Abstract
Patients who sustain a traumatic brain injury (TBI) are known to have a significantly quicker fracture healing time than patients with isolated fractures, but the underlying mechanism has yet to be identified. In this study, we found that the upregulation of miRNA-26a-5p induced by TBI correlated with a decrease in phosphatase and tensin homolog (PTEN) in callus formation. In vitro, overexpressing miRNA-26a-5p inhibited PTEN expression and accelerated osteoblast differentiation, whereas silencing of miRNA-26a-5p inhibited osteoblast activity. Reduction of PTEN facilitated osteoblast differentiation via the PI3K/AKT signaling pathway. Through luciferase assays, we found evidence that PTEN is a miRNA-26a-5p target gene that negatively regulates the differentiation of osteoblasts. Moreover, the present study confirmed that preinjection of agomiR-26a-5p leads to increased bone formation. Collectively, these results indicate that miRNA-26a-5p overexpression may be a key factor governing the improved fracture healing observed in TBI patients after the downregulation of PTEN and PI3K/AKT signaling. Upregulation of miRNA-26a-5p may therefore be a promising therapeutic strategy for promoting fracture healing.
Keywords: traumatic brain injury, fracture, miRNA, PTEN
Introduction
Patients with fractures and traumatic brain injuries (TBIs) are commonly observed in trauma settings. Those with both TBI and bone fracture have long been believed to heal more quickly than those with an isolated bone fracture. Researchers have suggested that there is an association between TBI, rapid callus formation, and heterotopic ossification in over 20% of cases.1, 2 In an in vivo study, Locher et al.3 suggested that TBIs can result in increased callus formation and higher mineral density compared with normal bone healing. The mechanisms underlying this phenomenon, however, remain unexplained.
Bone remodeling is an essential process in fracture healing, with osteoblasts being the primary mediators of bone formation.4 In recent decades, many researchers have shown that osteoblast differentiation is triggered by up- or downregulation of the hormones and growth factors that influence osteoblast differentiation and proliferation.5, 6, 7, 8, 9 MicroRNAs (miRNAs) are also believed to be significant regulators of osteoblast differentiation and bone development.10, 11 New evidence regarding the relationship between miRNAs and osteoblast differentiation has been identified.12 An in vitro study has also indicated that miR-208a-3p suppresses the differentiation of osteoblasts by ACVR1 targeting.13 Many relevant miRNAs, however, have thus far been assessed only in vitro. A wide range of factors influence osteoblast differentiation and miRNA expression, including cytokines (e.g., RANKL and BMP), signal transduction proteins (e.g., Wnt), and transcription factors (e.g., FOXO1 and Runx2).14, 15, 16, 17, 18 Several miRNAs are aberrantly expressed in TBI, playing various roles during the brain-injury-repair process.19, 20 MiR-26a-5p is a small noncoding RNA that has been reported to be upregulated after TBI. It also plays several widely reported roles in human health and disease.21 Li et al.22 reported that miR-26a is essential for the regulation of bone growth in individuals with osteoporosis, and administration of miRNA-26a treatment is sufficient to improve mesenchymal-stem-cell-associated osteogenesis. At present, however, we are not aware of any studies that assessed the effects of miRNA-26a on the osteogenic differentiation capabilities of MC3T3-E1 cells.
The gene, phosphatase and tensin homolog deleted on human chromosome 10 (PTEN), codes for a tumor-suppressor PIP phosphatase protein important for phosphatidylinositol (PI) (3,4,5) P3 dephosphorylation, thereby suppressing PI3K signaling.23, 24 The pathological role of PTEN in humans has been well described. Specifically, it has been linked to the D3 dephosphorylation of the inositol ring of PI 3,4,5-trisphosphate, PI 3,4-diphosphate, PI 3-phosphate, and inositol 1,3,4,5-tetrakisphosphate.25, 26, 27, 28, 29, 30 The physiological role of PTEN, as well as the miRNAs that serve as its upstream regulators, are still poorly understood. With respect to bone remodeling, evidence from an in vitro study investigating the endogenous role of PTEN in osteoblasts suggests that bone mass can be maintained by promoting the degradation of PTEN.30, 31 Downregulation of PTEN in bone cells increases endogenous bone mass by PI3K and AKT activation.30
In this study, we sought to assess the underlying mechanisms governing the phenomenon wherein TBI accelerates bone healing in patients with fracture, with a particular focus on miRNA-26a-5p, which is markedly elevated in patients with TBI. In addition, we also aimed to directly target PTEN and to test miRNA-26a-5p and its argonaute miRNA (agomiRNA) for their osteogenic potential in MC3T3-E1 cells.
Results
Serum and Callus miRNA-26a-5p Levels Are Increased after TBI
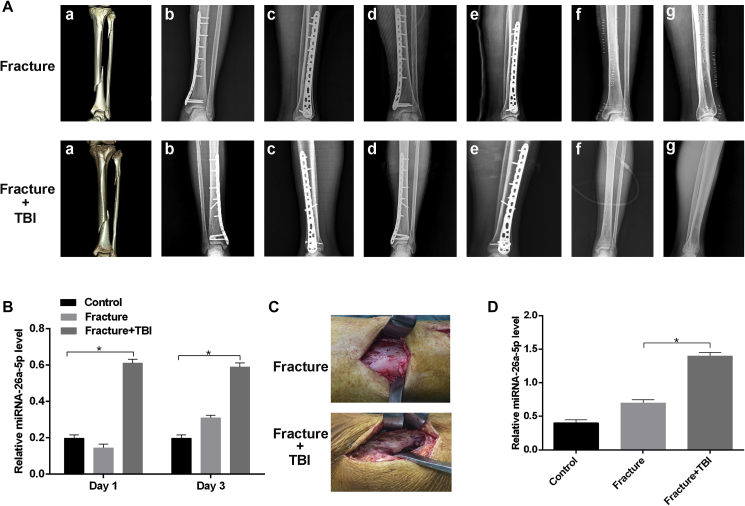
Clinical data indicated that fractures in patients with TBI healed noticeably faster than those in patients with an isolated fracture (Figure 1A). As miRNA-26a-5p has been linked to osteogenesis and bone formation,32 we evaluated miRNA-26a-5p expression in the serum of 18 patients in three groups (control, fracture, and fracture+TBI) by real-time qPCR. To minimize the influence of confounding factors, such as age and gender, all serum specimens were collected from male patients between 30 and 45 years of age. Serum miRNA-26a-5p levels in the fracture+TBI group were significantly increased relative to the isolated-fracture group at 24 and 72 h after injury (Figure 1B). Calluses were collected from patients during surgery and were analyzed by real-time qPCR, revealing that miRNA-26a-5p expression in samples from the fracture+TBI group were significantly increased relative to that in the fracture group (Figures 1C and 1D).
Figure 1.
Overexpression of miRNA-26a-5p in TBI Patients
(A) The fracture+TBI group showed a faster fracture healing rate than the isolated-fracture group. (a) preoperative three-dimensional CT image; anteroposterior (b) and lateral (c) radiographic images at 1 month after surgery; anteroposterior (d) and lateral (e) radiographic images 3 months after surgery; and anteroposterior (f) and lateral (g) radiographic images 1-year after surgery. (B) Relative expression of miRNA-26a-5p was higher in serum samples of the fracture+TBI group than the other groups. (C) The calluses at the fracture site were more visible in fracture+TBI group when compared with those in the fracture group. (D) The fracture+TBI group showed a higher level of miRNA-26a-5p than the other groups. Data are means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. n = 6 patients in each group.
