Abstract
Screening of an inhibitor library targeting kinases and epigenetic regulators identified several molecules having anti-proliferative synergy with BET bromodomain inhibitors (JQ1, OTX015) in triple-negative breast cancer (TNBC). GSK2801, an inhibitor of BAZ2A/B bromodomains, of the imitation switch chromatin remodeling complexes, and BRD9, of the SWI/SNF complex, demonstrated synergy independent of BRD4 control of P-TEFb-mediated pause-release of RNA polymerase II. GSK2801 or RNAi knockdown of BAZ2A/B with JQ1 selectively displaced BRD2 at promoters/enhancers of ETS-regulated genes. Additional displacement of BRD2 from rDNA in the nucleolus coincided with decreased 45S rRNA, revealing a function of BRD2 in regulating RNA polymerase I transcription. In 2D cultures, enhanced displacement of BRD2 from chromatin by combination drug treatment induced senescence. In spheroid cultures, combination treatment induced cleaved caspase-3 and cleaved PARP characteristic of apoptosis in tumor cells. Thus, GSK2801 blocks BRD2-driven transcription in combination with BET inhibitor and induces apoptosis of TNBC.
Keywords: BET bromodomain, BAZ2A, BAZ2B, drug synergy, triple-negative breast cancer
INTRODUCTION
Inhibitors of the bromodomain (BD) and extraterminal domain (BET) epigenetic readers have shown pre-clinical promise in multiple types of cancer, notably in acute myelogenous leukemia (AML) where BET proteins play a critical role in maintaining transcription of MYC (1–3). BRD2/3/4 interact with acetyl-lysine residues on histone tails and non-histone proteins via their BDs. Their regulatory function is in part attributed to the ability of BRD4 to associate with and recruit the positive transcription elongation factor (P-TEFb) complex via interactions involving its unique C-terminal domain (CTD). P-TEFb is a multi-subunit complex composed of CDK9 and its regulatory subunit cyclin T1. P-TEFb promotes productive gene transcription in concert with additional transcriptional coactivators (Mediator complex) and chromatin modifying enzymes (CREB binding protein (CBP)/p300). BRD4 is the best characterized BET family member, which recruits and activates the P-TEFb complex leading to transcriptional elongation through CDK9-catalyzed phosphorylation of serine 2 in the CTD of RNA polymerase II (Pol II) (4,5).
In addition to inhibition of tumor growth as single agents, BET bromodomain inhibitors (BETi) have been shown to block adaptive resistance in combination with inhibitors targeting receptor tyrosine kinases and the MEK-ERK and PI3K pathways (6–9). Inhibition of these signaling networks results in genome-wide enhancer formation involving the seeding of BRD4, MED1, H3K27 acetylation, and CBP/p300 that drives P-TEFb-dependent transcriptional adaptation (6). BETi blocks enhancer remodeling and prevents P-TEFb-dependent transcriptional adaptive reprogramming, making the action of the kinase inhibitors more durable and actually preventing or reversing resistance (6).
Pre-clinical models of breast cancer are generally sensitive to inhibition of BET BD-dependent transcription (7,10,11), specifically triple-negative breast cancer (TNBC) (12), which is a heterogeneous and aggressive disease defined by the absence of targetable receptors for estrogen and progesterone, and HER2 (13,14). Clinical trials are currently testing the efficacy of BETi in treating TNBC, other solid tumors and hematologic malignancies (ClinicalTrials.gov). Both BRD2 and BRD4 are overexpressed in basal-like TNBC, but the functional distinctions in the mechanisms by which BRD2 and BRD4 regulate transcription independent of P-TEFb activation remain poorly defined.
We performed dose-dependent drug synergy screens against inhibitors targeting the BET BD to define novel combinatorial strategies that selectively enhanced growth inhibition by BETi in TNBC. Of particular interest was the novel drug synergy we discovered between inhibitors targeting additional families of bromodomains encoded in CBP/p300, BAZ2A/B, and BRD9. GSK2801, an inhibitor of the BDs of BAZ2A (TIP5), BAZ2B and BRD9 (bromodomain-containing protein 9) showed little or no growth inhibition as a single agent, yet combined treatment with BETi resulted in strong growth inhibition of TNBC. BRD9, whose BD binds GSK2801 with a lower affinity than the BD of BAZ2A/B, is a member of the SWI/SNF regulatory complex that regulates chromatin remodeling and transcription. Bromodomain adjacent to zinc finger domain (BAZ2) proteins function as regulatory subunits which pair with one of two ATPases, SMARCA1 (SNF2L) or SMARCA5 (SNF2H), to form the core of imitation switch chromatin remodeling complexes (ISWI) (15). BAZ2A binds SMARCA5 to form the nucleolar remodeling complex (NoRC) which maintains a pool of heterochromatic ribosomal DNA (rDNA) (16). NoRC function is important not only to temper the amount of rDNA copies accessible for transcription, of which there are hundreds in a given mammalian cell, but also to maintain genomic stability of these highly repetitive regions. This protective function of the NoRC also extends to centromeres and telomeres (17,18). Little is known about the function of BAZ2B; it was recently reported that BAZ2B forms stable complexes with both ATPases making it a novel ISWI regulatory subunit (19).
The TCGA pan-cancer dataset indicates BAZ2A and BAZ2B are not frequently mutated across cancer with the exception of uterine corpus endometrial carcinoma where both BAZ2A and BAZ2B are mutated in greater than 10% of tumors. Increased expression of BAZ2A/B mRNA is observed across several tumor types including thyroid carcinoma and AML. A super-enhancer enriched for H3K27Ac was recently identified proximal to the BAZ2B gene in AML (20). BAZ2B is consistently highly expressed in AML with no change in copy number, suggesting epigenetic mechanisms of BAZ2A/B overexpression. BRD9 is also not frequently mutated in the TCGA pan-cancer dataset but is significantly amplified in lung cancers. BRD9 has also been implicated in driving AML growth as a subunit of the SWI/SNF complex via regulation of MYC transcription (21).
Our screening results provide a series of synergistic drug combinations to achieve durable inhibition of TNBC cell proliferation with BET or CBP/p300 BD inhibitors. This data highlights a unique mechanism of synergy between the BAZ2/BRD9 BD inhibitor GSK2801 and BETi. Drug synergy was not observed between GSK2801 and other inhibitors of P-TEFb transcriptional activation including CBP/p300 and CDK9 inhibitors, indicating a mechanism of synergy unrelated to pause-release of Pol II. Instead, loss of BRD2 from chromatin following combined GSK2801 and BETi blocked ETS-regulated gene transcription in the nucleoplasm as well as nucleolar transcription of rRNA. These findings support distinct functional roles of BRD2 in regulating nucleoplasmic and nucleolar transcription relative to other BET family proteins. Furthermore, these results reveal unique adaptive mechanisms of BRD2 on chromatin in response to BETi which are amenable to co-inhibition of BAZ2/BRD9 bromodomains to induce apoptosis in 3D-spheroid cultures of TNBC.
MATERIALS AND METHODS
Cell culture
HCC1806, WHIM12, and MDA-MB-468 cells were maintained in RPMI-1640 medium (Gibco, ThermoFisher Scientific) supplemented with 10% FBS, 1,000 Units/mL Penicillin and 1mg/mL Streptomycin. MDA-MB-231 and SUM-159 cells were maintained in DMEM/F12 1:1 medium (Gibco, ThermoFisher Scientific) supplemented with 5% FBS, 5 μg/mL insulin, 1 μg/mL hydrocortisone, 1,000 Units/mL penicillin and 1mg/mL streptomycin. SUM-149(+) and WHIM2 cells were maintained in HuMEC medium with defined media supplements (Gibco, ThermoFisher Scientific) and supplemented with 5% FBS, 1,000 Units/mL penicillin and 1mg/mL streptomycin. SUM-149(+) cells used in this study contain EpCAM+ /CD45f+ cells which were isolated via fluorescence activated cell sorting from the heterogeneous parental SUM-149 cell line.
Cell line authentication
All cell lines were obtained from the Lineberger Comprehensive Cancer Center as well as collaborating labs. Whim2 and Whim12 cell lines derived from PDXs have been whole-exome sequenced and RNA-sequenced for reference. All other cell lines used in this study have been authenticated by the Johns Hopkins Genetics Core Resources Facility using their short-tandem repeat (STR) testing. Cell lines are annually tested in the lab for Mycoplasma. Cell lines were passaged no longer than one month for all experiments performed.
Compounds
Information on compounds used in screening and subsequent validation experiments is listed in Supplementary Data File 1. BI-9564 (Cat. No. S8113) was obtained from Selleckchem and BAZ2-ICR (Cat. No. SML1276) was obtained from Sigma-Aldrich.
