Summary
The Nrd1-Nab3-Sen1 (NNS) complex carries out the transcription termination of noncoding RNAs (ncRNAs) by RNA polymerase II (Pol II) in yeast, although the detailed interactions among its subunits remain obscure. Here we have identified three sequence motifs in Sen1 that mediate direct interactions with the Pol II CTD interaction domain (CID) of Nrd1, determined the crystal structures of these Nrd1 interaction motifs (NIMs) bound to the CID, and characterized the interactions in vitro and in yeast. Removal of all three NIMs abolishes NNS complex formation and gives rise to ncRNA termination defects.
Graphical Abstract
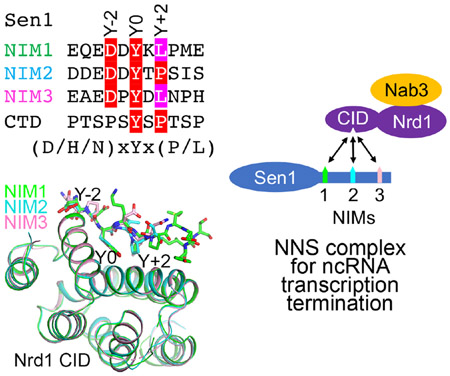
eTOC blurb
Zhang et al. report the identification of three Nrd1-interacting motifs (NIMs) of Sen1 and the crystal structures of their complexes with the Pol II C-terminal domain interaction domain (CID) of Nrd1. These interactions are important for transcription termination of non-coding RNAs in yeast.
Introduction
Nrd1 and Nab3 can form a complex through their dimerization domains and bind to RNA cooperatively through their RNA recognition modules (RRMs) (Bacikova et al., 2014; Conrad et al., 2000; Franco-Eschevarria et al., 2017; Lunde et al., 2011) (Fig. 1a). The helicase Sen1 is speculated to remove nascent RNA from Pol II and trigger termination by collapsing the transcription bubble (Arndt and Reines, 2015; Mischo and Proudfoot, 2013). Nrd1 also contains a CID that has a preference for Ser5P C-terminal domain (CTD) of Pol II (Kubicek et al., 2012; Vasiljeva et al., 2008), while Sen1 has a preference for Pol II Ser2P CTD (Chinchilla et al., 2012). NNS couples tightly with the TRAMP complex for 3’-end trimming of ncRNAs and degradation of unstable RNAs (LaCava et al., 2005; Vanacova et al., 2005; Vasiljeva and Buratowski, 2006; Wyers et al., 2005), which is mediated by the interaction between Nrd1 CID and Trf4, a subunit of the TRAMP complex (Kim et al., 2016; Tudek et al., 2014). The Nrd1 CID also interacts with Mpp6, a cofactor of the nuclear exosome (Kim et al., 2016). Besides ncRNA transcription termination (Kim et al., 2006), Sen1 also functions in R-loop resolution (Chan et al., 2014; Mischo et al., 2011) and pre-mRNA transcription termination (Beggs et al., 2012; Creamer et al., 2011). However, how Sen1 interacts with Nrd1 and Nab3 to form the NNS complex is currently not well understood, and this has been a long-standing question in the field.
Figure 1.
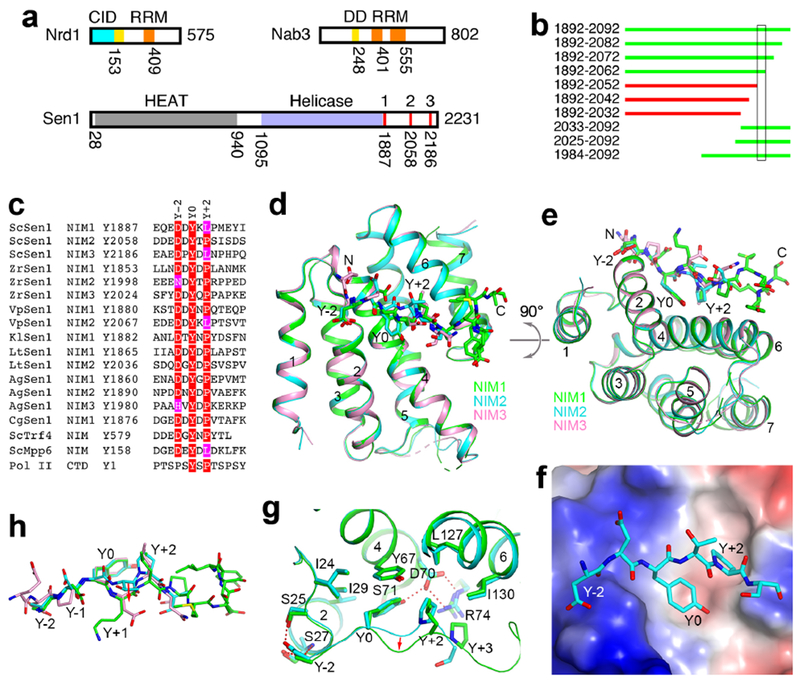
Identification of three NIMs in yeast Sen1. (a). Domain organizations of yeast Nrd1, Nab3 and Sen1. The domains are given different colors, CID (cyan), DD (yellow), RRM (orange), HEAT repeats (gray), helicase (light blue). The three NIMs are in red. The N-terminal domain of Sen1 is expected to be all helical, suggesting that it has a HEAT-like structure. There may be a second RRM in Nab3 based on secondary structure considerations. (b). Various segments of Sen1 can (green) or cannot (red) pull down Nrd1 CID. The segment that is crucial for the interaction is boxed. (c). Alignment of 15 NIMs found in seven fungal species. Also shown is the NIM in Trf4 and the Pol II CTD consensus. Residues in the consensus motif are highlighted in red and magenta. (d). Overlay of the structures of CID in complex with NIM1 (green), NIM2 (cyan), and NIM3 (pink). (e). Overlay of the structures, viewed after a rotation of 90° around the horizontal axis. (f). Electrostatic surface of CID in the complex with NIM2 (stick models). (g). Detailed interactions between NIM1 (green) and NIM2 (cyan) and the CID. Hydrogen-bonding interactions are indicated with the dashed lines (red). The conformational changes of the Y+1 and Y+2 residues are indicated with the red arrow. (h). Overlay of the binding modes of the three NIM peptides. The structure figures were produced with PyMOL (www.pymol.org). See also Fig. S1, S2, S3 and S4.