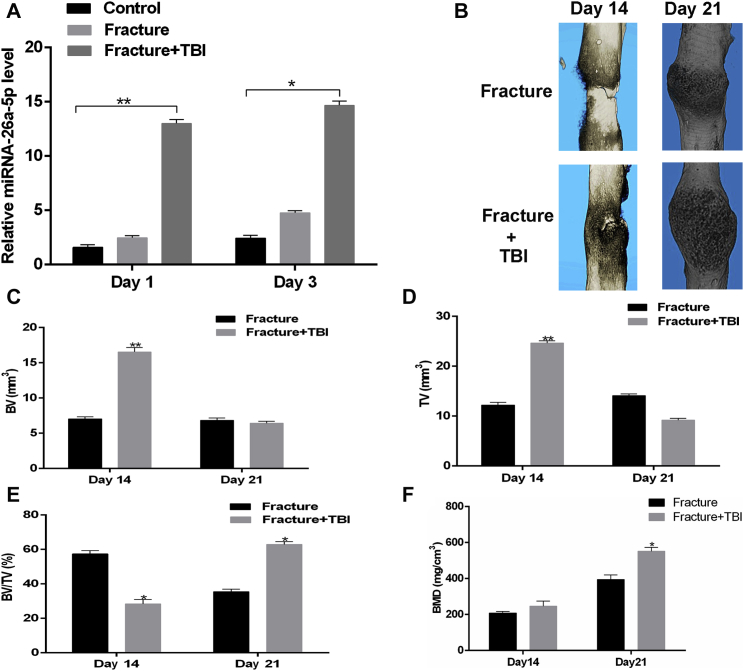
Next, mouse models of isolated fracture and fracture+TBI were developed and were used to assess miRNA-26a-5p levels by real-time PCR. Serum miRNA-26a-5p levels in the fracture+TBI model animals significantly increased relative to that in the fracture-only animals at 24 and 72 h after injury (Figure 2A). Quantitative micro-computed tomography (mCT) was used to assess key parameters within the fractured femur bone, such as trabecular bone volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th), and callus volume was further quantified by mCT. Total and callus bone volumes were increased in the fracture+TBI animals relative to those in fracture-only animals on post-fracture day 14, with a significant increase in bone mineral density (BMD) in the fracture+TBI group relative to the fracture group on post-fracture day 21 (Figures 2B–2F).
Figure 2.
Overexpression of miRNA-26a-5p in TBI Mouse Models
(A) Relative expression of miRNA-26a-5p was higher in fracture+TBI group than in the control group or fracture group at 1 and 3 days after injury. (B) mCT image comparison of fracture healing between fracture and fracture+TBI group at 14 and 21 days after injury. (C–F) BV (C) and TV (D) of the callus, BV/TV (E), and BMD (F) on days 14 and 21 after the operation, as measured by mCT, with n = 6 mice aged 6 weeks per group. Data are means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
miRNA-26a-5p Promotes Matrix Mineralization and Osteoblastogenesis
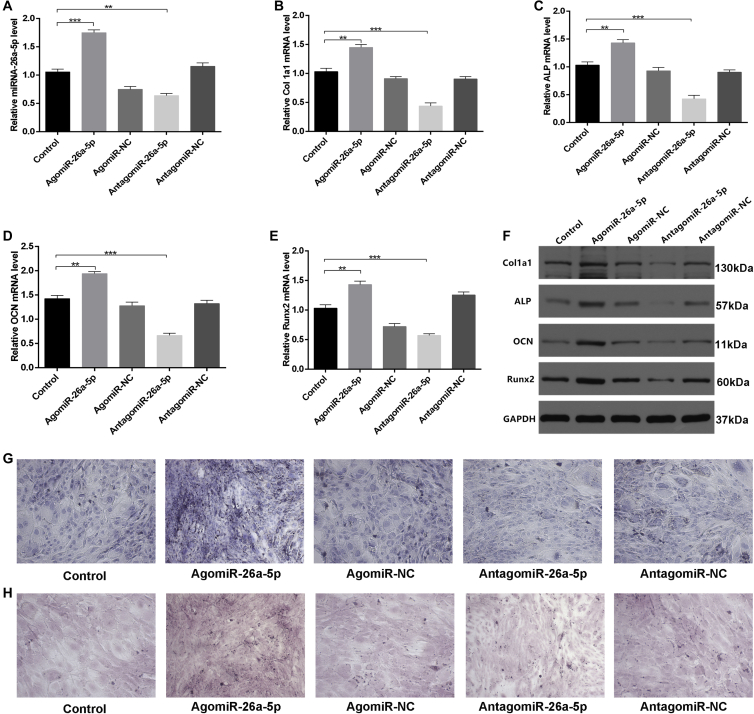
We next assessed the effect of miR-26a-5p on osteoblastogenesis by treating MC3T3-E1 cells with agomiR-negative control (agomiR-NC), agomiR-26a-5p, antagomiR-negative control (antagomiR-NC), antagomiR-26a-5p, or Lipofectamine 3000 only, as a control. We found that miR-26a-5p was upregulated in cells treated with agomiR-26a-5p (Figure 3A). In vitro, the effects of miR-26a-5p were next evaluated by investigating osteoblast activity and the expression of genes linked with bone formation, including alkaline phosphatase (ALP), Col1a1, OCN, and Runx2. After the cells were transfected with the above constructs for 48 h, qPCR and western blot analysis were used to assess these bone formation markers, revealing a significant increase in all the markers in the agomiR-26a-5p group, relative to the other groups (Figures 3B–3F). The relative intensity of western blots of the ALP, Col1a1, OCN, and Runx2 proteins in MC3T3-E1 cells treated with agomiR-NC, agomiR-26a-5p, antagomiR-NC, antagomiR-26a-5, or the corresponding controls for 48 h can be seen in Figure S1.
Figure 3.
miRNA-26a-5p Accelerates Osteoblast Activity and Matrix Mineralization
(A) miRNA-26a-5p was upregulated in agomiR-26a-5p-treated MC3T3-E1 cells after transfection for 48 h with the Lipofectamine control or 200 μM agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, or antagomiR-NC. (B–E) qPCR was used to assess the expression of osteoblast differentiation genes, including Col1a1 (B), ALP (C), OCN (D), and Runx2 (E) in MC3T3-E1 cells treated for 48 h with Lipofectamine control, agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, or antagomiR-NC. (F) Western blot analysis of ALP, Col1a1, OCN, and Runx2 protein levels in MC3T3-E1 cells treated for 48 h with Lipofectamine control, agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, or antagomiR-NC. (G) ALP staining in MC3T3-E1 transfected for 48 h with Lipofectamine control, agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, or antagomiR-NC. Scale bar, 10 mm. (H) Alizarin red-mediated calcium staining in MC3T3-E1 cells after transfection for 21 days with Lipofectamine control, agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, or antagomiR-NC. Scale bar, 10 mm. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Furthermore, to assess how miRNA-26a-5p influences extracellular matrix mineralization, we transfected cells as above, and after continuous culturing for 21 days, higher mineral deposition was evident in the agomiR-26a-5p group relative to other groups, especially when compared to antagomiR-26a-5p (Figures 3G and 3H). Collectively, our results indicated that miRNA-26a-5p positively regulates osteoblastogenesis and thereby accelerates ALP activity and mineralization.