Synergy screening
The optimal dose range for JQ1, OTX015 or CPI-637 was determined for each cell line screened across half log doses. JQ1 and OTX015 screens were performed in 6 × 6 dose response matrices. MDA-MB-231, SUM-149(+), and WHIM12 cell lines were treated with 3 nM – 1 μM JQ1. HCC1806, WHIM2, and MDA-MB-468 cell lines were treated with 10 nM – 3 μM JQ1. MDA-MB-231 cells were dosed with 10 nM – 3 μM OTX015. All CPI-637 screens were performed in 5 × 6 dose response matrices. MDA-MB-231, HCC1806, WHIM2, and WHIM12 cells were dosed with 100 nM – 10 μM CPI-637 in half-log doses. Cells were seeded in 384 well plates using a BioTek microplate dispenser. The following day cells were dosed with drug using a Beckman Coulter Biomek FX instrument. The screening library was tested for growth inhibition alone or in combination across six doses of the screening library: 10 nM, 100 nM, 300 nM, 1 μM, 3 μM, and 10 μM. Bortezomib (1 μM) was included as a positive control or 0.1% DMSO as a negative control for growth inhibition on each plate. Plates were incubated at 37°C for 96 hours and lysed by adding 10 μl Cell Titer-Glo reagent (Promega, Cat. No. G7570) to 50 μl cell media. Luminescence was measured using a PHERAstar FS instrument and growth inhibition was calculated relative to DMSO treated wells.
Drug synergy analysis
Drug synergy scores were generated using the SynergyFinder package 1.6.1 (22). Bliss, Loewe, highest single agent (HSA), and zero interaction potency (ZIP) scores were calculated without baseline correction and using default parameters with the exception that Emin was specified as 0 and Emax as 100. Synergy was assessed across individual doses of each library compound to generate six possible scores per compound. To be considered a hit, a given compound had to generate a positive mean synergy score for at least one dose tested in five out of six (for JQ1 screening data) or three out of four (for CPI-637 screening data) cell lines screened. Hits were then ranked by the mean of all synergy scores produced in the drug combination matrix. Synergy scores represent the percent growth inhibition induced by a drug combination which exceeded the expected growth inhibition. Expected growth inhibition was calculated based on the effect of each drug as a single agent.
Chromatin immunoprecipitation and library preparation
ChIP experiments were performed as described in (6). Briefly, approximately 1×107 cells per IP were crosslinked 10 minutes at room temperature in 1.1% formaldehyde. Nuclear extracts were sonicated 15 cycles (30-second pulse, 30-second cooling) using a Bioruptor Pico (Diagenode). Samples were tumbled overnight at 4°C with 10 μg antibody conjugated to protein A Dynabeads (ThermoFisher Scientific). Following reverse-crosslinking and RNase/proteinase K treatment, DNA was eluted from beads and purified using Qiagen MinElute PCR purification columns. ChIP-seq libraries were prepared using the KAPA HyperPrep Kit according to the manufacturer’s instructions. For each set of experimental samples an equal amount of DNA was used to generate libraries (between 10–50 ng depending on experiment). Samples were indexed using Illumina TruSeq indexed adapters. Dual size selection was performed following 12–18 cycles PCR amplification. Samples were 12-plexed and single end 75 bp reads were generated using an Illumina NextSeq-500. Raw and processed ChIP-seq datasets have been uploaded to GEO (accession # GSE116919)
RNA-seq
Total RNA (4ug) was isolated from cells using the Qiagen RNeasy Plus Kit. Sequencing libraries were prepared using the KAPA stranded mRNA-seq kit according to the manufacturer’s instructions with the exception that libraries were amplified 10 cycles by PCR. Samples were indexed using Illumina TruSeq indexed adapters. Samples were 12-plexed and run on an Illumina NextSeq-500 to produce 75-cycle single end reads.
RNA-seq analysis
TNBC patient and cell line datasets used in Figure 1C–D are deposited in dbGaP (phs001405.v1.p1) and GEO (accession no. GSE87424) respectively. Data was generated and processed as cited in (6). Briefly datasets were aligned to the human reference genome (hg19_M_rCRS) using MapSplice (23), sorted and indexed using SAMtools v1.2 (24), and transcript abundance estimates were generated using the RSEM expectation-maximization algorithm (25).
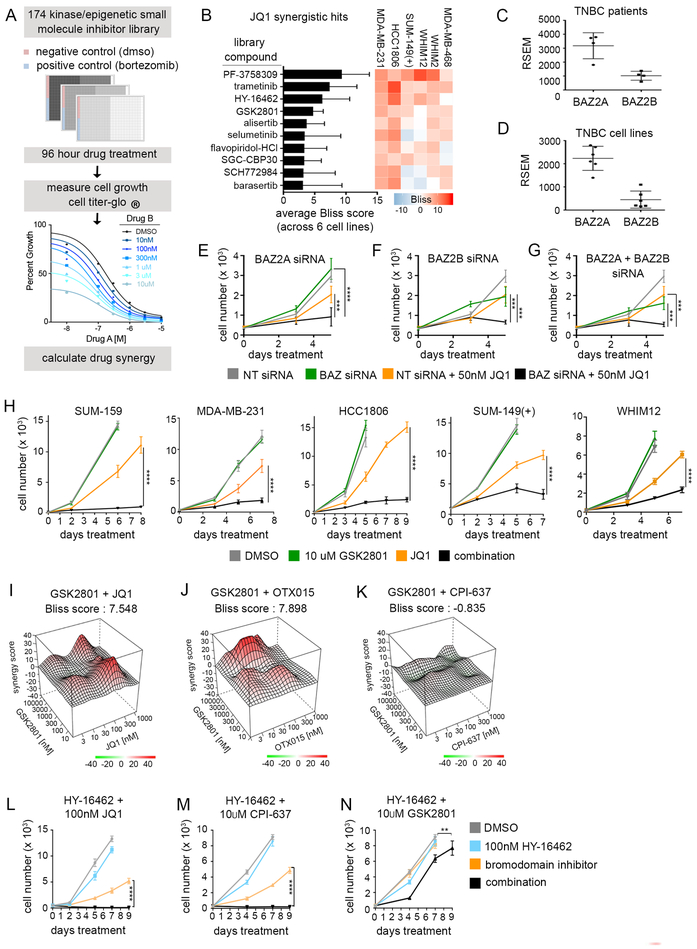
Figure 1: Drug synergy screens against BET and p300 BD inhibitors in TNBC cell lines.
(A) Schematic for screening approach. Cells were plated in 384 well plates and treated 96 hours with 6 × 6 dose concentrations of library compound in combination with either JQ1 (BETi) or CPI-637 (p300i). Cell viability was measured with Cell Titer-Glo and drug synergy was quantified using Synergy Finder. (B) Drug synergy rankings using Bliss scoring for JQ1 screens (performed in MDA-MB-231, HCC1806, SUM-149(+), MDA-MB-468, WHIM2, and WHIM12). Synergy scores represent the percent inhibition observed following combination treatment which exceeded the expected growth inhibition as calculated by the bliss independence model. Values were generated by first calculating the mean Bliss score across the full drug synergy matrix for each cell line (36 possible dose combinations). (C-D) RNA-seq reads of BAZ2A and BAZ2B expression in primary TNBC patient samples (C) vs. 6 TNBC cell lines (D) used for screening. (E-G) MDA-MB-231 growth curves transfected with non-targeting (NT) or BAZ2 siRNAs in combination with 50 nM JQ1. Error bars represent +/− SD, n = 6. P-values were calculated using two-tailed t-tests. (H) Growth curves across multiple TNBC cell lines with 10μM GSK2801. JQ1 doses were determined based on the relative sensitivity of each cell line. MDA-MB-231: 30nM JQ1, SUM159: 100nM JQ1, SUM-149(+) and WHIM12: 300nM JQ1, HCC1806: 500nM JQ1. Error bars represent +/− standard deviation, n = 6. P-values comparing JQ1 vs. combination drug treatment were calculated using two-tailed t-tests. (I-K) 3D drug interaction landscapes produced using SynergyFinder between GSK2801 and JQ1/OTX015 or CPI-637 in the MDA-MB-231 cell line following 96-hour drug treatment. (L-N) MDA-MB-231 growth curves with 100 nM HY-16462 in combination with 100 nM JQ1, 10 μM CPI-637, or 10 μM GSK2801. Error bars represent +/− SD, n = 6. P-values comparing BDi vs. combination drug treatment were calculated using two-tailed t-tests.
For drug-treated datasets used in the rest of the manuscript, QC-passed reads were aligned to the human reference genome (hg38) using STAR 2.4.2a (26) and reads were translated to transcriptome coordinates using Salmon 0.60 (27). Isoform data were collated to single gene IDs using the R package biomaRt (28), and abundance estimates were upper quartile normalized using R. Raw and processed RNA-seq data have been uploaded to GEO (accession no. GSE116919).
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was run on drug-treated RNA-seq datasets according to instructions at http://software.broadinstitute.org/gsea/index.jsp. All Hallmark, GO and Oncogenic Signature gene sets were analyzed using the following parameters: number of permutations: 1000, permutation type: gene_set, enrichment statistic: weighted, metric for ranking genes: Signal2Noise, gene set max size: 500, gene set min size: 15.