Results and discussion
It was reported that the C-terminal segment (CTS) of S. cerevisiae Sen1 (ScSen1) (residues 1876-2231) interacts strongly with Nab3 (Nedea et al., 2008). The CTS has few predicted secondary structures and no conservation among Sen1 homologs. However, we did not observe any interactions for purified K. lactis Sen1 CTS with Nab3 and Nrd1 constructs that cover the region from the dimerization domain (DD) to the C-terminus (Figs. 1a, S1a-1b). Surprisingly, we did observe a complex between a fragment of ScSen1 CTS (residues 1892-2092) and full-length Nrd1-Nab3 (Fig. S1c). Comparing the constructs used in these experiments, we hypothesized that the CID of Nrd1 mediated the formation of the complex. To confirm this hypothesis, we showed that deletion of the CID abolished complex formation (Fig. S1d). We then purified Nrd1 CID and demonstrated the direct interaction with purified Sen1 CTS by gel filtration (Fig. S1e). We also demonstrated interactions between the two proteins by pull-down experiments after co-expression in E. coli (Fig. S2a).
To map the region of ScSen1 CTS that is required for interaction with Nrd1 CID, we coexpressed various fragments of the CTS residues 1892-2092 with the CID, and discovered that Sen1 residues 1892-2062 could pull down CID but residues 1892-2052 could not (Figs. 1c, S2a, S2b). Residues 2052-2062 of Sen1 have the sequence DDDEDDYTPSI, and we realized that it is remarkably similar to the Nrd1 interaction motif (NIM) in Trf4 (Fig. 1c) (Kim et al., 2016; Tudek et al., 2014), suggesting that ScSen1 also contains an NIM. The Tyr and Pro residues in Trf4 are important for binding the Nrd1 CID (Tudek et al., 2014), and these residues are also present in the NIM of ScSen1.
A multiple sequence alignment of fungal Sen1 sequences aligned residues 2052-2062 of ScSen1 with DEDEDDYKLPT of V. polyspora Sen1 (VpSen1), suggesting that this could be another NIM. Surprisingly, we also found this motif in the CTS of ScSen1, near residue Tyr1887 (Fig. 1c) and just after the helicase domain (Fig. 1a), and confirmed its interaction with Nrd1 CID (Figs. S2c, S2d). Our crystal structure of its complex with the CID indicates that the Leu residue (rather than the Pro) is important for binding the CID (see below). Based on this information, we re-examined the ScSen1 CTS and found another sequence motif, near residue Tyr2186 (Fig. 1c), which we also confirmed by pull-down experiments (Fig. S2e). Therefore, ScSen1 has three NIMs in its CTS (Fig. 1a), and we have named them NIM1, NIM2, and NIM3 (Fig. 1c).
We then examined the CTS sequences of 12 fungal Sen1 and identified 15 NIMs in 7 fungal species, with a consensus motif (D/H/N)xYx(P/L), where x is frequently a hydrophilic residue (Fig. 1c). The Y-2 residue is predominantly Asp, while the Y+2 residue is predominantly Pro. Some species have three NIMs, while others have only one or two. Five of the fungal species examined, such as C. albicans, D. hansenii and S. pombe, do not appear to have an NIM in the CTS.
We have determined the crystal structures of Nrd1 CID in complex with each of the NIMs in ScSenl, at up to 2.0 Å resolution (Table 1). The structures of CID in the three complexes are essentially identical, with rms distance of 0.2-0.3 Å for equivalent Cα atoms between any pair of them (Figs. 1d, 1e). The NIMs bind at the same surface depression (Fig. 1f) as that for Pol II CTD (Kubicek et al., 2012) and Trf4 NIM (Kim et al., 2016; Tudek et al., 2014) (Fig. S3). Clear electron density for the conserved (D/H/N)xYx(P/L) motif was observed in the complexes (Fig. S3). The remaining residues of the NIM peptides were mostly disordered, except the C-terminal residues of NIM1, which are stabilized by crystal packing. Therefore, residues outside of the conserved motif do not have strong interactions with the CID.
Table 1.