miRNA-26a-5p Directly Targets PTEN
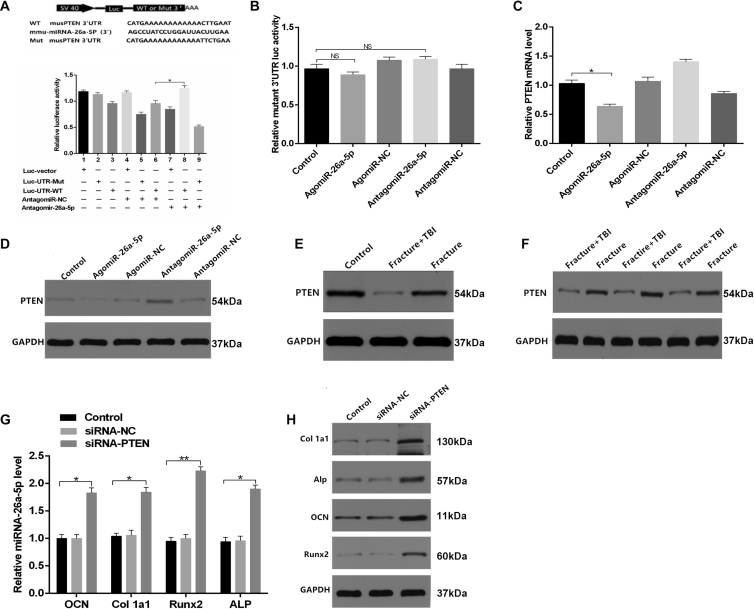
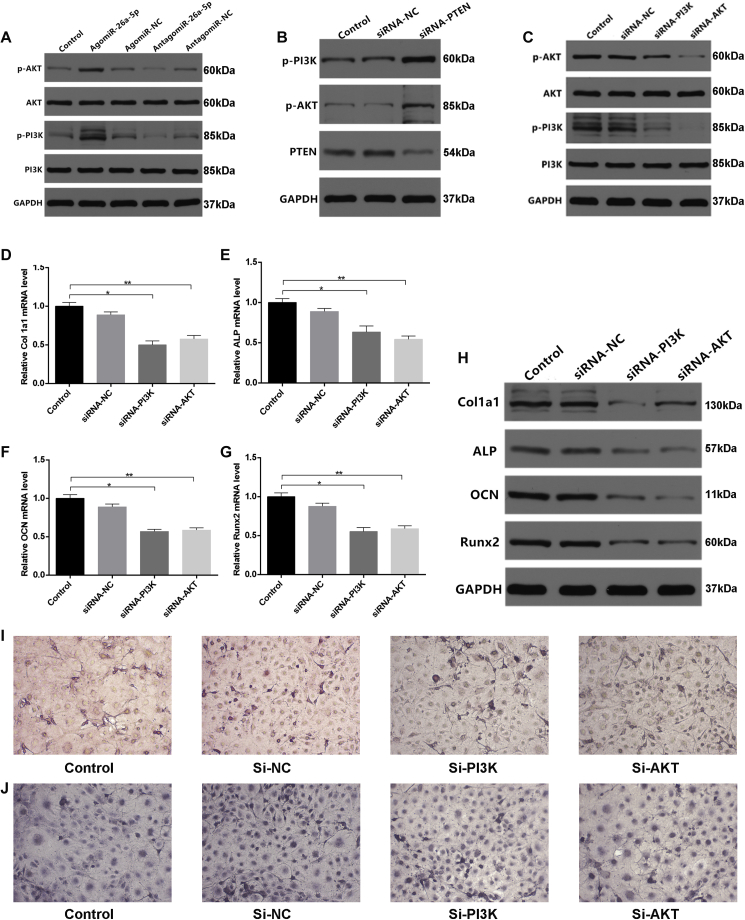
To investigate whether miRNA-26a-5p serves to target PTEN directly, we used either a wild-type (WT) PTEN 3′ UTR or a mutant-type (Mut) PTEN 3′ UTR construct fused to a luciferase reporter (Figure 4A). Using these constructs, we found that antagomiR-26a-5p, but not antagomiR-NC, substantially increased WT PTEN 3′ UTR reporter activity (Figure 4A), whereas it failed to enhance the activity of the mutated 3′ UTR PTEN reporter (Figure 4B). WT PTEN 3′ UTR reporter activity rose significantly after a reduction in endogenous miRNA-26a-5p levels following antagomiR-26a-5p treatment of these MC3T3-E1 cells (Figures 4A and 4B). When cells were transfected with the miRNA constructs for 48 h, we observed lower relative PTEN mRNA levels in the agomiR-26a-5p group, compared with the other groups (Figures 4C and 4D). In order to explore the connection between miRNA-26a-5p and PTEN, we thoroughly examined the expression of PTEN in the context of osteoblast differentiation. Calluses from patients and animal models of fracture or TBI+fracture were collected, and the relative PTEN mRNA levels were assessed by western blot analysis. We observed that there was a significant downregulation of PTEN in both patients and mice with fracture+TBI, relative to their fracture-only counterparts (Figures 4E and 4F), and we calculated the relative intensity of the western blots (Figures S2A–S2D).
Figure 4.
miRNA-26a-5p Targets PTEN to Functionally Inhibit Osteoblast Activity In Vitro
(A) Luciferase reporter constructs containing either a WT PTEN 3′ UTR (WT 3′ UTR) or this same region after site-directed mutagenesis (3′ UTR-Mut). Luc, luciferase. (B) Endogenous miRNA-26a-5p effects in MC3T3-E1 cells on WT PTEN 3′ UTR (luc-UTR), the PTEN 3′ UTR mutant (luc-UTR-Mut), as assessed after antagomiR-NC or antagomiR-26a-5p treatment. (C and D) qPCR (C) and western blot analysis (D) were used after control, agomiR-26a-5p, agomiR-NC, and antagomiR-26a-5p, and antagomiR-NC transfection to assess the expression of PTEN. (E) PTEN expression in calluses of control, fracture, and fracture+TBI patient groups. (F) PTEN expression in calluses of fracture and fracture+TBI mouse groups. (G and H) qPCR (G) and western blot analysis (H) were used after control, siRNA-NC, and siRNA-PTEN transfection to assess Col1a1, ALP, OCN, and Runx2 expression, with a scrambled siRNA used as the negative control (siRNA-NC). Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
To investigate whether osteoblast differentiation is PTEN dependent, we next assessed how PTEN siRNA affects osteoblastogenesis. PCR and western blot analysis revealed that siRNA-PTEN upregulated ALP, Col1a1, Runx2, and OCN (Figures 4G and 4H). Taken together, our results confirmed that miRNA-26a-5p directly targets the 3′ UTR of PTEN in the context of the differentiation of osteoblasts.
PI3K/AKT Signaling Is Involved in PTEN-Regulated Osteoblast Differentiation
In order to further clarify the relationship between PTEN and PI3K/AKT signaling, PI3K, phosphorylated PI3K (p-PI3K), AKT, and phosphorylated AKT (p-AKT) levels were assessed by western blot analysis in cells transfected with the various miRNA constructs, and the relative intensity of the western blots was calculated (Figures S3A–S3C). We found p-PI3K and p-AKT levels in the agomiR-26a-5p group to be higher than in other groups (Figure 5A). To explore how PTEN influences PI3K/AKT signaling, we used PTEN siRNA, PI3K siRNA, and AKT siRNA constructs and investigated their effects on PI3K/AKT signaling. The PTEN siRNA, PI3K siRNA, and AKT siRNA groups all exhibited significant inhibition of the phosphorylation of PI3K and AKT (Figures 5B and 5C).
Figure 5.