ChIP-seq analysis
ChIP-seq data sets were aligned using Bowtie v1.1.2 (29) to the human reference genome (hg19) or to a single copy of the ribosomal DNA repeat (GenBank: U13369.1). Alignments were performed using the following parameters: -v 2 – m 1. All analysis was performed using RefSeq (30) hg19 human gene annotations. Read density was normalized to the total number of million mapped reads (rpm). Data was normalized by subtracting read density of the input chromatin from the ChIP read density. HCC1806 and SUM-159 input datasets were used from (6), GEO accession no. GSE87424. MACS 2.2.2.20160309 (31) and HMCan v1.28 (32) were used to call enriched regions. We found that MACS was not calling peaks in the regions with high CNV and therefore used HMCan to call peaks in these regions. MACS was run using default settings. HMCan was run with narrow peak calling configuration file with no blacklisted regions. Peaks within 12.5 kb of each other were stitched as described (33). For comparative analysis across drug treatments, union peaks were defined which represent a collection of peak regions across all the treatment datasets in a project. Peak classification was performed as described in (6) and python code generated in the laboratory for ChIP-seq analysis is available at https://github.com/darshansinghunc/chippeakanalysis.
A peak in dataset A was considered overlapped by dataset B if 40% of the length of the peak in dataset A was covered by a peak in dataset B.
ChIP-seq data visualization
For ribosomal DNA alignments, reads were normalized to the total number of reads mapped to the hg19 reference genome per sample. Read counts were recorded in 250 bp bins and data was plotted using R. For fold-change plots a floor of 5 rpm was created in order to avoid artificially high fold-change values.
ChIP-seq density tracks in Figure 3L–O and Figure S4F–G were created by normalizing data to the total number of mapped reads. Read counts were plotted using Python.
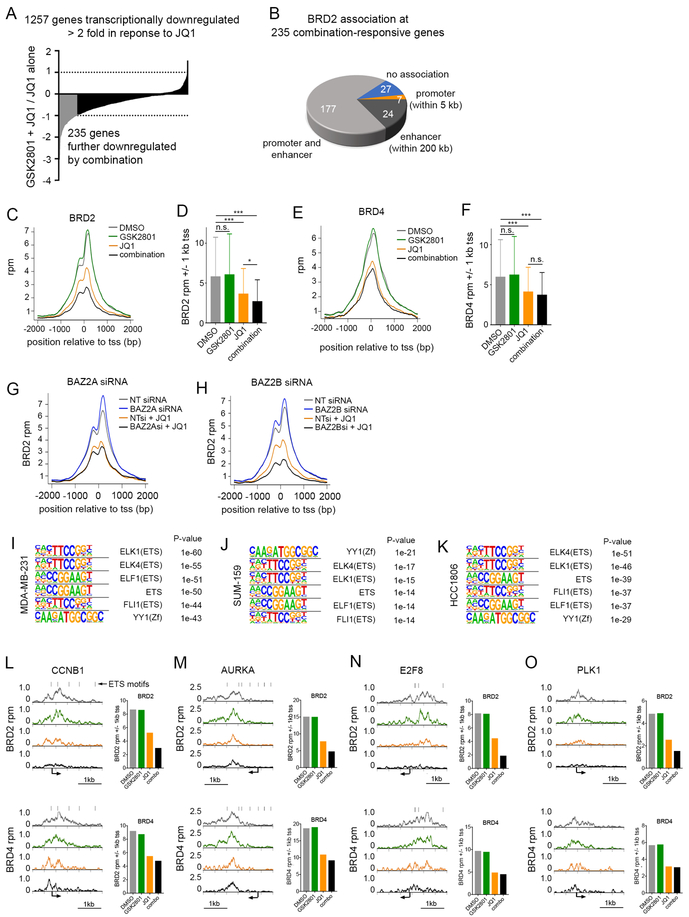
Figure 3: BRD2 is displaced from ETS-regulated gene promoters and enhancers following combination treatment with GSK2801 and JQ1.
(A) Long tail plot of MDA-MB-231 genes downregulated ≥ 2-fold following 72-hour treatment with 100 nM JQ1. Values represent mRNA fold-change between treatment with 10 μM GSK2801 + JQ1 vs JQ1 alone. Genes highlighted in grey were downregulated ≥ 2-fold in combination treated cells relative to JQ1 alone. (B) Association of BRD2 peaks with “combination-responsive” genes in DMSO treated MDA-MB-231 cells. (C-F) Metagene plots of BRD2 (C) and BRD4 (E) density at transcription start sites (tss) of “combination-responsive” genes. MDA-MB-231 cells were treated 48 hours with DMSO, 10 μM GSK2801, 100 nM JQ1 or the combination. Bar plots represent total read counts of BRD2 (D) or BRD4 (F) +/− 1 kb of the tss. P-values were calculated via two-tailed, paired t-tests (G-H) Metagene plots of BRD2 density at tss of “combination-responsive” genes following transfection with BAZ2A (G) or BAZ2B (H) siRNA. MDA-MB-231 cells were treated 48 hours with and without 100 nM JQ1 following transfection. (I-K) Motif analysis of BRD2 binding in DMSO treated samples of MDA-MB-231 (I), SUM-159 (J), and HCC1806 (K) cell lines. (L-O) BRD2 and BRD4 ChIP-seq density tracks following 48 hours treatment with DMSO, 10 μM GSK2801, 100 nM JQ1 or the combination in MDA-MB-231 cells. Black bars denote ETS binding motifs. Bar plots represent total read counts of BRD2 or BRD4 +/− 1 kb of the tss
Box-and-whisker plots for ChIP-seq datasets were generated using peaks with greater than or equal to 5 rpm density in at least one treatment condition. Paired t tests were performed to measure statistical differences between samples.
All metagene plots and heatmaps were generated using HOMER (34). 25 bp bins were used to generate all plots surrounding +/− 2 kb of the annotated tss or peak center. Heat maps were visualized using Java Treeview (35)
Motif Enrichment Analysis
HOMER was employed to define motifs enriched at BRD2-occupied genomic loci. All BRD2 peaks greater than or equal to 5 rpm in DMSO control samples were used for each cell line analyzed. Default parameters were used to search a 200 bp window surrounding peak centers.
Tumor spheroids
Tumor cells expressing red fluorescent protein were plated in low-adhesion round-bottom 96 well plates as mono-cultures or were co-cultured with reduction mammoplasty fibroblasts (RMFs) expressing green fluorescent protein. For SUM-149(+) spheroids 5,000 tumor cells were seeded per well or mixed at a 1:1 ratio with RMFs. For WHIM12 spheroids 1,000 tumor cells were mixed with 2,000 RMFs per well. Spheroids were given 48 hours to form before beginning drug treatment. At endpoint cell fluorescence was imaged using an Evos FL Auto 2 instrument. Following imaging, spheroids were lysed in Cell Titer-Glo 3D (Promega, Cat. No. G9681) by adding 50 μl reagent to 150 μl cell media. Plates were rotated at 150 rpm for 30 mins before luminescence was read using a PHERAstar FS.
RESULTS
High-throughput identification of synergistic drug combinations with BETi in TNBC
A 175 compound, small molecule inhibitor library targeting protein kinases and epigenetic modifiers was used in 6 × 6 dose response matrices to screen for drug synergy in combination with BD inhibitors (Figure 1A). Library compounds were selected based on target diversity as well as indication of clinical success and when suitable FDA-approval (Supplementary Data File 1). We screened six TNBC cell lines representing TNBC basal-like and claudin-low molecular subtypes (36,37) against JQ1, a BETi. The cell lines displayed a range of baseline sensitivities to JQ1 (IC50s ranged from 37.4 nM – 1.1 μM) (Figure S1A). Synergy was assessed using multiple computational models (22) including highest single agent (HSA), the Loewe additivity model (Loewe), the Bliss independence model (Bliss) and the zero interaction potency model (ZIP) (38) (Supplementary Data File 2). Common hits identified in all cell lines across all synergy models (Figure 1B; Figure S1B) were validated with an additional BETi, OTX015 (39), in the MDA-MB-231 cell line (Supplementary Data File 3).
Inhibition of previously identified targets: MEK1/2-ERK1/2 (trametinib and selumetinib (MEKi), SCH772984 (ERKi)), P-TEFb (HY-16462 and flavopiridol (CDK9i), SGC-CBP30 (CBP/p300i)) (6) and Aurora kinase (alisertib and barasertib) (11), validated the synergy screen (Figure 1B; Figure S1C–F). Among the other top hits was PF-3758309 that selectively targets PAK4 with higher potency than other PAK family members. PF-3758309 caused cell shape changes and diminished adherence in 2D culture of multiple TNBC cell lines. Western blots showed no reduction in pERK1/2 or pAKT following combination PAK/BET BD inhibition (Figure S1G), suggesting loss of cell number in 2D screens was due to cytoskeletal changes. PF-3758309 was not further pursued since clinical trials for this compound have been terminated and other PAKi have not progressed in patient trials despite significant numbers of studies in pre-clinical models (ClinicalTrials.gov).
Of particular interest from the screen hits was the novel drug synergy observed between BETi and GSK2801, an inhibitor of BAZ2A and BAZ2B bromodomains with partial activity against the BRD9 bromodomain. BAZ2A and BAZ2B are members of the bromodomain adjacent to zinc finger domain (BAZ) protein family and contain a PHD domain and homologous BD (40). We observed similar expression levels by RNA sequencing (RNA-seq) of BAZ2A and BAZ2B between primary TNBC patient tumors (dbGaP: phs001405.v1.p1) and cell lines (Figure 1C–D). BAZ2A is expressed more highly than BAZ2B in TNBC cell lines at an average RSEM read count of 2,233 compared to 451, respectively. Knockdown via RNAi of BAZ2A and B in combination with JQ1 resulted in significant growth inhibition (Figure 1E–G; Figure S1H), consistent with the screening results. We further validated synergistic growth inhibition with combination GSK2801 and JQ1 in a panel of TNBC cell lines (Figure 1H).