Data collection and refinement statistics
| CID+NIM1 | CID+NIM2 | CID+NIM3 | |
|---|---|---|---|
| Data collection | |||
| Space group | C2221 | C2221 | C2221 |
| Cell dimensions | |||
| a, b, c (Å) | 103.4, 109.1, 86.7 | 98.9, 102.5, 115.5 | 98.7, 103.0, 115.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å)1 | 50-2.1 (2.2-2.1) | 50-2.0 (2.1-2.0) | 50-2.8 (3.0-2.8) |
| No. of observations | 185,949 | 175,380 | 48,822 |
| No. of unique reflections | 27,909 | 39,419 | 14,439 |
| Rmerge (%) | 8.4 (67.9) | 8.0 (48.7) | 14.7 (48.8) |
| I/σI | 16.8 (5.4) | 12.2 (3.1) | 7.3 (2.8) |
| CC1/2 | |||
| Completeness (%) | 96.2 (96.7) | 98.1 (96.9) | 97.1 (97.2) |
| Redundancy | 6.7 (6.9) | 4.4 (4.3) | 3.4 (3.4) |
| Refinement | |||
| Resolution range (Å) | 46-2.1 (2.2-2.1) | 45-2.0 (2.1-2.0) | 45-2.8 (2.9-2.8) |
| No. of reflections | 27,890 | 39,414 | 14,426 |
| Rwork (%) | 18.6 (23.0) | 17.9 (25.2) | 23.7 (29.1) |
| Rfree (%) | 20.4 (28.6) | 22.6 (29.2) | 30.0 (31.3) |
| No. atoms | |||
| Protein | 2,428 | 3,561 | 3,560 |
| Cl | 0 | 2 | 2 |
| Water | 126 | 260 | – |
| B-factors | |||
| Protein | 42.7 | 35.4 | 40.9 |
| Cl | – | 23.5 | 31.2 |
| Water | 45.0 | 40.2 | – |
| RMS deviations | |||
| Bond lengths (Å) | 0.006 | 0.007 | 0.008 |
| Bond angles (°) | 0.73 | 0.76 | 1.10 |
| Ramachandran plot statistics (%) | |||
| Most favored region | 98.29 | 98.85 | 97.92 |
| Additional allowed region | 1.37 | 1.15 | 2.08 |
| PDB entry code | 6O3W | 6O3X | 6O3Y |
The numbers in parentheses are for the highest resolution shell.
The NIM assumes a mostly extended conformation and has three major contacts with the CID, via the Y0, Y-2 and Y+2 residues (Fig. 1e). The Y0 residue of the NIM has a central role in the interaction with CID. Its side chain aromatic ring interacts with the side chains of Ile29, Tyr67 and the Y+2 residue, while its hydroxyl group is hydrogen-bonded to the side chain of Asp70, which is also involved in a bidentate interaction with Arg74 (Fig. 1g). The Y+2 residue contacts a hydrophobic patch formed by Leu127, Ile130, Tyr67 and the Y0 residue of the NIM, and the Asp70-Arg74 ion pair forms one side of the binding site for this residue. With Leu at the Y+2 position, a change in the backbone structure of the Y+1 and Y+2 residues is necessary to make room for this bulkier side chain compared to Pro (Figs. 1g, 1h). The binding modes of NIM1 and NIM3, which both have Leu at the Y+2 position, are similar.
The Y-2 residue has hydrogen-bonding interactions with Ser25 and Ser27. It caps the N-terminus of helix a2, and the Asp side chain can also have favorable interactions with the dipole of this helix. Residues N-terminal to Y-2 are mostly negatively charged in ScSen1 as well as many other NIMs (Fig. 1c). An electro-positive patch in this region of Nrd1 CID (Fig. 1f) could suggest favorable interactions, but these residues in the NIMs are not ordered in the structures. In addition, K. lactis Sen1 has no negatively charged residues in this region, but it can interact with CID (Fig. S2d).
The structural observations are supported by our mutagenesis and fluorescence anisotropy binding assays. The Kd of the CID-NIM2 complex was 0.56±0.05 μM (Fig. 2a), similar to that of the CID-Trf4 NIM complex (Kim et al., 2016; Tudek et al., 2014). In comparison, the Kd for the Y2058F mutant of NIM2 was 30-fold higher, confirming the importance of the hydroxyl group of the Y0 residue for CID binding. Similarly, mutation of the Y-2 residue, D2056N, led to a 13-fold increase in the Kd, while mutations of the Y+2 residue, P2060A and P2060V, led to ~20-fold increase in the Kd (Fig. 2b). We also introduced mutations in the NIM binding site of Nrd1 CID and characterized their effects on the interaction. The Y67A mutant showed a 90-fold increase in the Kd for NIM2, and the D70A mutant had a 30-fold increase (Fig. 2c). In comparison, the D70N mutant showed only 3-fold higher Kd, indicating that the charge on this residue is not essential for NIM binding. These studies also indicate a strong hydrogen-bond between Y0 and Asp70, as the Y2058F mutant of the NIM (at the Y0 position) and the D70A mutant of CID both showed 30-fold higher Kd, corresponding to a ΔG of 2 kcal/mol and within range of the energy of a single hydrogen bond.
Figure 2.
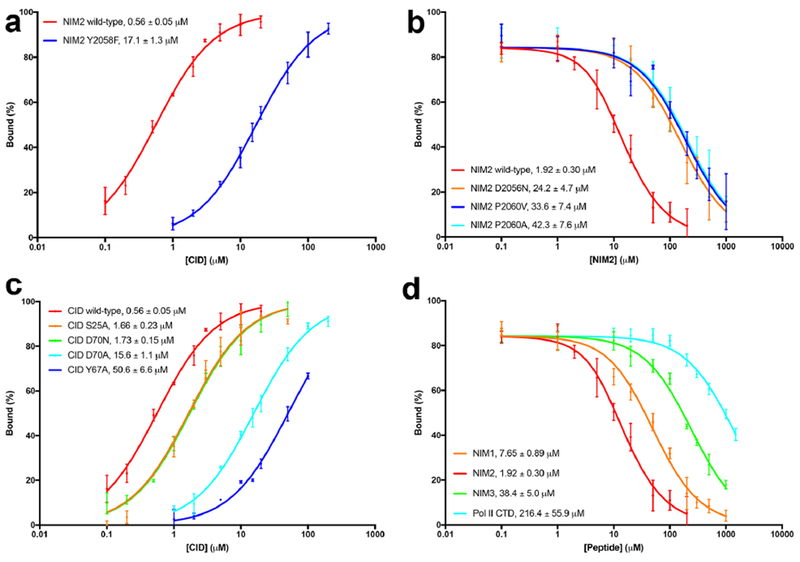
Binding affinity between Nrd1 CID and Sen1 NIMs. (a). Fluorescence anisotropy binding data for wild-type NIM2 and the Y2058F mutant. The observed Kd values are indicated. (b). Fluorescence anisotropy competition binding data for wild-type NIM2 and three mutants. (c). Fluorescence anisotropy binding data for NIM2 with wild-type and mutant CID. (d). Fluorescence anisotropy competition binding data for NIM1, NIM2, NIM3 and the Pol II Ser5P CTD. Error bars represent standard deviations from triplicate measurements.