Reduction of PTEN Facilitated Osteoblast Differentiation via Activating PI3K/AKT Signaling
(A) Western blot assessment of PI3K and AKT expression after transfection, using Lipofectamine control, agomiR-NC, agomiR-26a-5p, antagomiR-NC, or antagomiR-26a-5p transfection for 48 h. (B) Western blot assessment of PTEN, PI3K, and AKT levels after transfection with siRNA-NC, siRNA-PTEN or appropriate controls for 48 h. (C) Western blot analysis assessment of p-AKT, AKT, p-PI3K, and p-PI3K levels after transfection using control, 200 μM siRNA-NC, siRNA-PI3K, or siRNA-AKT for 48 h. (D–G) qPCR analysis of Col1a1 (D), ALP (E), OCN (F), and Runx2 (G) expression in MC3T3-E1 cells after transfection with control, siRNA-NC, siRNA-PI3K, or siRNA-AKT for 48 h. (H) Western blot analysis of Col1a1, ALP, OCN, and Runx2 levels after transfection with control, siRNA-NC, siRNA-PI3K, or siRNA-AKT for 48 h. (I) Alizarin red-mediated staining for calcium deposition in MC3T3-E1 cells after transfection with, siRNA-NC, siRNA-PI3K, or siRNA-AKT for 21 days. Scale bar, 10 mm. (J) ALP staining of MC3T3-E1 cells after transfection with control, siRNA-NC, siRNA-PI3K or siRNA-AKT for 48 h. Scale bar, 10 mm. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
To investigate whether osteoblastogenesis is dependent upon PI3K/AKT, we employed the PI3K- and AKT-targeting siRNAs to assess their effects on this process. We found that suppressing PI3K and AKT led to decreased ALP, Col1a1, Runx2, and OCN expression (Figures 5D–5H) when we calculated the relative intensity of the western blots (Figure S3D). Furthermore, to investigate how PI3K and AKT influence extracellular matrix mineralization, the cells were transfected with control siRNA or siRNAs targeting AKT or PI3K. After continuous cultivation for 21 days, lower mineral deposition was observed in the PI3K and AKT siRNA groups, relative to the control cells (Figure 5I). Similarly, PI3K and AKT siRNAs reduced ALP activity and staining in the PI3K and AKT siRNA groups (Figure 5J).
Local Administration of miRNA-26a-5p Accelerates Femur Fracture Healing in Mice
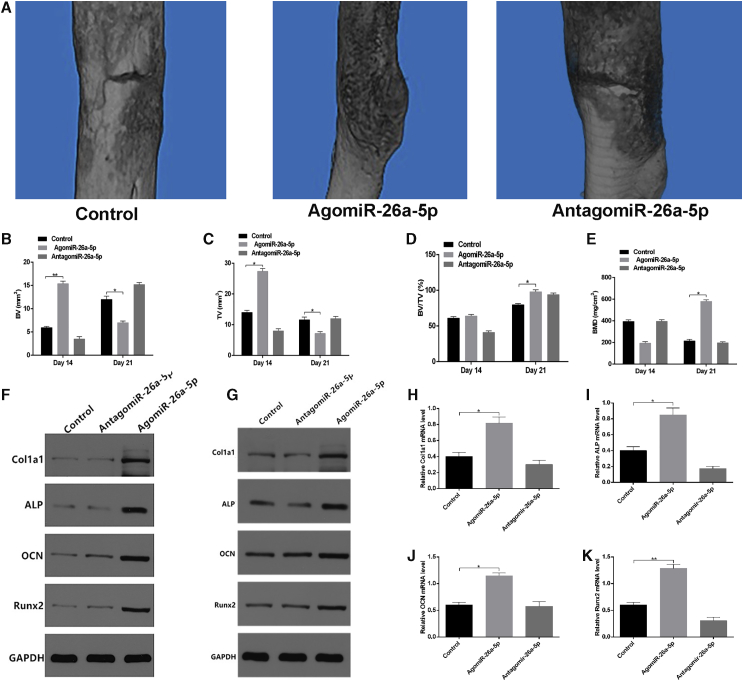
It has been reported that intraperitoneal injection of locked nucleic acid (LNA) can effectively knockdown specific miRNAs.33 We therefore administered PBS, agomiR-26a-5p, or antagomiR-26a-5p directly to the local fracture site in our model animals, to assess whether agomiR-26a-5p improves fracture healing. Local injection was performed at three time points (days 1, 3, and 7) after injury, and CT examination was performed at 14 and 21 days after injury. Mice were sacrificed after each mCT examination, bone calluses at the fractured position were harvested for western blot and PCR analyses, and the relative intensity of western blots was calculated (Figures S4A and S4B). Mice treated with agomiR-26a-5p exhibited a greater callus volume and reduced fracture gap relative to control and antagomiR-26a-5p animals (Figure 6A). By day 21, animals in the agomiR-26a-5p group, but not in the control or antagomiR-26a-5p groups, exhibited a lack of any visible boundary between the hardened callus and the cortical bone, with clear evidence of remodeling. Furthermore, in agomiR-26a-5p animals, there was a greater total and bone callus volume relative to control and antagomiR-26a-5p animals on post-fracture day 14, and the difference remained significant between agomiR-26a-5p and antagomiR-26a-5p animals on post-fracture day 21. In addition, agomiR-26a-5p animals exhibited significantly higher BMD than did the animals in the other two groups (Figures 6B–6E). In addition, bone formation marker genes were markedly upregulated in the agomiR-26a-5p group, compared with the other groups (Figures 6F–6K). These results indicate that miRNA-26a-5p acts as a positive regulator that accelerates fracture healing.
Figure 6.
Local Administration of AgomiR-26a-5p Enhanced Fracture Healing in Mice
PBS, agomiR-26a-5p, and antagomiR-26a-5p were delivered locally to the fracture site on days 1, 3, and 7 after fracture. (A) mCT image comparison of fracture healing between control, agomiR-26a-5p, and antagomiR-26a-5p groups at day 21 after injury. (B–E) BV (B) and TV (C) of the callus, BV/TV (D), and BMD (E) on days 14 and 21 after the operation were determined by mCT. n = 6 mice/group. (F and G) Western blot analysis of Col1a1, ALP, OCN, and Runx2 levels in calluses of the control, agomiR-26a-5p, and antagomiR-26a-5p groups on days 14 (F) and 21 (G) after fracture. (H–K) qPCR analysis of Col1a1 (H), ALP (I), OCN (J), and Runx2 (K) expression in calluses of control, agomiR-26a-5p, and antagomiR-26a-5p animals on day 21 after fracture. Data are means ± SD of triplicate experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
With the increasing incidence of traffic accidents, TBI and bone fracture are important public health concerns. There is clear value in identifying the association between TBI and bone fracture healing, as such research may provide novel insights allowing for improved bone formation and fracture healing. TBI is thought to promote the expression of pro-osteogenic genes and to drive increased bone mineral deposition.34, 35, 36 Several miRNAs are known to regulate the expression of osteogenic genes linked to bone remodeling, and they can further regulate human osteopathy.37, 38, 39, 40, 41 These miRNAs regulate target mRNA expression, and stimulating of miRNAs using agomiRNAs can thus modulate mRNA expression.42 Previous research has largely explored the role of biotic factors in bone remodeling, with few studies exploring the association between TBI-related miRNAs and bone healing.30, 31, 32, 33, 43, 44 Therefore, this study identified miRNA-26a-5p in osteoblasts as a key activator of bone formation in the context of TBI. The findings indicate that activating miRNA-26a-5p in osteoblasts in a therapeutic context may be of value, resulting in improved bone development and fracture healing.
Previous work has revealed that miRNA-26a-5p is expressed in chronic lymphocytic leukemia, lymphomas, and bone defects, indicating that miRNA-26a-5p participates in immunoreactivity and tissue regeneration.45, 46, 47 Several studies have indicated that TBI modulates the expression of many miRNAs that regulate cellular responses to this condition.48, 49, 50, 51 Because miRNA-26a-5p levels rise in TBI patients and the increase was validated in our study, we investigated the effects of miRNA-26a-5p in fracture healing. We ultimately clarified a novel mechanism wherein miRNA-26a-5p regulates PTEN expression in bone tissue. When PTEN expression is suppressed, it drives the expression of genes associated with osteoblastogenesis and promotes bone formation. PTEN expression is controlled at multiple levels in a cell-type, context-dependent manner.24, 25, 30 We found that miRNA-26a-5p expression modulates PTEN expression, indicating that PTEN is likely a functional miRNA-26a-5p target, explaining its ability to modulate bone formation.