In addition to binding the BDs of BAZ2A/B, GSK2801 has reported binding activity for the bromodomain of BRD9 (41), a member of the SWI/SNF chromatin remodeling complex. AlphaScreen assays confirmed GSK2801 interacted with the BRD9 BD. In contrast, a second BAZ2A/B inhibitor, BAZ2-ICR (42), showed no interaction with the BRD9 BD (Figure S2A–C). Neither inhibitor displayed activity against the BET BD (Figure S2D). Dose-dependent growth curves revealed that 3 μM GSK2801, which inhibits BAZ2A/B BDs but not the BRD9 BD, produced partial growth suppression in combination with JQ1 compared to 10 μM GSK2801 (Figure S2E). Using RNAi targeting BRD9 in combination with JQ1 produced enhanced growth suppression (Figure S2F–G). Thus, inhibition of BRD9 by GSK2801 contributed to growth inhibition observed with combination drug treatment (JQ1+GSK2801). Combining the selective BAZ2 BD inhibitor BAZ2-ICR with the selective BRD9 inhibitor BI-9564 elicited complete growth suppression in combination with JQ1, as seen with GSK2801 and JQ1 (Figure S2H). Western blots showed a BRD9-dependent reduction of c-MYC levels relative to JQ1 alone (Figure S2I), consistent with the role of BRD9 maintaining MYC expression and rapid cell proliferation as a member of the SWI/SNF complex (21).
Synergy screens were also performed with CPI-637, an inhibitor of the CBP/p300 BD, in order to assess the overlap of therapeutic vulnerabilities to those found with BETi (Supplementary Data File 4). Reports of the significant activity of kinase inhibitors on BET BDs (43–45) led us to perform AlphaScreen assays across four doses (10nM, 100nM, 1uM, and 10uM) of our screening library to control for binding against the BET and CBP/p300 BDs (Supplementary Data File 5). None of the screen hits displayed off-target activity at previously defined synergistic doses against either the BET or CBP/p300 BD (Figure S2J), which might otherwise account for drug synergy. We observed significant overlap between the BETi and CBP/p300i screening datasets, suggesting BET protein function is tightly linked to CBP/p300 acetyltransferase activity (Figure S2K). Uniquely, GSK2801 only displayed growth inhibition in combination with BETi, JQ1 and OTX015, and not the CBP/p300i CPI-637 (Figure 1I–K). Furthermore, GSK2801 did not synergize with inhibition of CDK9, another member of the P-TEFb complex, compared to p300i and BETi (Figure 1L–N). This indicates a mechanism of synergy between GSK2801 and BETi which is independent of P-TEFb-mediated transcriptional activation controlled by BRD4.
Selective BRD2, 4 and 9 displacement from chromatin following treatment with BETi (JQ1) and BAZ2A/B bromodomain inhibitor (GSK2801)
To gain a more comprehensive understanding of how bromodomain proteins respond to GSK2801 and JQ1 treatment, we quantified BRD2, BRD4 and BRD9 on chromatin via chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq). Both BRD2 and BRD4, whose BDs bind JQ1, were significantly lost from chromatin following single agent JQ1 treatment (Figure 2A–B). There is a large degree of overlap between BET proteins at baseline with 38% of BRD4 peaks overlapping with BRD2 and almost all BRD2 peaks (94%) overlapping with BRD4 peaks. Despite this co-occupancy of chromatin peaks, relative to JQ1 treatment alone, only BRD2 but not BRD4 showed an enhanced loss from chromatin with JQ1 + GSK2801 combination drug treatment (Figure 2A–B). Classification of BRD2 peaks showed GSK2801-dependent loss of BRD2 at both promoter and enhancer regions across the genome (Figure S3A–B). This indicates the interaction of GSK2801 with BAZ2A/B and BRD9 BDs causes dissociation of BRD2 but not BRD4 from chromatin in the context of JQ1 treatment.
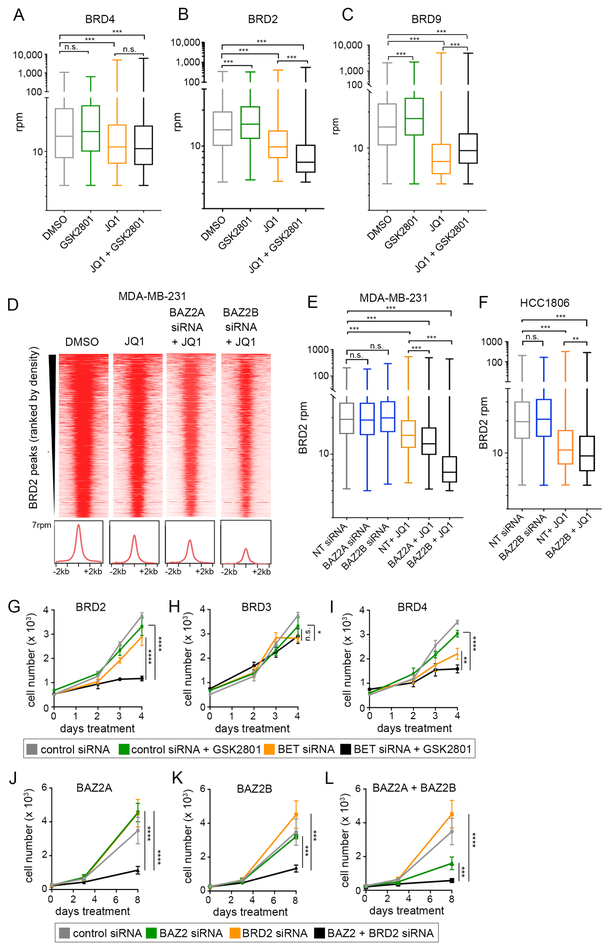
Figure 2: BRD2 is displace from chromatin following combination drug treatment.
(A-C) Levels of mapped BRD4 (A) BRD2 (B) and BRD9 (C) union peak density following 48 hours treatment with 100 nM JQ1, 10 μM GSK2801, or the combination in MDA-MB-231 cells. Statistical significance was measured via two tailed, paired t-tests. (D-E) BRD2 peak density following transfection with 25 nM non-targeting (NT), BAZ2A or BAZ2B siRNA. MDA-MB-231 cells were treated 48 hours following transfection +/− 100 nM JQ1. Statistical significance was measured via two-tailed, paired t-tests. (F) BRD2 density following transfection with 25 nM non-targeting (NT) or BAZ2B siRNA alone and in combination with 500 nM JQ1 treatment in the HCC1806 cell line. Statistical significance was measured via two-tailed, paired t-tests. (G-I) MDA-MB-231 growth curves transfected with GAPDH siRNA as a control or 25 nM BRD2 siRNA (G), 25 nM BRD3 siRNA (H), and 1nM BRD4 siRNA (I) alone and in combination with 10 μM GSK2801. Error bars represent +/− SD, n = 6. P-values were calculated using two-tailed t-tests. (J-L) MDA-MB-231 cells transfected with 25 nM non-targeting (NT) or BRD2 siRNA alone and in combination with 25 nM BAZ2A siRNA (J), BAZ2B siRNA (K) or BAZ2A + BAZ2B siRNAs (L). Error bars represent +/− SD, n = 6. P-values were calculated using two-tailed t-tests.
ChIP-seq also was used to determine co-occupancy on chromatin between BRD9 and BET proteins. We observed overlap between BRD4 and BRD9 independent of BRD2, but overlap between BRD2 and BRD9 was only observed when BRD4 was also present (Figure S3C). This suggests BRD9 is in chromatin complexes that include BET proteins, most notably BRD4. BRD9 was lost from chromatin following BETi despite the specificity of JQ1 for BET bromodomains and not the BRD9 bromodomain (Figure 2C). We did not observe enhanced loss of BRD9 from chromatin following the addition of BAZ2i to the JQ1 treatment. This contrasts with BRD2, whose dissociation from chromatin is greater with the combination of GSK2801 + JQ1 than seen with JQ1 alone (Figure 2B–C).
We observed a modest, but significant, increase in binding of BET proteins and BRD9 to chromatin across our ChIP experiments with single agent GSK2801 treatment (Figure 2A–C). The majority of these peaks were classified at promoter regions and did not correlate with regions most responsive to JQ1 treatment. BAZ2A, BAZ2B and BRD9 are members of chromatin remodeling complexes and therefore, inhibition of their bromodomains may affect accessibility of chromatin and result in the increased chromatin occupancy of BRD2 and BRD9 that we observed following single agent GSK2801 treatment.
ChIP-seq in an additional TNBC cell line, SUM-159 also showed significant loss of BRD2 from chromatin following combination treatment (GSK2801 + JQ1) relative to JQ1 alone (Figure S3D), consistent with BAZ2A/B proteins having a selective regulation of BRD2 relative to BRD4 and BRD9. Importantly, knockdown of either BAZ2A or BAZ2B phenocopied treatment with GSK2801 in combination with JQ1, with greater displacement measured by ChIP-seq of BRD2 relative to JQ1 treatment alone (Figure 2D–E). Loss of BRD2 chromatin occupancy following combination BAZ2B RNAi/JQ1 treatment relative to JQ1 alone was confirmed in the HCC1806 cell line (Figure 2F; Figure S3E).
