repair the heart. Curr. Opin. Biotechnol. 2015;34:225–231. doi: 10.1016/j.copbio.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gadde S., Rayner K.J. Nanomedicine Meets microRNA: Current Advances in RNA-Based Nanotherapies for Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2016;36:e73–e79. doi: 10.1161/ATVBAHA.116.307481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong S.B., Katwadi K., Kwek X.Y., Ismail N.I., Chinda K., Ong S.G., Hausenloy D.J. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin. Ther. Targets. 2018;22:247–261. doi: 10.1080/14728222.2018.1439015. [DOI] [PubMed] [Google Scholar]
- 21.Small E.M., Frost R.J.A., Olson E.N. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E., Olson E.N. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu S., Huang M., Li Z., Jia F., Ghosh Z., Lijkwan M.A., Fasanaro P., Sun N., Wang X., Martelli F. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122(11, Suppl):S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutcheson R., Terry R., Chaplin J., Smith E., Musiyenko A., Russell J.C., Lincoln T., Rocic P. MicroRNA-145 restores contractile vascular smooth muscle phenotype and coronary collateral growth in the metabolic syndrome. Arterioscler. Thromb. Vasc. Biol. 2013;33:727–736. doi: 10.1161/ATVBAHA.112.301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong D., Yoo J., Lee P., Kepreotis S.V., Lee A., Wahlquist C., Brown B.D., Kho C., Mercola M., Hajjar R.J. miR-25 Tough Decoy Enhances Cardiac Function in Heart Failure. Mol. Ther. 2018;26:718–729. doi: 10.1016/j.ymthe.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karakikes I., Chaanine A.H., Kang S., Mukete B.N., Jeong D., Zhang S., Hajjar R.J., Lebeche D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meloni M., Marchetti M., Garner K., Littlejohns B., Sala-Newby G., Xenophontos N., Floris I., Suleiman M.S., Madeddu P., Caporali A., Emanueli C. Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol. Ther. 2013;21:1390–1402. doi: 10.1038/mt.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey R., Velasquez S., Durrani S., Jiang M., Neiman M., Crocker J.S., Benoit J.B., Rubinstein J., Paul A., Ahmed R.P. MicroRNA-1825 induces proliferation of adult cardiomyocytes and promotes cardiac regeneration post ischemic injury. Am. J. Transl. Res. 2017;9:3120–3137. [PMC free article] [PubMed] [Google Scholar]
- 29.Quattrocelli M., Crippa S., Montecchiani C., Camps J., Cornaglia A.I., Boldrin L., Morgan J., Calligaro A., Casasco A., Orlacchio A. Long-term miR-669a therapy alleviates chronic dilated cardiomyopathy in dystrophic mice. J. Am. Heart Assoc. 2013;2:e000284. doi: 10.1161/JAHA.113.000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y., Shen Z., Miao L., Xin X., Lin S., Zhu Y., Guo W., Zhu Y.Z. miRNA-30 family inhibition protects against cardiac ischemic injury by regulating cystathionine-γ-lyase expression. Antioxid. Redox Signal. 2015;22:224–240. doi: 10.1089/ars.2014.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das S., Bedja D., Campbell N., Dunkerly B., Chenna V., Maitra A., Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE. 2014;9:e96820. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng S., Zhao Q., Zhen L., Zhang C., Liu C., Wang G., Zhang L., Bao L., Lu Y., Meng L. Neonatal Heart-Enriched miR-708 Promotes Proliferation and Stress Resistance of Cardiomyocytes in Rodents. Theranostics. 