PI3K/AKT signaling is a cell-growth-regulatory pathway involved in many diseases.52, 53 Previous studies54, 55 suggested that this pathway is essential for the differentiation of bone progenitor cells. Fan et al.54 suggested that tetrahydroxystilbene glucoside (TSG) can promote the differentiation of MC3T3-E1 cells via activation of PI3K/AKT signaling. In addition, the importance of PTEN as a regulator of PI3K/AKT signaling in osteoblasts has been validated in another study.30 Herein, we found that downregulation of PTEN activated PI3K/AKT signaling, leading to the overexpression of osteoblast-specific gene transcription.
In the present study, we focused our research only on middle-aged males, in order to exclude the effects of sex or age on the expression of miRNAs. Future studies should therefore extend these findings to female patients and individuals of different ages. In addition, we were not able to collect calluses from all patients on the same day after injury, and the expression of miRNA-26a-5p levels in the calluses may have been affected by the different times of surgery and the differences between individual patients. Even so, our study suggests the potential value of the therapeutic activation of miRNA-26a-5p in osteoblasts as a means of accelerating fracture healing. More work is needed to investigate the extent of miRNA-26a-5p dysfunction in bone formation in the context of TBI.
Conclusions
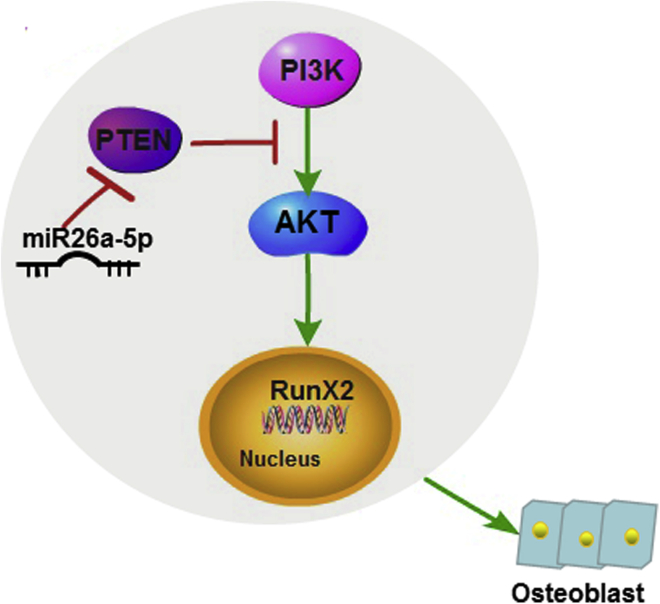
This study indicates not only that the stimulation of miRNA-26a-5p using agomiR-26a-5p increases osteoblast differentiation in MC3T3-E1 cells, but it highlights the potential signaling pathways involved in regulating fracture healing in the context of TBI (Figure 7). Moreover, treatment with agomiR-26a-5p decreased PTEN expression and increased bone formation marker gene expression, suggesting that bone architecture is also improved by this treatment. Overall, this allows us to conclude that the activation of miRNA-26a-5p and the subsequent decrease in PTEN expression by agomiR-26a-5p represents a promising therapeutic approach for accelerating fracture healing.
Figure 7.
A Diagrammatic Sketch of miR-26a-5p-Mediated Osteoblast Differentiation
miR-26a-5p is upregulated in bone and reduces PTEN translation, thereby activating the PI3K-AKT signaling pathway, subsequently inducing RunX2 expression and increasing bone formation.
Materials and Methods
Human Serum and Bone Tissue Preparation
Serum from 18 male patients (including 6 without fractures, 6 with fractures patients, and 6 with fracture+TBI) were obtained from the Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, via venous blood draw. Twelve calluses were obtained from the patients who had fractures or fractures+TBI. The clinical information of those patients is presented in Table S1; their mean age was 38.78 ± 6.02 years. Admitted patients underwent conventional X-ray imaging of fractures, and the Glasgow Coma Scale (GCS) was used to identify patients with a TBI, which was ultimately diagnosed on the basis of a GCS score of 9 to 12, together with CT findings. Those in the fracture-only group had GCS scores of 13 or higher. Patients with previous bone-related or nervous system diseases, malignant disease, open type III fractures, diabetes, autoimmune diseases, or other chronic inflammatory disease were excluded, as were patients with a history of prolonged use of steroids, non-steroidal anti-inflammatory drugs, or immunosuppressant agents or of bisphosphonate therapy. The Committees of Clinical Ethics in the Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China), approved all studies, and informed consent was obtained from all participants.
Femoral Fracture Models
A total of 18 male C57BL/6J mice (age 6 weeks) were from The Center of Experimental Animals, Tongji Medical College, Huazhong University of Science and Technology. We used 10% chloral hydrate (0.3 mL/100 g body weight) for anesthesia. The Institutional Animal Care and Use Committee at Tongji Medical College, Huazhong University of Science and Technology, approved all animal studies. A mouse femoral fracture model was created surgically by longitudinal incision and blunt separation of the underlying muscles without removal of the periosteum, as described in a previous study.39 A diamond disk was used to cut the femur, producing a transverse osteotomy in the mid-diaphysis region. The fractures were then stabilized with a 23G intramedullary needle. After 14 days, half of the mice were sacrificed, and the calluses at the fracture location were harvested for subsequent analysis. On day 21 after the operation, the remaining mice were sacrificed, and the callus was harvested for subsequent analysis.
Fracture Concomitant with TBI Mouse Models
Six of the 18 mice were modeled as fractures concomitant with TBI. A weight-drop device was used to model TBI.56 The mice were anesthetized and placed under this device on a platform, with the skull being exposed by a 1.0 cm midline longitudinal scalp incision made with a power micro drill. Once a 1.5 mm impact area was exposed, a 24 newton weight was dropped onto this area from 5 cm above. Thereafter, the wound was closed, and the animals were returned to their cages.
Blood Collection
Blood samples were collected via the orbital canthus vein, 1 and 3 days after surgery for determination of miRNA-26a-5p levels. miRNA-26a-5p levels were analyzed by qPCR.
mCT Analysis
The fracture site was scanned with a Bruker SkyScan 1176 scanner mCT system to provide images at 2,400 views, 5 frames/view, 37 kV, and 121 mA, and these images were then analyzed with Bruker mCT evaluation software, to facilitate segmentation and 3-dimensional morphometric analysis and to measure density and distance parameters (Bruker, Germany). The soft tissues were removed thoroughly from the bone, and ethanol was used to fix the femurs. After the scanning, the calluses were preserved at −80°C for miRNA detection by PCR and western blot analysis. Parameters measured were as follows: Tb.N, BV/TV per tissue volume, average cortical thickness (Ct.Th), cortical area fraction (Ct.Ar/Tt.Ar), cortical bone area (Ct.Ar), Tb.Sp., Tb.Th, total cross-sectional area (Tt.Ar), and BMD.