Growth regulation in response to BRD2, BRD3 or BRD4 knockdown by siRNA
Knockdown of BET family proteins via RNAi in MDA-MB-231 cells revealed loss of BRD2 expression alone had little effect on growth but induced strong growth inhibition in combination with GSK2801 (Figure 2G). BRD3 loss of expression had no effect on growth in the presence or absence of GSK2801 (Figure 2H). BRD4 knockdown caused a partial growth arrest alone and the addition of GSK2801 modestly enhanced growth arrest (Figure 2I). In an additional cell line, SUM-149(+), we observed a significant growth inhibition with BRD2 knockdown alone and complete growth inhibition following the addition of GSK2801 (Figure S3F). We confirmed combined growth regulation between BRD2 and each BAZ protein via RNAi knockdown. Co-knockdown of BRD2 and BAZ2A or BAZ2B enhanced growth inhibition of TNBC, with strong growth inhibition observed when all three proteins were lost (Figure 2J–L), confirming inhibition of both BAZ2A and BAZ2B contribute to drug synergy with BETi. The strong growth inhibition observed with RNAi knockdown of BAZ2A/B contrasts with the partial growth inhibition observed using a selective BAZ2A/B BD inhibitor, BAZ2-ICR, in combination with JQ1 (Figure S2H). The reason for this is unclear, except for the difference between loss of the protein by knockdown versus the small molecule BD inhibition of BAZ2A/B in the absence of BRD9 BD inhibition. Consistent with our screening data, knockdown of BRD2 did not enhance the anti-proliferative effect of an inhibitor targeting the BD of CBP/p300 (SGC-CBP30) and a CDK9 inhibitor (HY-16462) in MDA-MB-231 cells (Figure S3G–I). Cumulatively, the findings demonstrate that BRD2, BAZ2A/B and BRD9 control a chromatin network regulating cell growth independent of P-TEFb.
Displacement of BRD2 from chromatin following combination drug treatment occurs at genomic loci associated with ETS transcription factors
We sought to understand the functional consequence of BRD2 loss from chromatin in response to combination drug treatment by assessing the correlation between changes in BRD2 chromatin occupancy and gene transcription. RNA-seq revealed 1,257 genes were transcriptionally downregulated ≥ two-fold in response to JQ1 in MDA-MB-231 cells. Most JQ1-responsive genes were further downregulated following the addition of GSK2801, including 235 genes downregulated two-fold or greater relative to JQ1 treatment alone (Figure 3A). We next measured the proximity of BRD2 peaks to the 235 “combination transcriptionally-responsive” genes. Promoter and putative enhancer BRD2 peaks were defined within 5 kb or 200 kb of the gene promoter respectively. Of the 235 combination-responsive genes, 117 had both a promoter and enhancer BRD2 peak, 7 only had measurable BRD2 at their promoter, 24 had only BRD2 enhancers and 27 had no measurable BRD2 peaks within 200 kb of their promoter (Figure 3B). Metagene plots of BRD2 and BRD4 at the transcription start site (tss) of combination-responsive genes showed both BET proteins were lost following JQ1 treatment, but only BRD2 was significantly lost with the addition of GSK2801 relative to JQ1 alone (Figure 3C–F). Similar plots of enhancers showed consistent results with BRD2 density lost following combination drug treatment but not BRD4 density (Figure S4A–B). Waterfall plots measuring the fold change in BRD2 and BRD4 density following combination treatment relative to JQ1 revealed a higher resolution view of BET protein dynamics at the tss of individual responsive genes. BRD2 reads were lost at the majority of tss while BRD4 displayed a larger range of responses which did not mirror those of BRD2 (Figure S4C). In some cases, BRD4 was lost from the promoter of responsive genes, however in these cases BRD2 was often lost to a greater degree. We were able to validate BAZ2A/B-dependent displacement of BRD2 at the tss of combination-responsive genes using RNAi knockdown of BAZ2A/B proteins in the presence of JQ1 (Figure 3G–H). Similar to the global analysis (Figure 2D–E), BAZ2B knockdown resulted in greater displacement of BRD2 from transcription start sites of responsive genes compared to BAZ2A suggesting BAZ2B is primarily responsible for BRD2 regulation at these genes. This analysis validates a BAZ2A/B-dependent loss of BRD2 at genomic loci proximal to transcriptionally responsive genes following co-inhibition of BAZ2 and BET BDs.
Consensus binding sequence analysis of BRD2 ChIP peaks using Homer software determined significant enrichment of binding motifs for class 1 ETS transcription factors and the structural protein YY1 across three TNBC cell lines: MDA-MB-231, SUM-159, and HCC1806 (Figure 3I–K). Consistent with our results, a recent study found enrichment of ETS binding motifs at BRD2-occupied loci in T cells as well as motifs for the structural protein CTCF (46). ETS binding motifs were also enriched at a subset of genomic loci where BRD2 is significantly lost following combination JQ1 + GSK2801 drug treatment (Figure S4D–E). Of the 235 responsive genes, 85 contained an ETS binding motif within 200 bp of the tss. We observed almost complete loss of BRD2 from multiple ETS-regulated genes critical for cell cycle progression including cyclin B1, Aurora kinase A, E2F8, PLK1, LMNB1 and MAPK13 following combination drug treatment (Figure 3L–O; Figure S4F–G) however, BRD4 density was maintained or only slightly decreased relative to JQ1 treatment alone. These genes were transcriptionally repressed with JQ1 and further repressed with addition of GSK2801 (Figure S4H). Gene Set Enrichment Analysis (GSEA) of RNA-seq data from MDA-MB-231, HCC1806, and SUM-159 cells (Supplementary Data File 6) showed loss of transcriptional programs which are regulated by ETS transcription factors including c-MYC targets, core serum response, E2F targets and G2M checkpoint in response to combination drug treatment (Figure S4I). Cell cycle analysis via propidium iodide staining revealed G1 arrest in response to JQ1, which was enhanced with combination drug treatment (Figure S4J–K). These data indicate BRD2 regulates transcriptional programs necessary to drive cell growth, specifically those regulated by ETS transcription factors.
BRD2 is localized in the nucleolus and nucleoplasm of TNBC cells
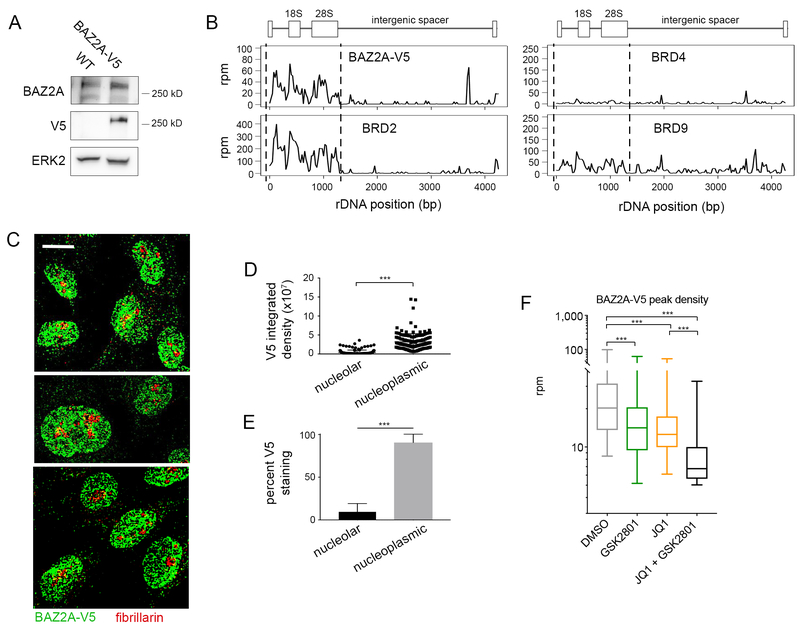
Further inspection of GSEA revealed an enrichment of ribosome biogenesis and rRNA processing gene sets in untreated samples relative to cells treated with combination GSK2801 + JQ1 (Figure 4A). Additionally, we observed enrichment of chromatin silencing gene sets in combination-treated cells, specifically silencing at ribosomal DNA. All stages of rRNA transcription, processing and ribosome biogenesis occur in the nucleolus. Notably, transcription of rDNA is the rate-limiting step of ribosome biogenesis and therefore serves as a rheostat of translational activity in cells. Silencing of rDNA regulates replicative senescence during aging (47,48), and nucleolar repression is sufficient to initiate and maintain senescence in tumor cells (49,50). We tested the localization of each BET protein to the nucleolus via immunofluorescence (Figure 4B). BRD2 was the only BET protein which localized to both the nucleoplasm and nucleolus. BRD3 was largely present in the nucleoplasm and excluded from the nucleolus, and BRD4 was present in both the cytoplasm and nucleoplasm but also excluded from the nucleolus (Figure 4B and Figure S5A–B). Quantification of staining confirmed BRD2 was significantly greater in the nucleolus among the BET proteins (Figure 4C). Furthermore, BRD2 displayed greater staining density in the nucleolus when compared to its nucleoplasmic staining which was not seen with BRD3 or BRD4 (Figure 4D).