The Kd of the CID-NIM1 complex (7.6 μM) was 4-fold higher than that of the NIM2 complex, while the Kd of the CID-NIM3 complex (38 μM) was 20-fold higher (Fig. 2d). NIM1 and NIM3 have identical residues at the Y-2, Y0 and Y+2 positions (Fig. 1c), although the Leu residue at Y+2 in NIM3 assumes a different conformation in the complex compared to NIM1 (Fig. 1h), which may be related to its 5-fold difference in affinity for the CID. The Kd of the Mpp6 peptide, which also has a Leu at the Y+2 position (Fig. 1c), is 13.6 μM (Kim et al., 2016). Together with the data on the P2060A and P2060V mutants, our observations suggest that several hydrophobic residues could be tolerated at the Y+2 position. In comparison, the Pol II Ser5P CTD peptide studied here had the weakest affinity for the CID, with a Kd of 216±56 μM, comparable to the value reported previously (Tudek et al., 2014). The binding mode of the CTD peptide has substantial differences to the NIMs (Fig. S3d). The CTD also contains multiple repeats, which could provide increased avidity for the interaction with Nrd1.
We showed that mutation of the Tyr residue in all three NIMs in Sen1 CTS (Y1887A, Y2058A, Y2186A) was able to abolish the interaction with CID, while single-site mutants were still able to interact with CID (Fig. S4). We also confirmed that other regions of Sen1, such as its helicase and HEAT repeat domains, could not interact with Nrd1 CID, and that Nrd1 lacking the CID could not mediate the formation of the NNS complex (Fig. S4). We examined the amino acid sequences of these other regions of ScSen1 and did not find occurrences of the consensus motif. Overall, these data indicate that, under the conditions tested here, Sen1 interacts with Nrd1 solely through the NIM-CID contacts, and that Sen1 does not interact with Nab3.
We next characterized the Sen1-Nrd1 interaction in yeast. Deletion of the CTS had no effect on growth (Fig. 3a), consistent with earlier data (Chen et al., 2014), nor did mutating the NIM tyrosines to alanine, individually or in combination (data not shown). A Nrd1 mutant lacking the CID also grows normally (Vasiljeva et al., 2008), suggesting that none of the Nrd1 CID interactions are essential under laboratory growth conditions. However, the Nrd1 CID deletion does show NNS termination defects in a sensitive reporter assay (Steinmetz and Brow, 1998). When the SNR13 terminator is placed in an intron upstream of the Cup1 coding region, NNS read-through produces a functional Cup1 mRNA that confers resistance to copper in the growth medium. We used this reporter in cells with a chromosomal Sen1 mutation (E1597K) and tested whether our Sen1 constructs complement the defect (reverting cells back to copper sensitivity) or were also defective (copper resistance). Interestingly, the 1-1884 construct lacking the entire CTS fully complements a SEN1 deletion or the E1597K point mutant for overall growth rate, but is not effective at preventing NNS read-through at the reporter (Fig. 3b). In contrast, the smaller deletions that retain one (1-2055) or two (1-2183) NIMs completely restore termination and copper sensitivity. A NIM1 point mutant in the context of the 1-2055 construct apparently terminates effectively on this reporter as well, so we cannot definitively assign the function to the NIM. Nonetheless, we can conclude that Sen1 region 1884-2055 containing NIM1 does contribute to Sen1 function.
Figure 3.
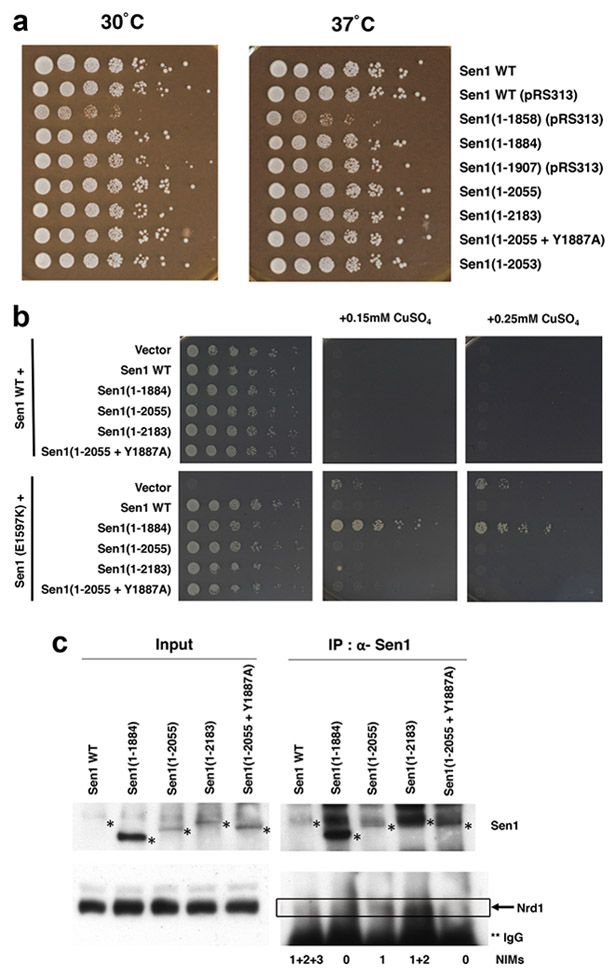
Characterization of the effects of the Sen1-Nrd1 interaction in yeast. (a). Sen1 CTS is not required for growth. The indicated Sen1 mutants were created by plasmid shuffling and tested for growth by serial dilution spotting assay on rich YPD media. The Sen1(1-1858) mutant is missing the C-terminal region of the helicase domain, which may explain its growth phenotype. (b). Various C-terminal truncations of Sen1 were transformed into SEN1 wild-type or E1597K point mutant cells and tested for the ability to prevent NNS read-through at the Cup1 reporter. NNS termination defects result in copper resistance. The 1-1884 construct of Sen1, which lacks all three NIMs, could not prevent read-through. (c). Sen1 NIMs are crucial for interaction with Nrd1 in yeast. Cell extracts from the indicated Sen1 strains were immunoprecipitated with anti-Sen1 antibody and probed for the presence of Nrd1 and Sen1 proteins. Positions of Sen1 proteins are marked with a single asterisk, and positions of Nrd1 are marked with the box. Nrd1 is partially obscured by its proximity to IgG heavy chain on the blot, marked with two asterisks.