2017;7:1953–1965. doi: 10.7150/thno.16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kheirolomoom A., Kim C.W., Seo J.W., Kumar S., Son D.J., Gagnon M.K.J., Ingham E.S., Ferrara K.W., Jo H. Multifunctional Nanoparticles Facilitate Molecular Targeting and miRNA Delivery to Inhibit Atherosclerosis in ApoE(-/-) Mice. ACS Nano. 2015;9:8885–8897. doi: 10.1021/acsnano.5b02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesizza P., Prosdocimo G., Martinelli V., Sinagra G., Zacchigna S., Giacca M. Single-Dose Intracardiac Injection of Pro-Regenerative MicroRNAs Improves Cardiac Function After Myocardial Infarction. Circ. Res. 2017;120:1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 35.Gomes R.S., das Neves R.P., Cochlin L., Lima A., Carvalho R., Korpisalo P., Dragneva G., Turunen M., Liimatainen T., Clarke K. Efficient pro-survival/angiogenic miRNA delivery by an MRI-detectable nanomaterial. ACS Nano. 2013;7:3362–3372. doi: 10.1021/nn400171w. [DOI] [PubMed] [Google Scholar]
- 36.Li R.Q., Wu Y., Zhi Y., Yang X., Li Y., Xua F.J., Du J. PGMA-Based Star-Like Polycations with Plentiful Hydroxyl Groups Act as Highly Efficient miRNA Delivery Nanovectors for Effective Applications in Heart Diseases. Adv. Mater. 2016;28:7204–7212. doi: 10.1002/adma.201602319. [DOI] [PubMed] [Google Scholar]
- 37.Xue X., Shi X., Dong H., You S., Cao H., Wang K., Wen Y., Shi D., He B., Li Y. Delivery of microRNA-1 inhibitor by dendrimer-based nanovector: An early targeting therapy for myocardial infarction in mice. Nanomedicine (Lond.) 2018;14:619–631. doi: 10.1016/j.nano.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Pulkkinen K.H., Ylä-Herttuala S., Levonen A.-L. Heme oxygenase 1 is induced by miR-155 via reduced BACH1 translation in endothelial cells. Free Radic. Biol. Med. 2011;51:2124–2131. doi: 10.1016/j.freeradbiomed.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Song M.A., Paradis A.N., Gay M.S., Shin J., Zhang L. Differential expression of microRNAs in ischemic heart disease. Drug Discov. Today. 2015;20:223–235. doi: 10.1016/j.drudis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouzet F., Bachelet-Violette L., Alsac J.-M., Suzuki M., Meulemans A., Louedec L., Petiet A., Jandrot-Perrus M., Chaubet F., Michel J.B. Radiolabeled fucoidan as a p-selectin targeting agent for in vivo imaging of platelet-rich thrombus and endothelial activation. J. Nucl. Med. 2011;52:1433–1440. doi: 10.2967/jnumed.110.085852. [DOI] [PubMed] [Google Scholar]
- 41.Saboural P., Chaubet F., Rouzet F., Al-Shoukr F., Azzouna R.B., Bouchemal N., Picton L., Louedec L., Maire M., Rolland L. Purification of a low molecular weight fucoidan for SPECT molecular imaging of myocardial infarction. Mar. Drugs. 2014;12:4851–4867. doi: 10.3390/md12094851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachelet L., Bertholon I., Lavigne D., Vassy R., Jandrot-Perrus M., Chaubet F., Letourneur D. Affinity of low molecular weight fucoidan for P-selectin triggers its binding to activated human platelets. Biochim. Biophys. Acta. 2009;1790:141–146. doi: 10.1016/j.bbagen.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Bonnard T., Yang G., Petiet A., Ollivier V., Haddad O., Arnaud D., Louedec L., Bachelet-Violette L., Derkaoui S.M., Letourneur D. Abdominal aortic aneurysms targeted by functionalized polysaccharide microparticles: a new tool for SPECT imaging. Theranostics. 2014;4:592–603. doi: 10.7150/thno.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silva A.K.A., Letourneur D., Chauvierre C. Polysaccharide nanosystems for future progress in cardiovascular pathologies. Theranostics. 2014;4:579–591. doi: 10.7150/thno.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M., Bachelet-Violette L., Rouzet F., Beilvert A., Autret G., Maire M., Menager C., Louedec L., Choqueux C., Saboural P. Ultrasmall superparamagnetic iron oxide nanoparticles coated with fucoidan for molecular MRI of intraluminal thrombus. Nanomedicine (Lond.) 2015;10:73–87. doi: 10.2217/nnm.14.51. [DOI] [PubMed] [Google Scholar]