Western Blot Analysis
Calluses or cells were lysed with lysis buffer (AS1004; Aspen, South Africa) with 1% protease inhibitor (AS1008; Aspen, South Africa) on ice. The protein fractions were collected and underwent SDS-PAGE separation, followed by transfer onto a nitrocellulose membrane (IPVH00010; Millipore, USA). Membranes were blocked using 5% skim milk and then probed at 4°C overnight with primary antibodies, followed by horseradish-peroxidase (HRP)-conjugated secondary antibodies (AS1058; Aspen, South Africa). A chemiluminescence detection system (LiDE110; Canon, Japan ) was used to visualize protein based on provided directions. Antibodies were as follows: anti-collagen I (1:500, ab34710; Abcam, USA, ), anti-ALP (1:1,000, ab95462; Abcam, USA), anti-osteocalcin (1:500, ab93876; Abcam, USA), anti-RunX2 (1:500, ab23981; Abcam, USA), anti-PTEN (1:2,000, 9188; CST, USA, ), anti-p-AKT (1:1,000, 4060; CST, USA), anti-AKT (1:2,000, 9272; CST, USA), anti-p-PI3K (1:500, ab182651; Abcam, USA), anti-PI3K (1:2,000, 4292; Abcam, USA), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10,000, ab37168; Abcam, USA). All experiments were conducted in triplicate.
miRNA Extraction and qPCR
RNAlater (76104; QIAGEN, Germany) was used to preserve callus samples prior to miRNA extraction. TRIzol (15596018; Invitrogen, USA) was used for RNA extraction, based on provided protocols. cDNA was generated through the one-step Prime Script miRNA cDNA synthesis kit, and SYBR Premix Ex TaqII (RR820A; TaKaRa, Japan) was used for amplification of equivalent cDNA amounts. A Thermal Cycler C-1000 Touch system (CFX Manager, 10021377; Bio-Rad, USA) was used to perform qPCR analysis, with the U6 small nuclear RNA (snRNA) or GAPDH mRNA used for normalization. The data are given as fold change over the appropriate controls. All primer sequences are shown in Table 1.
Table 1.
miRNAs and mRNA Primer Sequences
| microRNA or Gene Name | Primer Sequence (5′ to 3′) |
|---|---|
| Mmu-miR-26a-5p-forward | GGTTCAAGTAATCCAGGATAGGCT |
| Mmu-miR-26a-5p-reverse | CTCAACTGGTGTCGTGGAGTC |
| Mmu-miR-U6-forward | CTCGCTTCGGCAGCACAT |
| Mmu-miR-U6-reverse | AACGCTTCACGAATTTGCGT |
| M-PTEN-forward | TGAGAGACATTATGACACCGCC |
| M-PTEN-reverse | TTACAGTGAATTGCTGCAACATG |
| M-ALP-forward | TGACTACCACTCGGGTGAACC |
| M-ALP-reverse | TGATATGCGATGTCCTTGCAG |
| M-COL1A1-forward | CTGACTGGAAGAGCGGAGAG |
| M-COL1A1 | CGGCTGAGTAGGGAACACAC |
| M-OCN-forward | TTCTGCTCACTCTGCTGACCC |
| M-OCN-reverse | CTGATAGCTCGTCACAAGCAGG |
| M-Runx2-forward | CGCCACCACTCACTACCACAC |
| M-Runx2-reverse | TGGATTTAATAGCGTGCTGCC |
| M-GAPDH-forward | TGAAGGGTGGAGCCAAAAG |
| M-GAPDH-reverse | AGTCTTCTGGGTGGCAGTGAT |
Cell Culture and Transfection
MC3T3-E1 cells, which are mouse osteoblasts, were a kind donation from the Huazhong University of Science and Technology (Wuhan, China). The α-minimal essential medium (α-MEM; SH30265.01B; Hyclone, USA) used for their growth contained 10% fetal bovine serum (FBS, 10099141; Gibco, USA) and 1% penicillin and streptomycin (SV30010; Hyclone, USA). The cells were grown at 37°C with 5% CO2 at 95% humidity and were used for up to eight passages. Transfection of agomiR-26a-5p, agomiR-NC, antagomiR-26a-5p, and antagomiR-NC (GenePharma Shanghai) at 200 μM was conducted using Lipofectamine 3000 (L3000001; Thermo Fisher Scientific, USA) based on provided directions. Lipofectamine 3000 was also used to transfect cells with miRNAs or siRNA oligos. Cells were transfected with PTEN siRNA, PI3K siRNA, and AKT siRNA (RIBOBIO, Guangzhou) at 50 nM.
ALP Staining
A 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) ALP color-development kit (C3206; Beyotime, China) was used according to the provided directions to assess ALP staining. Briefly, after the cells were washed twice with PBS, they were fixed with 10% formalin for 15 min. The BCIP/NBT liquid substrate was then applied to the cells for 24 h. The samples were prepared in the dark at room temperature. The color change was observed with a charge-coupled device (CCD) microscope, and the stained cell cultures were imaged with a scanner (V600; Epson, Japan). The samples were analyzed in triplicate.
Alizarin Red Staining
MC3T3-E1 cells were grown in osteogenic medium supplemented with 100 nM dexamethasone, 50 mM ascorbic acid, and 10 mM b-glycerophosphate (HUXMA-90021; Cyagen, USA) in six-well plates to induce osteoblast mineralization. The cells were fixed in 10% formalin at room temperature for 15 min, after which they were washed in 2 mL PBS and stained with 1 mL 0.5% alizarin red staining solution at room temperature for 15 min. The cells were rinsed with distilled water for 5 min with shaking, and red mineralized nodules were assessed with a CCD microscope and scanned (V600; Epson, Japan). All the staining experiments were repeated three times.
Luciferase Reporter Assay
The mouse PTEN 3′ UTR region, which contained a miR-26a-5p binding sequence, was amplified by PCR from murine genomic DNA and subcloned into the pGL3 vector (E1741; Promega, USA). A QuikChange Site-Directed Mutagenesis Kit (210518; Stratagene, USA) was used to mutate this sequence. MC3T3-E1 cells (2.5 × 105 cells per well) were transiently transfected in 24-well plates with Lipofectamine 3000 (L3000001 Thermo Fisher Scientific, USA). Both 100 ng luciferase constructs and 10 ng pRL-TK (E2241; Promega, USA) Renilla luciferase plasmid were transfected into the cells, and a dual luciferase reporter assay system (E1910; Promega, USA) was then used, according to the directions provided. A luminometer (Glomax, Promega) was used for quantification, with firefly luciferase activity being normalized to that of Renilla luciferase.
Therapeutic Stimulation of miR-26a-5p in the Fracture Group
Mice were injected locally, using PBS (control group), agomiR-26a-5p (agomiR-26a-5p group), and antagomiR-26a-5p (antagomiR-26a-5p group), at the fracture sites. A volume of 0.1 mL per injection was administered on days 1, 3, and 7 after fracture induction. agomiR-26a-5p and antagomiR-26a-5p were purchased from GenePharma Shanghai. After the experiments were completed, the animals were euthanized, bones and calluses were isolated, and miR-26a-5p expression was analyzed.
Statistical Analysis
Data are means ± SD, and Prism 6.0 (GraphPad Software, La Jolla, CA) was used for all analyses, unless otherwise noted. One-way analysis of variance with Tukey’s post hoc test was applied to compare three or more groups, and two-tailed Student’s test was applied to compare data between two groups. p < 0.05 was deemed statistically significant.
Author Contributions
Y. X., B.M., and G.L. conceived and designed the experiments. Y.X., B.M., G.L., C.Y., L.C., H.X., Q.W., L.H., Y.L., W.Z., and Y.S. performed the experiments. Y.X., B.M., G.L., and H.X. analyzed the data. P.Z., J.L., and Q.W. contributed reagents, materials, and analysis tools. C.Y. and A.C.P. performed statistical analyses. Y.X. and B.M. wrote the paper. A.C.P. edited the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Our work was supported by the National Science Foundation of China (81772345), the Healthy Commission Key Project of Hubei Province (WJ2019Z009), the Science and Technology Department of Hubei Province (2016CFB424), and the Development Center for Medical Science and Technology, National Health and Family Planning Commission of the People’s Republic of China (ZX-01-C2016024).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.06.001.