We were also able to show that removing or mutating NIMs from the Sen1 CTS had a profound effect on the interaction with Nrd1, based on co-immunoprecipitation experiments (Fig. 3c). A Sen1 mutant lacking all three NIMs showed essentially no binding to Nrd1. A mutant with one NIM showed weaker binding compared to one with two NIMs, and mutation of the Tyr residue to Ala in this NIM essentially blocked binding to Nrd1. These results confirm the observations from the crystal structures and demonstrate the importance of all three NIMs for Sen1-Nrd1 interactions. Removal of all three NIMs is required to abolish binding between Sen1 and Nrd1.
As we and others have previously noted (Mischo et al., 2018), normal levels of full-length Sen1 are extremely low and difficult to detect by Western blotting. Correspondingly, little Nrd1 was detected upon immunoprecipitation (Fig. 3 c). Certain N-terminal truncations stabilize Sen1 by removing degradation-promoting sequences (DeMarini et al., 1995; Mischo et al., 2018). We found that the C-terminal truncations of Sen1 also produced higher levels of Sen1 protein, suggesting additional degradation signals in the CTS. On the other hand, the low levels of Sen1 compared to Nrd1 suggest that each Sen1 could be engaged by more than one Nrd1 in cells, which is supported by our co-immunoprecipitation data (Fig. 3c). It remains to be seen whether these Nrd1 molecules can coordinate their functions for the NNS complex, but such cooperativity would be consistent with the observation that efficient NNS terminators typically require multiple Nrd1 and Nab3 bindings sites (Carroll et al., 2007).
In conclusion, we have identified three NIMs in the CTS of budding yeast Sen1 and elucidated the molecular basis for their recognition by Nrd1 CID. The identification of one of these NIMs (NIM2) in ScSen1 was independently reported recently (Han et al., 2018). Although neither the Sen1 NIMs nor Nrd1 CID is essential for supporting viability, deletions do show termination defects in a reporter assay. The conservation of these regions suggests these interactions are very likely to promote NNS function. A recent report demonstrates that Tyr1 in the CTD is required for termination by the NNS pathway (Collin et al., 2019). The Nrd1 CID now has four known binding partners, Sen1, Trf4 (Tudek et al., 2014), Mpp6 (Kim et al., 2016) and Pol II CTD (Kubicek et al., 2012; Vasiljeva et al., 2008), and other motifs that bind this CID could also be possible. In fact, the crystal structure of free Nrd1 CID (Vasiljeva et al., 2008) contains a packing contact that places a YLNA motif from another molecule in the crystal, from helix α7 that has partially unwound, in the binding site, with the Ala residue occupying the Y+2 position (Fig. S3e). Further studies will shed light on whether additional motifs could actually support interactions between Nrd1 CID and other proteins, and how the Sen1-Nrd1 interactions mediate the functions of the NNS complex.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Liang Tong (ltong@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial E. coli BL21 Star (DE3) cells (Novagen) were used for protein expression in LB media. The cells were induced with 0.4 mM IPTG and grown at 16 °C for 16-20 h. Yeast strains were grown in YPD media and plated on YPD agar.
METHOD DETAILS
Protein expression and purification
S. cerevisiae Nrd1 CID (residues 6-151) was sub-cloned into the pET28a vector (Novagen). The recombinant protein, with an N-terminal hexa-histidine tag, was over-expressed in E. coli BL21 Star (DE3) cells (Novagen), which were induced with 0.4 mM IPTG and allowed to grow at 16 °C for 16–20 h. The soluble protein was purified by nickel-charged immobilizedmetal affinity chromatography, ion-exchange chromatography and gel filtration chromatography. The purified protein was concentrated in a buffer containing 20 mM Tris (pH 7.0), 150 mM NaCl, 10 mM DTT, and stored at −80 °C. The His-tag was not removed prior to crystallization.
Protein crystallization
Crystals of Nrd1 CID-Sen1 NIM complex were grown at 20 °C with the hanging-drop vapor diffusion method. Nrd1 CID protein solution was at 20 mg/ml concentration, and the protein was mixed with Sen1 NIM peptides (GL Biochem) at a molar ratio of 1:10. The reservoir solution contained 0.1 M sodium citrate (pH 4.0), 1 M lithium chloride and 20% (w/v) PEG 6000. Fully-grown crystals were obtained one day after set-up. The crystals were cryo-protected in the crystallization solution supplemented with 15% (v/v) ethylene glycol and flash-frozen in liquid nitrogen for data collection at 100 K.
Data collection and processing
X-ray diffraction data sets were collected at a wavelength of 0.979 Å on a Pilatus-6MF pixel array detector at the 24-ID-C beamline of the Advanced Photon Source (APS). The diffraction images were processed and scaled with XDS (Kabsch, 2010).
Structure determination and refinement.