- 46.Juenet M., Varna M., Aid-Launais R., Chauvierre C., Letourneur D. Nanomedicine for the molecular diagnosis of cardiovascular pathologies. Biochem. Biophys. Res. Commun. 2015;468:476–484. doi: 10.1016/j.bbrc.2015.06.138. [DOI] [PubMed] [Google Scholar]
- 47.Chauvierre C., Labarre D., Couvreur P., Vauthier C. Novel polysaccharide-decorated poly(isobutyl cyanoacrylate) nanoparticles. Pharm. Res. 2003;20:1786–1793. doi: 10.1023/b:pham.0000003376.57954.2a. [DOI] [PubMed] [Google Scholar]
- 48.Chauvierre C., Labarre D., Couvreur P., Vauthier C. Radical emulsion polymerization of alkylcyanoacrylates initiated by the redox system dextran-cerium(IV) under acidic aqueous conditions. Macromolecules. 2003;36:6018–6027. [Google Scholar]
- 49.Chauvierre C., Manchanda R., Labarre D., Vauthier C., Marden M.C., Leclerc L. Artificial oxygen carrier based on polysaccharides-poly(alkylcyanoacrylates) nanoparticle templates. Biomaterials. 2010;31:6069–6074. doi: 10.1016/j.biomaterials.2010.04.039. [DOI] [PubMed] [Google Scholar]
- 50.Lira M.C.B., Santos-Magalhães N.S., Nicolas V., Marsaud V., Silva M.P.C., Ponchel G., Vauthier C. Cytotoxicity and cellular uptake of newly synthesized fucoidan-coated nanoparticles. Eur. J. Pharm. Biopharm. 2011;79:162–170. doi: 10.1016/j.ejpb.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Fink S.L., Cookson B.T. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoelzle M.K., Svitkina T. The cytoskeletal mechanisms of cell-cell junction formation in endothelial cells. Mol. Biol. Cell. 2012;23:310–323. doi: 10.1091/mbc.E11-08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paszek E., Czyz J., Woźnicka O., Jakubiak D., Wojnarowicz J., Łojkowski W., Stepień E. Zinc oxide nanoparticles impair the integrity of human umbilical vein endothelial cell monolayer in vitro. J. Biomed. Nanotechnol. 2012;8:957–967. doi: 10.1166/jbn.2012.1463. [DOI] [PubMed] [Google Scholar]
- 54.Matuszak J., Baumgartner J., Zaloga J., Juenet M., da Silva A.E., Franke D., Almer G., Texier I., Faivre D., Metselaar J.M. Nanoparticles for intravascular applications: physicochemical characterization and cytotoxicity testing. Nanomedicine (Lond.) 2016;11:597–616. doi: 10.2217/nnm.15.216. [DOI] [PubMed] [Google Scholar]
- 55.Verderio P., Avvakumova S., Alessio G., Bellini M., Colombo M., Galbiati E., Mazzucchelli S., Avila J.P., Santini B., Prosperi D. Delivering colloidal nanoparticles to mammalian cells: a nano-bio interface perspective. Adv. Healthc. Mater. 2014;3:957–976. doi: 10.1002/adhm.201300602. [DOI] [PubMed] [Google Scholar]
- 56.Juenet M., Aid-Launais R., Li B., Berger A., Aerts J., Ollivier V., Nicoletti A., Letourneur D., Chauvierre C. Thrombolytic therapy based on fucoidan-functionalized polymer nanoparticles targeting P-selectin. Biomaterials. 2018;156:204–216. doi: 10.1016/j.biomaterials.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 57.Barman B., Bhattacharyya S.N. mRNA Targeting to Endoplasmic Reticulum Precedes Ago Protein Interaction and MicroRNA (miRNA)-mediated Translation Repression in Mammalian Cells. J. Biol. Chem. 2015;290:24650–24656. doi: 10.1074/jbc.C115.661868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Detzer A., Sczakiel G. Phosphorothioate-stimulated uptake of siRNA by mammalian cells: a novel route for delivery. Curr. Top. Med. Chem. 2009;9:1109–1116. doi: 10.2174/156802609789630884. [DOI] [PubMed] [Google Scholar]