Figure 4: BRD2 is the only BET protein localized to the nucleolus.
(A) GSEA of MDA-MB-231, HCC1806, and SUM-159 RNA-seq datasets treated 72 hours with 10 μM GSK2801 and 100 nM JQ1 (MDA-MB-231 and SUM-159) or 500 nM JQ1 (HCC1806). Orange values represent ribosome biogenesis or rRNA processing gene signatures. Positive enrichment scores represent gene sets enriched in DMSO and negative enrichment scores represent gene sets enriched in GSK2801 + JQ1 treated samples. (B) Immunofluorescent staining of BET proteins with fibrillarin (nucleolar marker) and Hoechst (nuclear marker) (C-D) Quantification of nucleolar vs nucleoplasmic staining intensity of BET proteins. A one-way analysis of variance (ANOVA) was conducted to compare intensity of BET protein staining, p-value < 2e-16 for (C) and (D). Post hoc comparisons were made using the Tukey HSD test.
Dual suppression of BAZ2/BRD9 and BRD2 bromodomains suppresses rDNA transcription
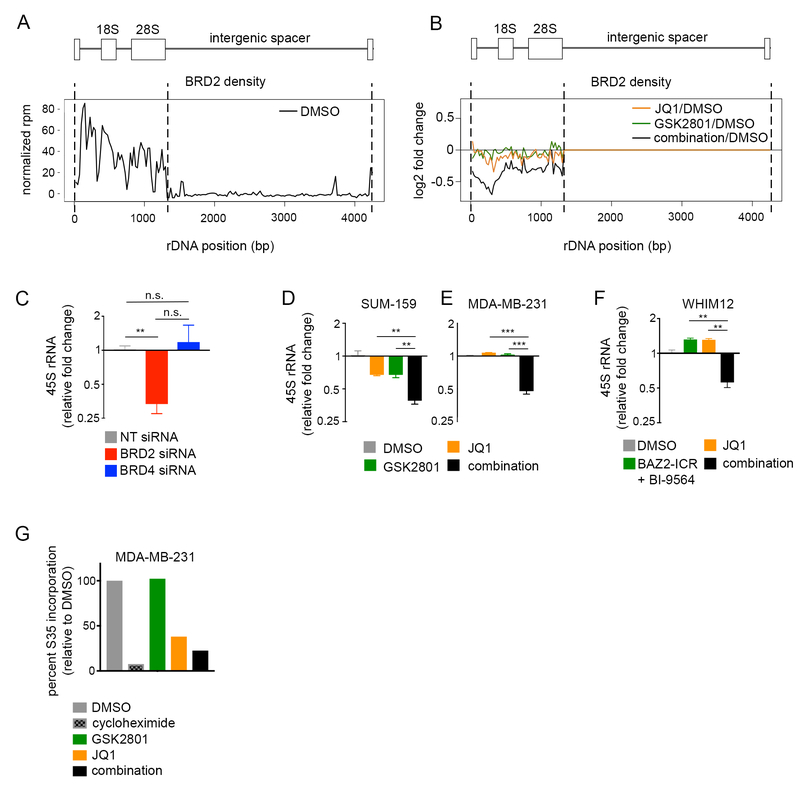
Consistent with our immunofluorescence data, BRD2 but not BRD4 ChIP density aligned to the coding region of the rDNA repeat (Figure 5A; Figure S6A–B). We observed modest changes in BRD2 occupancy at rDNA following single agent treatment with either JQ1 or GSK2801, however, BRD2 was significantly lost from the rDNA coding region following combination drug treatment with greatest loss at the rDNA promoter and start of the 18S exon (Figure 5B; Figure S6B). Knockdown of BRD2, but not BRD4, resulted in transcriptional loss of the 45S rRNA precursor observed by qPCR (Figure 5C). Interestingly, rDNA is transcribed by RNA polymerase I and so these data may explain, in part, why we observed a mechanism of drug synergy between GSK2801 and JQ1 independent of P-TEFb complex regulation, which exclusively regulates pause-release of RNA polymerase II. Indeed, combination drug treatment resulted in loss of the 45S rRNA specifically in combination-treated samples across multiple cell lines (Figure 5D–F). We performed S35 methionine labeling experiments to quantify global translation in response to drug in MDA-MB-231 cells. Although these results largely mirrored cell growth, we saw marked loss of S35 methionine incorporation following combination drug treatment (Figure 5G). These data support a novel role of BRD2 in positively regulating rRNA transcription and suggest loss of rRNA, and subsequent translation, during combination drug treatment contributes to synergistic growth inhibition.
Figure 5: Displacement of BRD2 from the rDNA repeat coincides with transcriptional repression of rRNA.
(A) Alignment of BRD2 ChIP-seq reads to the rDNA repeat in MDA-MB-231 cells. (B) Response of BRD2 density on rDNA in response to 48-hour treatment with DMSO, 10 μM GSK2801, 100 nM JQ1 or the combination in MDA-MB-231 cells. (C) qPCR measuring 45S rRNA following 96-hour transfection with non-targeting (NT), BRD2, or BRD4 siRNA in MDA-MB-231 cells. Error bars represent SD, n = 3. P-values were calculated using two-tailed t-tests. (D-E) qPCR measuring 45S rRNA in SUM-159 (D) and MDA-MB-231 (E) cells following 96-hour treatment with DMSO, 10 μM GSK2801, 100 nM JQ1 or the combination. Error bars represent SD, n = 3. P-values were calculated using two-tailed t-tests. (F) qPCR measuring 45S rRNA in the WHIM12 cell line following 96-hour treatment with 500nM JQ1 +/− 10 μM BAZ2-ICR and 300 nM BI-9564. Error bars represent SD, n = 3. P-values were calculated using two-tailed t-tests. (G) S35 labeling of protein production in MDA-MB-231 cells following 72-hour treatment with 10 μM GSK2801, 100 nM JQ1, or the combination. 100 μg/ml cycloheximide treatment was included as a positive control.
BAZ2A is present in the nucleolus and nucleoplasm and is co-regulated with BET BD proteins
Due to the lack of commercial antibodies with selective specificity for BAZ proteins needed for ChIP, we used CRISPR to engineer a C-terminal in-frame V5 epitope tag in the BAZ2A gene in MDA-MB-231 cells (Figure S7A–B). The presence of multiple alternative transcription start sites and lack of C-terminal sequence required for guide RNA design prohibited us from successfully tagging BAZ2B. Immunoblotting confirmed BAZ2A-V5 was expressed at similar levels as BAZ2A protein in parental cells (Figure 6A). ChIP-seq using a V5 antibody revealed BAZ2A-V5 bound the coding region of rDNA in a similar pattern as seen with BRD2 but not BRD4 or BRD9 (Figure 6B). The presence of BAZ2A-V5 in the nucleolus is consistent with the published function of BAZ2A (TIP5) as a member of the nucleolar remodeling complex. These findings are consistent with BAZ2A being associated with transcriptional complexes containing BRD2 within the nucleolus.
Figure 6: BAZ2A is co-regulated with BRD2 in the nucleolus and nucleoplasm.
(A) Western blot of MDA-MB-231 cell lysate expressing wildtype BAZ2A vs. BAZ2A-V5. (B) Alignment of BAZ2A-V5, BRD2, BRD4, and BRD9 ChIP-seq reads to the rDNA repeat in MDA-MB-231 cells. (C) Representative images of immunofluorescent staining of BAZ2A-V5 with fibrillarin (nucleolar marker) (D-E) Quantification of nucleolar vs nucleoplasmic staining intensity of V5 in approx. 200 individual cells. (F) Levels of BAZ2A-V5 union peak density following 48 hours treatment with 100 nM JQ1, 10 μM GSK2801, or the combination in MDA-MB-231 cells.
Immunofluorescent imaging confirmed BAZ2A-V5 staining in the nucleolus with significantly stronger staining density of BAZ2A-V5 in the nucleoplasm (Figure 6C–D). Quantification of staining revealed that, on average, only 10% of total BAZ2A-V5 staining in the nucleus was restricted to the nucleolus (Figure 6E). Quantification of global BAZ2A-V5 chromatin occupancy revealed a significant loss of BAZ2A-V5 from chromatin following inhibition of its bromodomain by GSK2801 (Figure 6F). We also observed significant global displacement of BAZ2A following treatment with the BETi JQ1, suggesting BAZ2A is co-regulated with BET BD proteins such as BRD2 on chromatin.