The structures were solved by molecular replacement with the program Phaser-MR in PHENIX (Adams et al., 2002; McCoy et al., 2007). The crystal structure of S. cerevisiae Nrd1 CID (PDB code 3CLJ) (Vasiljeva et al., 2008) was used as the search model. Manual model rebuilding was carried out with Coot (Emsley and Cowtan, 2004). The structure refinement was performed with the program PHENIX, with translation, libration, and screw-rotation (TLS) parameters. The data processing and refinement statistics are summarized in Table 1.
Fluorescence anisotropy assay.
Nrd1 CID mutants were expressed and purified using the same protocol as that for the wild-type CID. NIM1, NIM2, NIM3, NIM2 mutants, Pol II Ser5p CTD and N-terminally 5-carboxyfluorescein (5-FAM) labeled NIM2 peptides were obtained for fluorescence anisotropy (GL Biochem). For direct binding assays, 5 nM 5-FAM labeled NIM2 peptide was titrated with wild-type and mutants of Nrd1 CID at increasing concentrations. For competition assays, complex of 5 nM 5-FAM labeled NIM2 peptide and 3 μM wild-type Nrd1-CID was titrated with unlabeled NIM1, NIM2, NIM3, NIM2 mutants and Pol II Ser5p CTD peptides at increasing concentrations. The measurements were conducted on a Synergy Neo2 Hybrid Multi-Mode Reader (Biotek). Samples were excited at 485 nm and emissions were recorded at 528 nm. Measurements were performed in triplicates at room temperature in 20 mM Tris (pH 7.0), 200 mM NaCl, 5 mM DTT. The binding curves were fitted by GraphPad Prism (GraphPad Software, La Jolla) using hyperbolic equation for direct binding and the analytical model for competitive binding (Wang, 1995).
Mixing assay.
Mixtures of purified proteins were incubated on ice for 1 h and separated on Superose 6 Increase 10/300 GL column (GE Healthcare) in 20 mM Tris (pH 7.0), 200 mM NaCl, 5 mM DTT.
Pull-down assay
Various regions of S. cerevisiae Sen1 CTS and its mutants and K. lactis Sen1 CTS were sub-cloned into the pET28a-SUMO vector unless specified otherwise. The recombinant Sen1 proteins would contain an N-terminal hexa-histidine tag followed by a SUMO tag unless specified otherwise. The CTS construct covering residues 1892-2042 contained two SUMO tags due to a cloning artifact. The plasmid was co-transformed with one for un-tagged S. cerevisiae Nrd1 CID (residues 6-151) or K. lactis Nrd1 CID (residues 1-155) and over-expressed in E. coli BL21 Star (DE3) cells (Novagen). The culture was induced with 0.4 mM IPTG and allowed to grow at 16 °C for 16–20 h. The soluble protein was incubated with nickel-charged resin for 1 h at 4 °C, washed with a buffer containing 20 mM Tris (pH 7.0), 200 mM NaCl, 20 mM imidazole and eluted with a buffer containing 20 mM Tris (pH 7.0), 200 mM NaCl, 150 mM imidazole. The eluate was mixed with protein dye and run on an SDS-PAGE gel.
Yeast genetics
For complementation testing (Fig. 3a), yeast strains were derived by plasmid shuffling as previously described (Mischo et al., 2018). Briefly, the plasmids listed below were transformed into YSB3181 (ura3Δ0, leu2Δ0, trp1Δ::LEU2/KanR, his3Δ1, met15Δ0, sen1Δ::KanMX, [pRS416 + −700 Sen1]), selecting for the plasmid marker using synthetic complete media lacking the appropriate amino acid (HIS3/histidine for pRS313 or TRP1/tryptophan for pRS414). The pRS416+/−700 Sen1 plasmid carrying a URA3 marker was then removed by selection on 5-fluoro-orotic acid media. For the growth assay shown in Fig. 3a, three-fold serial dilutions of post-selection cells were spotted on YPD plates. Cells were grown at 30 °C for three days. The following strains carry the listed plasmids expressing the indicated Sen1 mutants:
YSB3514: Sen1 pRS414 + −700 Sen1 (WT) (Mischo et al., 2018)
YSB3515: pRS313-Sen1 (WT) (Chen et al., 2014)
YSB3516: pRS313-Sen1(1-1858) (Chen et al., 2014)
YSB3517: pRS414-Sen1(1-1884)
YSB3518: pRS313-Sen1(1-1907) (Chen et al., 2014)
YSB3519: pRS414-Sen1(1-2055)
YSB3520: pRS414-Sen1(1-2183)
YSB3521: pRS414-Sen1-Y1887A YSB3522: pRS414-Sen1(1-2053)
YSB3523: pRS414-Sen1(1-2055 + Y1887A)
Plasmids with no citation were created for this study by inverted PCR using pRS414 + − 700 Sen1 as template. Deletion junctions and point mutations were verified by sequencing.
To test for read-through at a Sen1-dependent terminator, isogenic yeast strains 46a/YF1313 (MATa, ura3-, leu2-, trp1-, his3-, lys2, ade2, cup1Δ, SEN1) and nrd2-1/YF1517 (MATa, ura3-, leu2-, trp1-, his3-, lys2, ade2, cup1Δ, sen1(E1597K)), both from (Steinmetz and Brow, 1996), were transformed with reporter plasmid pGAC24-SNR13(125-232) (Steinmetz et al., 2001). This reporter expresses an ACT1-CUP1 fusion gene that contains the SNR13 terminator within an intron. Cells with defects in NNS termination produce read-through transcripts that can express functional Cup1 protein, conferring resistance to copper. The sen1(E1597K) mutant (also known as nrd2-1) grows slowly but is copper resistant. The two yeast strains were transformed with the plasmids described above and tested for complementation of sen1(E1597K) mutant phenotypes in nrd2-1/YF1517, or dominant negative effects in 46a/YF1313. Transformed cells were spotted on plates using a three-fold dilution series and grown for two (no copper) or five (plus copper) days.