Contributor Information
Bobin Mi, Email: mi19882@163.com.
Guohui Liu, Email: liuguohui@hust.edu.cn.
Supplemental Information
References
- 1.Tsitsilonis S., Seemann R., Misch M., Wichlas F., Haas N.P., Schmidt-Bleek K., Kleber C., Schaser K.D. The effect of traumatic brain injury on bone healing: an experimental study in a novel in vivo animal model. Injury. 2015;46:661–665. doi: 10.1016/j.injury.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 2.Perkins R., Skirving A.P. Callus formation and the rate of healing of femoral fractures in patients with head injuries. J. Bone Joint Surg. Br. 1987;69:521–524. doi: 10.1302/0301-620X.69B4.3611150. [DOI] [PubMed] [Google Scholar]
- 3.Locher R.J., Lünnemann T., Garbe A., Schaser K., Schmidt-Bleek K., Duda G., Tsitsilonis S. Traumatic brain injury and bone healing: radiographic and biomechanical analyses of bone formation and stability in a combined murine trauma model. J. Musculoskelet. Neuronal Interact. 2015;15:309–315. [PMC free article] [PubMed] [Google Scholar]
- 4.Ducy P., Schinke T., Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y., Li J., Sheng R., Deng P., Wang Y. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto T. [Recent advances in the regulation of bone remodeling] Nihon Rinsho. 2004;62(Suppl 2):37–46. [PubMed] [Google Scholar]
- 7.Yu B., Zhao X., Yang C., Crane J., Xian L., Lu W., Wan M., Cao X. Parathyroid hormone induces differentiation of mesenchymal stromal/stem cells by enhancing bone morphogenetic protein signaling. J. Bone Miner. Res. 2012;27:2001–2014. doi: 10.1002/jbmr.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balani D.H., Ono N., Kronenberg H.M. Parathyroid hormone regulates fates of murine osteoblast precursors in vivo. J. Clin. Invest. 2017;127:3327–3338. doi: 10.1172/JCI91699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Horst G., Farih-Sips H., Löwik C.W., Karperien M. Multiple mechanisms are involved in inhibition of osteoblast differentiation by PTHrP and PTH in KS483 Cells. J. Bone Miner. Res. 2005;20:2233–2244. doi: 10.1359/JBMR.050821. [DOI] [PubMed] [Google Scholar]
- 10.Zha X., Sun B., Zhang R., Li C., Yan Z., Chen J. Regulatory effect of microRNA-34a on osteogenesis and angiogenesis in glucocorticoid-induced osteonecrosis of the femoral head. J. Orthop. Res. 2018;36:417–424. doi: 10.1002/jor.23613. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y., Zheng Y., Jia L., Li W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-β1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells. 2015;33:3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z., Wang Y., Sun Z., Wang H., Zhou H., Zhang L., Zhang S., Cao X. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci. Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arfat Y., Basra M.A.R., Shahzad M., Majeed K., Mahmood N., Munir H. miR-208a-3p Suppresses Osteoblast Differentiation and Inhibits Bone Formation by Targeting ACVR1. Mol. Ther. Nucleic Acids. 2018;11:323–336. doi: 10.1016/j.omtn.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K.M., Park S.J., Jung S.H., Kim E.J., Jogeswar G., Ajita J., Rhee Y., Kim C.H., Lim S.K. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J. Bone Miner. Res. 2012;27:1669–1679. doi: 10.1002/jbmr.1604. [DOI] [PubMed] [Google Scholar]
- 15.Fushimi S., Nohno T., Nagatsuka H., Katsuyama H. Involvement of miR-140-3p in Wnt3a and TGFβ3 signaling pathways during osteoblast differentiation in MC3T3-E1 cells. Genes Cells. 2018;23:517–527. doi: 10.1111/gtc.12591. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X.H., Geng G.L., Su B., Liang C.P., Wang F., Bao J.C. MicroRNA-338-3p inhibits glucocorticoid-induced osteoclast formation through RANKL targeting. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15037674. gmr.15037674. [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Sun Q., Wan C., Li L., Zhang L., Chen Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J. Cell. Physiol. 2014;229:1494–1502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- 18.Tian L., Zheng F., Li Z., Wang H., Yuan H., Zhang X., Ma Z., Li X., Gao X., Wang B. miR-148a-3p regulates adipocyte and osteoblast differentiation by targeting lysine-specific demethylase 6b. Gene. 2017;627:32–39. doi: 10.1016/j.gene.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Su Y., Deng M.F., Xiong W., Xie A.J., Guo J., Liang Z.H., Hu B., Chen J.-G., Zhu X., Man H.-Y. MicroRNA-26a/Death-Associated Protein Kinase 1 Signaling Induces Synucleinopathy and Dopaminergic Neuron Degeneration in Parkinson’s Disease. Biol. Psychiatry. 2018;85:769–787. doi: 10.1016/j.biopsych.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redell J.B., Moore A.N., Ward N.H., 3rd, Hergenroeder G.W., Dash P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma. 2010;27:2147–2156. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellano L., Dabrowska A., Pellegrino L., Ottaviani S., Cathcart P., Frampton A.E., Krell J., Stebbing J. Sustained expression of miR-26a promotes chromosomal instability and tumorigenesis through regulation of CHFR. Nucleic Acids Res. 2017;45:4401–4412. doi: 10.1093/nar/gkx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Fan L., Hu J., Zhang L., Liao L., Liu S., Wu D., Yang P., Shen L., Chen J., Jin Y. MiR-26a Rescues Bone Regeneration Deficiency of Mesenchymal Stem Cells Derived From Osteoporotic Mice. Mol. Ther. 2015;23:1349–1357. doi: 10.1038/mt.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moses C., Nugent F., Waryah C.B., Garcia-Bloj B., Harvey A.R., Blancafort P. Activating PTEN Tumor Suppressor Expression with the CRISPR/dCas9 System. Mol. Ther. Nucleic Acids. 2019;14:287–300. doi: 10.1016/j.omtn.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashash D., Sayyadi M., Safaroghli-Azar A., Sheikh-Zeineddini N., Riyahi N., Momeny M. Small molecule inhibitor of c-Myc 10058-F4 inhibits proliferation and induces apoptosis in acute leukemia cells, irrespective of PTEN status. Int. J. Biochem. Cell Biol. 2019;108:7–16. doi: 10.1016/j.biocel.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Hamada K., Maeda Y., Mizutani A., Okada S. The Phosphatidylinositol 3-Kinase p110α/PTEN Signaling Pathway Is Crucial for HIV-1 Entry. Biol. Pharm. Bull. 2019;42:130–138. doi: 10.1248/bpb.b18-00801. [DOI] [PubMed] [Google Scholar]
- 26.Millis S.Z., Jardim D.L., Albacker L., Ross J.S., Miller V.A., Ali S.M., Kurzrock R. Phosphatidylinositol 3-kinase pathway genomic alterations in 60,991 diverse solid tumors informs targeted therapy opportunities. Cancer. 2019;125:1185–1199. doi: 10.1002/cncr.31921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neirijnck Y., Kühne F., Mayère C., Pavlova E., Sararols P., Foti M., Atanassova N., Nef S. Tumor Suppressor PTEN Regulates Negatively Sertoli Cell Proliferation, Testis Size, and Sperm Production In Vivo. Endocrinology. 2019;160:387–398. doi: 10.1210/en.2018-00892. [DOI] [PubMed] [Google Scholar]