Combination treatment with BAZ2/BRD9 and BET bromodomain inhibitors induces senescence and apoptosis
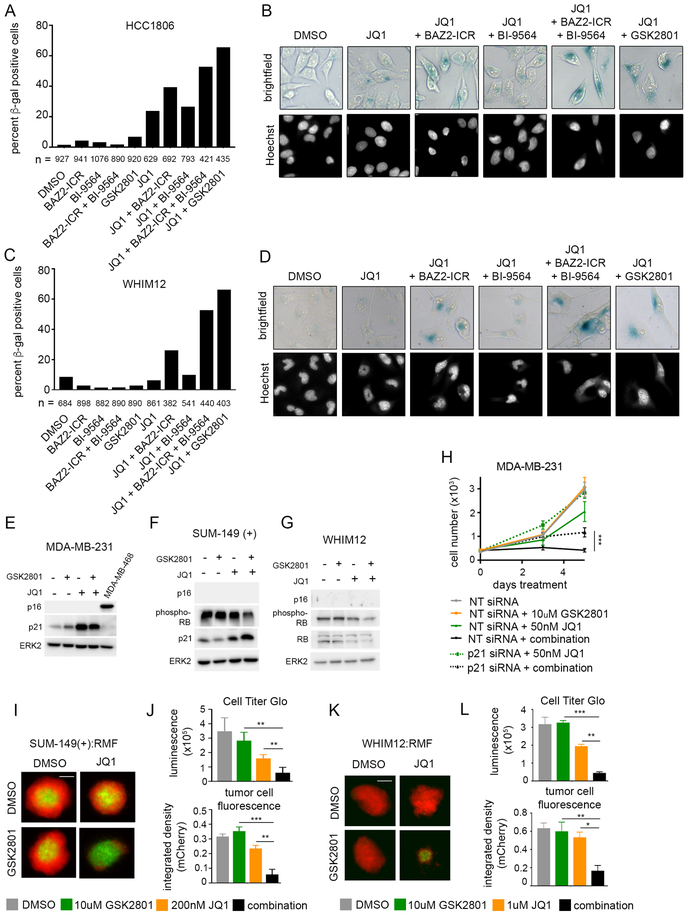
To further characterize the cellular phenotype in response to combination drug treatment, we performed beta-galactosidase (β-gal) staining of TNBC cell lines grown in 2D-cultures treated with drug for 96 hours. Combination drug treatment resulted in a higher percentage of β-gal positive cells relative to JQ1 alone (Figure 7A–D). Again, treatment with a selective BAZ2A/B BD inhibitor, BAZ2-ICR induced a partial response and it wasn’t until the addition of the BRD9 inhibitor, BI-9564, that we saw a similar percentage of β-gal positive cells compared to GSK2801. This confirms that although BAZ2A/B inhibition is contributing to synergistic growth inhibition, the effect of GSK2801 on BRD9 also contributes to drug synergy. Positive β-gal staining suggests that cells are entering a senescent state following combination drug treatment. We looked at which drivers of senescence signaling were responsible for the phenotype. All cell lines screened contain TP53 mutations common to TNBC, and most cell lines also contain deletion or transcriptional silencing of CDKN2A (p16) (Figure S8A). This left CDKN1A (p21) and RB1 the retinoblastoma (RB) tumor suppressor as candidate regulators of senescence. RNA-seq data showed induction of p21 transcript levels following a dose of JQ1 that induces modest β-gal staining (Figure S8B). Western blots in multiple cell lines confirmed induction of p21 protein levels and decrease in phospho-Rb in combination-treated samples (Figure 7E–G; Figure S8C–D). While knockdown of p21 via RNAi was able to rescue JQ1 growth inhibition, it only partially rescued growth inhibition of combination drug-treated cells (Figure 7H). These data suggest induction of p21 only partially regulates the senescent state of cells treated with JQ1 and GSK2801. It is therefore likely treatment with BAZ2A/B/BRD9i and BETi results in senescence via multiple mechanisms. First, inhibition of cell cycle gene transcription results in G1 arrest and induction of p21. Secondly, this growth arrest is likely enhanced by nucleolar suppression and translational silencing following loss of BRD2 from the rDNA promoter.
Figure 7: Combined GSK2801 and JQ1 treatment induces senescence and apoptosis in TNBC cell lines.
(A-D) Senescence-associated beta-galactosidase staining in HCC1806 (A-B) and WHIM12 (C-D) cell lines following 96-hour drug treatment. Cells were dosed with 10 μM GSK2801, 10 μM BAZ2-ICR, 1 μM BI-9564, and either 300 nM JQ1 (HCC1806) or 500 nM JQ1 (WHIM12). (E-G) Western blots of MDA-MB-231 (E), SUM-149(+) (F), and WHIM12 (G) cell lysates following 72-hour treatment with 10 μM GSK2801 and either 100 nM JQ1 (MDA-MB-231), 300 nM JQ1 (SUM-149(+)), or 500 nM JQ1 (WHIM12). (H) MDA-MB-231 cells transfected with 25 nM non-targeting (NT) or p21 siRNA alone and in combination with 10 μM GSK2801 and 50 nM JQ1. Error bars represent +/− SD, n = 6. (I-L) 3D spheroids with SUM-149(+) and WHIM12 cell lines co-cultured with reduction mammoplasty fibroblasts (RMF) treated ten days with GSK2801 and JQ1. Tumor cell fluorescence was measured at endpoint and cell viability was measured with Cell Titer-Glo 3D reagent, Error bars represent +/− SD, n = 3. P-values were calculated using two-tailed t-tests.
GSK2801 has poor pharmacokinetics in mice combined with a relatively high IC50 for the BAZ2/BRD9 BDs (41), so we tested combination drug treatment in 3D-spheroid models (Figure 7I–L; Figure S8E–F). Breast cancer cells expressing red fluorescent protein (RFP) were co-cultured with reduction mammoplasty fibroblasts (RMFs) expressing green fluorescent protein (GFP). We observed a dramatic reduction in breast tumor cell fluorescence following ten days combination treatment compared to either single agent alone in SUM-149(+) and WHIM12 cell lines (Figure 7I–L). This was consistent with ATP measurements using CellTiter-Glo 3D reagent, which showed diminished cell viability in the combination treated spheroids. There was no reduction in fluorescence from the normal breast fibroblast population following drug treatment, indicating a selective loss of tumor cell viability (Figure S8E–F). Monoculture spheroids established using only SUM-149(+) tumor cells displayed cleaved caspase-3 and cleaved PARP following 36 hours treatment with combination GSK2801 + 100nM or 300nM JQ1 (Figure S8G). Apoptosis was only present in spheroids treated with both drugs and not with either JQ1 or GSK2801 alone. These results were consistent with CellTiter-Glo measurements which showed dose-dependent reduction in cell viability with increasing doses of JQ1 in combination with GSK2801 (Figure S8H). These data highlight co-inhibition of BAZ2/BRD9 and BET BDs as an effective strategy to induce apoptosis in TNBC.
DISCUSSION
Ten BET BD inhibitors are currently in different phases of clinical trials for multiple tumor types including breast cancer (ClinicalTrials.gov). Despite some dose-dependent toxicity such as thrombocytopenia, the tolerability of different BETi in patients suggests that synergistic combination therapies may be practical. We previously showed the relevance of synergistic combination therapies with BETi in both HER2+ and TNBC pre-clinical models, demonstrating that BETi effectively blocks epigenetic transcriptional reprogramming making targeted kinase inhibitors, lapatinib and trametinib, more durable in their inhibition of tumor growth in vitro and in vivo (6,7). Our BETi screens identified inhibitors of MEK1/2-ERK1/2, CDK9, CBP/p300 and Aurora kinase as synergistic drug combinations with multiple TNBC cell lines. Trametinib and selumetinib are FDA-approved MEK1/2 inhibitors and there are multiple Aurora kinase inhibitors currently in all phases of clinical trials, alone and in different combinations with chemotherapy and targeted therapeutics (ClinicalTrials.gov). Therefore, BETi combination trials with MEK1/2 or Aurora kinase inhibitors could be initiated quickly because of the substantial knowledge of pharmacology and tolerability of each in patient trials.
The BDs of BAZ2/BRD9 were identified as novel targets for synergy with BETi to produce antiproliferative responses for multiple TNBC cell lines. The synergy seen with GSK2801 was regulated by a different mechanism than inhibitors targeting CBP/p300 and CDK9 of the P-TEFb transcriptional regulatory complex that is associated with BRD4. Instead, the uniqueness of this combination treatment is selective enhancement of BRD2 chromatin release in response to inhibition of BAZ2/BRD9 BDs.
While our data supports a mechanism of synergy by which BRD9 modulates cell growth through regulation of MYC expression, it is possible BRD9 is involved in regulation of BRD2 or BAZ2 proteins. ChIP data shows overlap between BRD2 and BRD9 peaks at regions where BRD2 is responsive to drug treatment. However, we do not observe the same displacement of BRD9 from chromatin following combination drug treatment as we see with BRD2. BRD9 is released from chromatin in response to JQ1 treatment alone, even though JQ1 does not bind the BRD9 BD, demonstrating the complexity of the BD encoded proteins and their interaction with chromatin.
Genes regulated by ETS transcription factors and the 45S rDNA promoter are selectively targeted by GSK2801 in combination with JQ1 for loss of BRD2, as defined by ChIP assay. Targeting of ETS regulated genes and ribosomal RNA transcription using GSK2801+JQ1 gave a strong antiproliferative response combined with the induction of pro-apoptotic caspase-3 activity and PARP cleavage in 3D cultures. To date, the most tractable inhibitor of the BAZ2A/B BD is GSK2801 that also binds the BRD9 BD. Inhibition of the BRD9 bromodomain clearly contributed to the antiproliferative action of the JQ1+GSK2801 combination treatment. Use of this tool compound enabled us to establish inhibition of BAZ2/BRD9 BDs as an effective means to block BRD2-driven transcription in combination with BETi in TNBC 2D-cultures and 3D-spheroid models. Why senescence is observed in 2D-cultures and apoptosis observed in 3D-cultures is unclear, but most likely is related to the adherence and cell interaction differences in the two models.