Immunoprecipitation experiments
Overnight cultures of yeast were inoculated into 50 ml YPD at OD595 of 0.3 and grown for about five hours to OD 1.0 at 30 °C. Cells were pelleted, washed, and re-suspended in nucleosome isolation buffer (0.1% Triton X-100, 10 mM MgCl2, 20 mM HEPES (pH 7.8), 250 mM sucrose) plus 150 mM NaCl, and protease (leupeptin, pepstatin A, PMSF, aprotinin) and phosphatase inhibitors (sodium fluoride and sodium vanadate) (van Werven et al., 2008). Cells were lysed by vortexing with glass beads for six 30 second intervals with cooling on ice in between. Debris was removed by microfuging at 13000 rpm for 20 minutes at 4 °C. After decanting the supernatant, 10 μl of 10 mg/ml RNaseA was added to make sure any interactions were not mediated by RNA. For immunoprecipitations, 2 mg of extracts were incubated with 2 μl of anti-Sen1 (Chen et al., 2014) (a gift from Dave Brow, University of Wisconsin) and 20 ml of Protein A-Sepharose beads overnight at 4 °C. Beads were gently pelleted and washed three times for five minutes with the same buffer used during immunoprecipitation.
Bound proteins were eluted into loading buffer, separated by SDS-polyacrylamide gel electrophoresis, and blotted at 45 volts to nitrocellulose overnight at 4 °C. After blocking, membranes were probed with anti-Sen1 or anti-Nrd1 (Steinmetz and Brow, 1998) (also provided by Dave Brow), using anti-rabbit IgG-HRP conjugate and ThermoFisher Pico Plus developer to image bands on X-ray film.
Replication
The fluorescence anisotropy binding assays were performed in triplicates.
Strategy for randomization
N/A
Blinding
N/A
Sample size estimation and statistical method of computation
N/A
Inclusion and exclusion criteria
No data were excluded in the fitting to the fluorescence anisotropy binding curves.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data for fluorescence anisotropy binding assays are shown as means ± SD based on three independent experiments.
DATA AND SOFTWARE AVAILABILITY
The accession number for the coordinates and structure factors of Nrd1 in complex with NIM1 is 6O3W, that with NIM2 is 6O3X, and that with NIM3 is 6O3Y.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Senl | Chen et al., 2014 (Dave Brow) | N/A |
| Anti-Nrd1 | Steinmetz and Brow, 1998 (Dave Brow) | N/A |
| Bacterial and Virus Strains | ||
| E. coli BL21 (DE3) Star | Novagen | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium citrate | Fisher Scientific | S279-3 |
| Lithium chloride | Sigma-Aldrich | 203637-50G |
| PEG 6000 | Sigma-Aldrich | 81260-1KG |
| ScSen1-NIM1: EQEDDYKLPMEYIT | GL Biochem | N/A |
| ScSen1-NIM2-FAM: DDEDDYTPSISD-[FAM] | GL Biochem | N/A |
| ScSen1-NIM2-Y2058F-FAM: DDEDDFTPSISD-[FAM] | GL Biochem | N/A |
| ScSen1-NIM2-P2060A: DDEDDYTASISD | GL Biochem | N/A |
| ScSen1-NIM2: DDEDDYTPSISD | GL Biochem | N/A |
| ScSen1-NIM2-P2060V: DDEDDYTVSISD | GL Biochem | N/A |
| ScSen1-NIM2-D2056N: DDENDYTPSISD | GL Biochem | N/A |
| ScSen1-NIM3: EAEDPYDLNPHPQ | GL Biochem | N/A |
| Pol II CTD: SPT(pS)PSYSPT(pS)PS | GL Biochem | N/A |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Nrd1 CID | Vasiljeva et al., 2008 | PDB: 3CLJ |
| Nrd1 CID in complex with Sen1 NIM1 | This paper | 6O3W |
| Nrd1 CID in complex with Sen1 NIM2 | This paper | 6O3X |
| Nrd1 CID in complex with Sen1 NIM3 | This paper | 6O3Y |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Yeast strains | ||
| YSB3181 (SEN1 shuffling strain) | Mischo et al, 2018 | N/A |
| nrd2-1/YF1517 | Steinmetz and Brow, 1996 (Dave Brow) | N/A |
| 46a/YF1313 | Steinmetz and Brow, 1996 (Dave Brow) | N/A |
| Oligonucleotides | ||
| Forward primer used to clone Nrd1-CID construct in pET28a C AGC GGC CTG GTG CCG CGC GGC AGC CAT ATG GAT TTT CAA AAT TTT GTA GCT ACC TTG G |
Eton Bioscience | N/A |
| Reverse primer used to clone Nrd1-CID construct in pET28a GTG GTG GTG GTG GTG CTC GAG TGC GGC CGC CTA AGC AAA GCA TTT TGA CCT GAT GGC |
Eton Bioscience | N/A |
| Forward primer used to clone Sen1 constructs in pET28a-SUMO GGC CTG GTG CCG CGC GGC AGC CAT ATG xxx xxx indicates coding-sequence-complementary region of variable length |
Eton Bioscience | N/A |
| Reverse primer used to clone Sen1 constructs in pET28a-SUMO GTG GTG GTG CTC GAG TGC GGC CGC CTA xxx xxx indicates coding-sequence-complementary region of variable length |
Eton Bioscience | N/A |
| Forward primer used to clone Nrd1-CID construct in pCDF GT TTA ACT TTA ATA AGG AGA TAT ACC ATG GGC GAT TTT CAA AAT TTT GTA GCT ACC TTG G |
Eton Bioscience | N/A |
| Reverse primer used to clone Nrd1-CID construct in pCDF C TGT TCG ACT TAA GCA TTA TGC GGC CGC CTA AGC AAA GCA TTT TGA CCT GAT GGC |
Eton Bioscience | N/A |
| Recombinant DNA | ||
| pET28a-Nrd1-CID | This study | N/A |
| pET28a-SUMO-Sen1 | This study | N/A |
| pCDF-Nrd1-CID | This study | N/A |
| Software and Algorithms | ||
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| PHENIX | McCoy, 2007; Adams, 2002 | https://www.phenix-online.org/ |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | Schrodinger LLC | https://pymol.org |
| GraphPad Prism | GraphPad Software | www.graphpad.com |
| Other | ||
Highlights.