- 59.Huang R.B., Mocherla S., Heslinga M.J., Charoenphol P., Eniola-Adefeso O. Dynamic and cellular interactions of nanoparticles in vascular-targeted drug delivery. Mol. Membr. Biol. 2010;27:312–327. doi: 10.3109/09687688.2010.522117. [DOI] [PubMed] [Google Scholar]
- 60.Singh S., Narang A.S., Mahato R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 61.Cao Y., Gong Y., Liu L., Zhou Y., Fang X., Zhang C., Li Y., Li J. The use of human umbilical vein endothelial cells (HUVECs) as an in vitro model to assess the toxicity of nanoparticles to endothelium: a review. J. Appl. Toxicol. 2017;37:1359–1369. doi: 10.1002/jat.3470. [DOI] [PubMed] [Google Scholar]
- 62.Ma X., Ma C., Zheng X. MicroRNA-155 in the pathogenesis of atherosclerosis: a conflicting role? Heart Lung Circ. 2013;22:811–818. doi: 10.1016/j.hlc.2013.05.651. [DOI] [PubMed] [Google Scholar]
- 63.Mito S., Ozono R., Oshima T., Yano Y., Watari Y., Yamamoto Y., Brydun A., Igarashi K., Yoshizumi M. Myocardial protection against pressure overload in mice lacking Bach1, a transcriptional repressor of heme oxygenase-1. Hypertension. 2008;51:1570–1577. doi: 10.1161/HYPERTENSIONAHA.107.102566. [DOI] [PubMed] [Google Scholar]
- 64.Watari Y., Yamamoto Y., Brydun A., Ishida T., Mito S., Yoshizumi M., Igarashi K., Chayama K., Ohshima T., Ozono R. Ablation of the bach1 gene leads to the suppression of atherosclerosis in bach1 and apolipoprotein E double knockout mice. Hypertens. Res. 2008;31:783–792. doi: 10.1291/hypres.31.783. [DOI] [PubMed] [Google Scholar]
- 65.Yano Y., Ozono R., Oishi Y., Kambe M., Yoshizumi M., Ishida T., Omura S., Oshima T., Igarashi K. Genetic ablation of the transcription repressor Bach1 leads to myocardial protection against ischemia/reperfusion in mice. Genes Cells. 2006;11:791–803. doi: 10.1111/j.1365-2443.2006.00979.x. [DOI] [PubMed] [Google Scholar]
- 66.Ma S., Tian X.Y., Zhang Y., Mu C., Shen H., Bismuth J., Pownall H.J., Huang Y., Wong W.T. E-selectin-targeting delivery of microRNAs by microparticles ameliorates endothelial inflammation and atherosclerosis. Sci. Rep. 2016;6:22910. doi: 10.1038/srep22910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown D.I., Griendling K.K. Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circ. Res. 2015;116:531–549. doi: 10.1161/CIRCRESAHA.116.303584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Connor P.M., Gutterman D.D. Resurrecting hope for antioxidant treatment of cardiovascular disease: focus on mitochondria. Circ. Res. 2010;107:9–11. doi: 10.1161/CIRCRESAHA.110.223321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seddon M., Looi Y.H., Shah A.M. Oxidative stress and redox signalling in cardiac hypertrophy and heart failure. Heart. 2007;93:903–907. doi: 10.1136/hrt.2005.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsutsui H., Kinugawa S., Matsushima S. Oxidative stress and heart failure. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2181–H2190. doi: 10.1152/ajpheart.00554.2011. [DOI] [PubMed] [Google Scholar]
- 71.Ollivier V., Parry G.C.N., Cobb R.R., de Prost D., Mackman N. Elevated cyclic AMP inhibits NF-kappaB-mediated transcription in human monocytic cells and endothelial cells. J. Biol. Chem. 1996;271:20828–20835. doi: 10.1074/jbc.271.34.20828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.