Triple-negative breast cancers do not contain significant mutations or copy number alterations in BAZ2A, BAZ2B or BRD9. Our study is the first to highlight BAZ2A/B and BRD9 as targetable vulnerabilities in TNBC in the context of BETi. Noncanonical functions of BAZ2A have been previously characterized in driving cancer phenotypes. Overexpression of BAZ2A in prostate cancer predicts disease recurrence and, in this context, BAZ2A was shown to interact with EZH2 and modulate expression of protein-coding genes in the nucleoplasm (51). BAZ2A also interacts with TCF7L2 to drive beta-catenin signaling and promote growth in hepatocellular carcinoma (52). There is less direct evidence for BAZ2B’s involvement in cancer but hypomethylation of the BAZ2B gene is associated with poor outcome in acute lymphoblastic leukemia (53), suggesting it may play a functional role in driving tumor growth and progression. The distinct functional roles of BAZ2A in cancer suggest inhibition of its BD may result in tumor-specific toxicity. Together our findings present co-inhibition of BAZ2A/B and BRD9 BDs in combination with BET protein BD inhibition as an effective strategy to block nucleoplasmic and nucleolar BRD2-regulated transcription for growth arrest, and induction of apoptosis of TNBC.
Supplementary Material
Implications:
Synergistic inhibition of bromodomains encoded in BAZ2A/B, BRD9 and BET proteins induces apoptosis of TNBC by a combinatorial suppression of ribosomal DNA transcription and ETS-regulated genes.
ACKNOWLEDGMENTS
All fluorescent imaging was performed at the Microscopy Services Laboratory at UNC Chapel Hill. Cell cycle analysis was performed at the Flow Cytometry Core Facility at UNC Chapel Hill. The UNC Flow Cytometry Core Facility and Microscopy Services Laboratory are supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
This study was funded by NIH grant CA058223 (G.L. Johnson), Susan G. Komen Foundation grant IIR12–225201 (G.L. Johnson), NIH grant GM116534 (S.M. Bevill) and the University Cancer Research Fund (G.L. Johnson).
Footnotes
Declaration of interests: The authors declare no competing interests.
REFERENCES
- 1.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell. 2011. September;146(6):904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012. October 4;120(14):2843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011. October;478(7370):524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hargreaves DC, Horng T, Medzhitov R. Control of Inducible Gene Expression by Signal-Dependent Transcriptional Elongation. Cell. 2009. July;138(1):129–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itzen F, Greifenberg AK, Bösken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014. July 8;42(12):7577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zawistowski JS, Bevill SM, Goulet DR, Stuhlmiller TJ, Beltran AS, Olivares-Quintero JF, et al. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer Discov. 2017;7(3):302–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015. April 21;11(3):390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou M-M, et al. Kinase and BET Inhibitors Together Clamp Inhibition of PI3K Signaling and Overcome Resistance to Therapy. Cancer Cell. 2015. June 8;27(6):837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014. October;514(7521):247–51. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Pena J, Serrano-Heras G, Montero JC, Corrales-Sanchez V, Pandiella A, Ocana A. In Silico Analysis Guides Selection of BET Inhibitors for Triple-Negative Breast Cancer Treatment. Mol Cancer Ther. 2016. August 1;15(8):1823–33. [DOI] [PubMed] [Google Scholar]
- 11.Sahni JM, Gayle SS, Bonk KLW, Vite LC, Yori JL, Webb B, et al. Bromodomain and Extraterminal Protein Inhibition Blocks Growth of Triple-negative Breast Cancers through the Suppression of Aurora Kinases. J Biol Chem. 2016. November 4;291(45):23756–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016. January;529(7586):413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider BP, Winer EP, Foulkes WD, Garber J, Perou CM, Richardson A, et al. Triple-Negative Breast Cancer: Risk Factors to Potential Targets. Clin Cancer Res. 2008. December 15;14(24):8010–8. [DOI] [PubMed] [Google Scholar]
- 14.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001. September 11;98(19):10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdel F, Rippe K. Chromatin remodelling in mammalian cells by ISWI-type complexes - where, when and why?: ISWI chromatin remodellers in mammalian cells. FEBS J. 2011. October;278(19):3608–18. [DOI] [PubMed] [Google Scholar]
- 16.Strohner R NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001. September 3;20(17):4892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, et al. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010. July 7;29(13):2135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postepska-Igielska A, Krunic D, Schmitt N, Greulich-Bode KM, Boukamp P, Grummt I. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013. June 25;14(8):704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oppikofer M, Bai T, Gan Y, Haley B, Liu P, Sandoval W, et al. Expansion of the ISWI chromatin remodeler family with new active complexes. EMBO Rep. 2017. October;18(10):1697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minzel W, Venkatachalam A, Fink A, Hung E, Brachya G, Burstain I, et al. Small Molecules Co-targeting CKIα and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models. Cell [Internet]. 2018. August [cited 2018 Aug 29]; Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867418309735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohmann AF, Martin LJ, Minder JL, Roe J-S, Shi J, Steurer S, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol. 2016;12(9):672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinforma Oxf Engl. 2017. August 1;33(15):2413–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010. October;38(18):e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009. August 15;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013. January;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017. April;14(4):417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005. August 15;21(16):3439–40. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007. January;35(Database issue):D61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashoor H, Hérault A, Kamoun A, Radvanyi F, Bajic VB, Barillot E, et al. HMCan: a method for detecting chromatin modifications in cancer samples using ChIP-seq data. Bioinforma Oxf Engl. 2013. December 1;29(23):2979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013. April 11;153(2):320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010. May 28;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004. November 22;20(17):3246–8. [DOI] [PubMed] [Google Scholar]
- 36.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012. September 23;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res [Internet]. 2010. October [cited 2018 May 11];12(5). Available from: http://breast-cancer-research.biomedcentral.com/articles/10.1186/bcr2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model. Comput Struct Biotechnol J. 2015;13:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C, et al. The BET Bromodomain Inhibitor OTX015 Affects Pathogenetic Pathways in Preclinical B-cell Tumor Models and Synergizes with Targeted Drugs. Clin Cancer Res Off J Am Assoc Cancer Res. 2015. April 1;21(7):1628–38. [DOI] [PubMed] [Google Scholar]
- 40.Jones MH, Hamana N, Nezu J i, Shimane M. A novel family of bromodomain genes. Genomics. 2000. January 1;63(1):40–5. [DOI] [PubMed] [Google Scholar]
- 41.Chen P, Chaikuad A, Bamborough P, Bantscheff M, Bountra C, Chung C, et al. Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B. J Med Chem. 2016. February 25;59(4):1410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drouin L, McGrath S, Vidler LR, Chaikuad A, Monteiro O, Tallant C, et al. Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B. J Med Chem. 2015. March 12;58(5):2553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciceri P, Müller S, O’Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014. April;10(4):305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dittmann A, Werner T, Chung C-W, Savitski MM, Fälth Savitski M, Grandi P, et al. The Commonly Used PI3-Kinase Probe LY294002 Is an Inhibitor of BET Bromodomains. ACS Chem Biol. 2014. February 21;9(2):495–502. [DOI] [PubMed] [Google Scholar]
- 45.Ember SW, Lambert QT, Berndt N, Gunawan S, Ayaz M, Tauro M, et al. Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics. Mol Cancer Ther. 2017. June;16(6):1054–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, et al. Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation. Mol Cell. 2017. March 16;65(6):1068–1080.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comai L The nucleolus: a paradigm for cell proliferation and aging. Braz J Med Biol Res Rev Bras Pesqui Medicas E Biol. 1999. December;32(12):1473–8. [DOI] [PubMed] [Google Scholar]
- 48.Guarente L Link between aging and the nucleolus. Genes Dev. 1997. October 1;11(19):2449–55. [DOI] [PubMed] [Google Scholar]
- 49.Nishimura K, Kumazawa T, Kuroda T, Katagiri N, Tsuchiya M, Goto N, et al. Perturbation of Ribosome Biogenesis Drives Cells into Senescence through 5S RNP-Mediated p53 Activation. Cell Rep. 2015. March;10(8):1310–23. [DOI] [PubMed] [Google Scholar]
- 50.Yang L, Song T, Chen L, Soliman H, Chen J. Nucleolar repression facilitates initiation and maintenance of senescence. Cell Cycle Georget Tex. 2015;14(22):3613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ICGC Project on Early Onset Prostate Cancer, Gu L, Frommel SC, Oakes CC, Simon R, Grupp K, et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat Genet. 2015. January;47(1):22–30. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Wu W, Ding H, Li Q, Xie K. The transcription factor 7 like 2‑binding protein TIP5 activates β‑catenin/transcription factor signaling in hepatocellular carcinoma. Mol Med Rep [Internet]. 2018. March 28 [cited 2018 Jun 25]; Available from: http://www.spandidos-publications.com/10.3892/mmr.2018.8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarrete-Meneses M del P, Pérez-Vera P. Epigenetic alterations in acute lymphoblastic leukemia. Bol Méd Hosp Infant México Engl Ed. 2017. July;74(4):243–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.