- 28.Malentacchi F., Turrini I., Sorbi F., Projetto E., Castiglione F., Fambrini M., Petraglia F., Pillozzi S., Noci I. Pilot investigation of the mutation profile of PIK3CA/PTEN genes (PI3K pathway) in grade 3 endometrial cancer. Oncol. Rep. 2019;41:1560–1574. doi: 10.3892/or.2018.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma X., Zhou J., Liu J., Wu G., Yu Y., Zhu H., Liu J. LncRNA ANCR promotes proliferation and radiation resistance of nasopharyngeal carcinoma by inhibiting PTEN expression. OncoTargets Ther. 2018;11:8399–8408. doi: 10.2147/OTT.S182573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Bruxvoort K.J., Zylstra C.R., Liu J., Cichowski R., Faugere M.C., Bouxsein M.L., Wan C., Williams B.O., Clemens T.L. Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc. Natl. Acad. Sci. USA. 2007;104:2259–2264. doi: 10.1073/pnas.0604153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye L., Lou F., Yu F., Zhang D., Wang C., Wu F., Li X., Ping Y., Yang X., Yang J. NUMB maintains bone mass by promoting degradation of PTEN and GLI1 via ubiquitination in osteoblasts. Bone Res. 2018;6:32. doi: 10.1038/s41413-018-0030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paquet J., Moya A., Bensidhoum M., Petite H. Engineered cell-free scaffold with two-stage delivery of miRNA-26a for bone repair. Ann. Transl. Med. 2016;4:204. doi: 10.21037/atm.2016.05.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elmén J., Lindow M., Schütz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjärn M., Hansen H.F., Berger U. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 34.Walker C., Wu X., Liu N.K., Xu X.M. Bisperoxovanadium mediates neuronal protection through inhibition of PTEN and activation of PI3K/AKT-mTOR signaling following traumatic spinal injuries. J. Neurotrauma. 2019 doi: 10.1089/neu.2018.6294. Published online March 28, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu L., Xing Q., Huang T., Zhou J., Liu T., Cui Y., Cheng T., Wang Y., Zhou X., Yang B. HDAC1 Silence Promotes Neuroprotective Effects of Human Umbilical Cord-Derived Mesenchymal Stem Cells in a Mouse Model of Traumatic Brain Injury via PI3K/AKT Pathway. Front. Cell. Neurosci. 2019;12:498. doi: 10.3389/fncel.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R., Liang Y., Wei S. The expressions of NGF and VEGF in the fracture tissues are closely associated with accelerated clavicle fracture healing in patients with traumatic brain injury. Ther. Clin. Risk Manag. 2018;14:2315–2322. doi: 10.2147/TCRM.S182325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeliger C., Balmayor E.R., van Griensven M. miRNAs Related to Skeletal Diseases. Stem Cells Dev. 2016;25:1261–1281. doi: 10.1089/scd.2016.0133. [DOI] [PubMed] [Google Scholar]
- 38.Eskildsen T., Taipaleenmäki H., Stenvang J., Abdallah B.M., Ditzel N., Nossent A.Y., Bak M., Kauppinen S., Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L., Holmstrøm K., Qiu W., Ditzel N., Shi K., Hokland L., Kassem M. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 2014;32:902–912. doi: 10.1002/stem.1615. [DOI] [PubMed] [Google Scholar]
- 40.Baek D., Lee K.M., Park K.W., Suh J.W., Choi S.M., Park K.H., Lee J.W., Kim S.H. Inhibition of miR-449a Promotes Cartilage Regeneration and Prevents Progression of Osteoarthritis in In Vivo Rat Models. Mol. Ther. Nucleic Acids. 2018;13:322–333. doi: 10.1016/j.omtn.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo F., Xie Y., Wang Z., Huang J., Tan Q., Sun X., Li F., Li C., Liu M., Zhang D. Adeno-Associated Virus-Mediated RNAi against Mutant Alleles Attenuates Abnormal Calvarial Phenotypes in an Apert Syndrome Mouse Model. Mol. Ther. Nucleic Acids. 2018;13:291–302. doi: 10.1016/j.omtn.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 43.Ma J., Shi M., Li J., Chen B., Wang H., Li B., Hu J., Cao Y., Fang B., Zhao R.C. Senescence-unrelated impediment of osteogenesis from Flk1+ bone marrow mesenchymal stem cells induced by total body irradiation and its contribution to long-term bone and hematopoietic injury. Haematologica. 2007;92:889–896. doi: 10.3324/haematol.11106. [DOI] [PubMed] [Google Scholar]
- 44.Lu L.L., Liu Y.J., Yang S.G., Zhao Q.J., Wang X., Gong W., Han Z.B., Xu Z.S., Lu Y.X., Liu D. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Haematologica. 2006;91:1017–1026. [PubMed] [Google Scholar]
- 45.D’Abundo L., Callegari E., Bresin A., Chillemi A., Elamin B.K., Guerriero P., Huang X., Saccenti E., Hussein E.M.A.A., Casciano F. Anti-leukemic activity of microRNA-26a in a chronic lymphocytic leukemia mouse model. Oncogene. 2017;36:6617–6626. doi: 10.1038/onc.2017.269. [DOI] [PubMed] [Google Scholar]
- 46.Li Y., Fan L., Liu S., Liu W., Zhang H., Zhou T., Wu D., Yang P., Shen L., Chen J., Jin Y. The promotion of bone regeneration through positive regulation of angiogenic-osteogenic coupling using microRNA-26a. Biomaterials. 2013;34:5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X., Li Y., Chen Y.E., Chen J., Ma P.X. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat. Commun. 2016;7:10376. doi: 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W.X., Sullivan P.G., Springer J.E. Mitochondria and microRNA crosstalk in traumatic brain injury. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;73:104–108. doi: 10.1016/j.pnpbp.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 49.Kumar A., Stoica B.A., Loane D.J., Yang M., Abulwerdi G., Khan N., Kumar A., Thom S.R., Faden A.I. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflammation. 2017;14:47. doi: 10.1186/s12974-017-0819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Zhang X., Chen X., Wang L., Yang G. Exosome Mediated Delivery of miR-124 Promotes Neurogenesis after Ischemia. Mol. Ther. Nucleic Acids. 2017;7:278–287. doi: 10.1016/j.omtn.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klaw M.C., Xu C., Tom V.J. Intraspinal AAV Injections Immediately Rostral to a Thoracic Spinal Cord Injury Site Efficiently Transduces Neurons in Spinal Cord and Brain. Mol. Ther. Nucleic Acids. 2013;2:e108. doi: 10.1038/mtna.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chai C., Song L.J., Han S.Y., Li X.Q., Li M. MicroRNA-21 promotes glioma cell proliferation and inhibits senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT signaling pathway. CNS Neurosci. Ther. 2018;24:369–380. doi: 10.1111/cns.12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Collins C.J., Vivanco J.F., Sokn S.A., Williams B.O., Burgers T.A., Ploeg H.L. Fracture healing in mice lacking Pten in osteoblasts: a micro-computed tomography image-based analysis of the mechanical properties of the femur. J. Biomech. 2015;48:310–317. doi: 10.1016/j.jbiomech.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan Y.S., Li Q., Hamdan N., Bian Y.F., Zhuang S., Fan K., Liu Z.J. Tetrahydroxystilbene Glucoside Regulates Proliferation, Differentiation, and OPG/RANKL/M-CSF Expression in MC3T3-E1 Cells via the PI3K/Akt Pathway. Molecules. 2018;23:E2306. doi: 10.3390/molecules23092306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine A.J., Feng Z., Mak T.W., You H., Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 56.Flierl M.A., Stahel P.F., Beauchamp K.M., Morgan S.J., Smith W.R., Shohami E. Mouse closed head injury model induced by a weight-drop device. Nat. Protoc. 2009;4:1328–1337. doi: 10.1038/nprot.2009.148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.