Three Nrd1 interaction motifs (NIMs) are identified in C-terminal segment of Sen1.
The NIMs are bound in the same pocket as the Pol II CTD phosphopeptide.
Removal of all three NIMs abolishes Nrd1-Nab3-Sen1 complex formation.
Removal of all three NIMs produces ncRNA termination defects.
Acknowledgments.
This research is supported by NIH grants R35GM118093 and S10OD012018 (to LT) and R01GM056663 (to SB). We thank Dave Brow (University of Wisconsin, Madison) for Sen1 and Nrd1 materials; S. Banerjee, K. Perry, R. Rajashankar, J. Schuermann, N. Sukumar for access to NE-CAT 24-ID-C beamline at the Advanced Photon Source. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, funded by the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by an NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests. The authors declare no competing interests.
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst D58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Arndt KM, and Reines D (2015). Termination of transcription of short noncoding RNAs by RNA polymerase II. Ann Rev Biochem 84, 381–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacikova V, Pasulka J, Kubicek K, and Stefl R (2014). Structure and semi-sequence- specific RNA binding of Nrd1. Nucl Acid Res 42, 8024–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs S, James TC, and Bond U (2012). The PolyA tail length of yeast histone mRNAs varies during the cell cycle and is influenced by Sen1p and Rrp6p. Nucl Acid Res 40, 2700–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, and Corden JL (2007). Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, and Hieter P (2014). Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet 10, e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Muller U, Sundling KE, and Brow DA (2014). Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human senataxin that elicit cerebellar ataxia. Genetics 198, 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla K, Rodriguez-Molina JB, Ursic D, Finkel JS, Ansari AZ, and Culbertson MR (2012). Interactions of Sen1, Nrd1, and Nab3 with multiple phosphorylated forms of the Rpb1 C-terminal domain in Saccharomyces cerevisiae. Eukaryotic Cell 11, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin P, Jeronimo C, Poitras C, and Robert F (2019). RNA polymerase II CTD tyrosine 1 is required for efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Epub. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, and Corden JL (2000). A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154, 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, and Corden JL (2011). Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet 7, e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Papa FR, Swaminathan S, Ursic D, Rasmussen TP, Culbertson MR, and Hochstrasser M (1995). The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol 15, 6311–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan KD (2004). Coot: model-building tools for molecular graphics. Acta Cryst D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Franco-Eschevarria E, Gonzalez-Polo N, Zorrilla S, Martinez-Lumbreras S, Santiveri CM, Campos-Olivas R, Sanchez M, Calvo O, Gonzalez B, and Perez-Canadillas J-M (2017). The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition. Nucl Acid Res 45, 10293–10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Jasnovidova O, Tudek A, Kubicek K, Libri D, Stefl R, and Porrua O (2018). Termination of non-coding transcription in yeast relies on both a CTD-interaction domain and a CTD-mimic in Sen1. bioRxiv, doi 10.1101/433045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Integration, scaling, space-group assignment and post-refinement. Acta Cryst D66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Heo DH, Kim I, Suh JY, and Kim M (2016). Exosome cofactors connect transcription termination to RNA processing by guiding terminated transcripts to the appropriate exonuclease within the nuclear exosome. J Biol Chem 291, 13229–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky AM, Moore C, and Buratowski S (2006). Distinct pathways for snoRNA and mRNA termination. Mol Cell 24, 723–734. [DOI] [PubMed] [Google Scholar]
- Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, and Stefl R (2012). Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Develop 26, 1891–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, and Tollervey D (2005). RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Horner M, and Meinhart A (2011). Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM. Nucl Acid Res 39, 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Chun Y, Harlen KM, Smalec BM, Dhir S, Churchman LS, and Buratowski S (2018). Cell-cycle modulation of transcription termination factor Sen1. Mol Cell 70, 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, and Proudfoot NJ (2011). Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, and Proudfoot NJ (2013). Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta 1829, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, and Nagy PL (2008). The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell 29, 577–587. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, and Brow DA (1996). Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol 16, 6993–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, and Brow DA (1998). Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci USA 95, 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, and Corden JL (2001). RNA-binding protein Nrd1 directs poly(A)-independent 3’-end formation of RNA polymerase II transcripts. Nature 413, 327–331. [DOI] [PubMed] [Google Scholar]
- Tudek A, Porrua O, Kabzinski T, Lidschreiber M, Kubicek K, Fortova A, Lacroute F, Vanacova S, Cramer P, Stefl R, et al. (2014). Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell 55, 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, and Timmers HT (2008). Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Develop 22, 2359–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, and Keller W (2005). A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3, e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, and Buratowski S (2006). Nrd1 interacts with the nuclear exosome for 3’processing of RNA polymerase II transcripts. Mol Cell 21, 239–248. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, and Meinhart A (2008). The Nrd1-Nab3- Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15, 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX (1995). An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett 360, 111–114. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour M-E, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. (2005). Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the coordinates and structure factors of Nrd1 in complex with NIM1 is 6O3W, that with NIM2 is 6O3X, and that with NIM3 is 6O3Y.


