Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive sarcomas typically developing in the context of neurofibromatosis type 1 (NF-1). With the exception of surgical resection, these tumors are resistant to all current therapies, and unresectable, recurrent, or metastatic tumors are considered incurable. Preclinical studies have identified several novel candidate molecular targets for therapeutic intervention, but, to date, targeted therapies have proven ineffective. Recent studies have identified recurrent mutations in polycomb repressive complex 2 (PRC2) core components, EED and SUZ12 in MPNST. These mutations result in global loss of the histone H3 lysine 27 trimethylation epigenetic mark, normally deposited by PRC2, and subsequent gain in acetylation at this residue. This altered chromatin state has been shown to promote MPNST malignancy; however, acetylation at this residue sensitizes MPNSTs to BRD4 and BET inhibition. Interestingly, the catalytic component of PRC2, enhancer of zeste homolog 2 (EZH2), is not mutated in MPNST, hinting that a non-canonical, PRC2-independent function of EZH2 may play a role in this cancer. This review examines the pathobiology of MPNST, the contribution of PRC2 subunits to this process, and the prospects for PRC2-related therapies for this cancer.
Introduction
Neurofibromatosis type 1 (NF-1) is an autosomal dominant cancer predisposition syndrome afflicting approximately one in 3,500 individuals worldwide (1), making it one of the most common genetic disorders. NF-1 patients exhibit a wide variety of symptoms, including skeletal malformities such as scoliosis (2) and tibial dysplasia (3), cognitive and behavioral impairments (4), and neoplasms that range from benign pigmented lesions to aggressive sarcomas known as malignant peripheral nerve sheath tumors (MPNSTs).
NF-1 arises through germline loss of function mutations in the neurofibromin 1 gene (NF1), first identified in 1990 (5–7). Its protein product, neurofibromin 1 (NF1), is 2,818 amino acids in length and possesses multiple functions, including Ras GTPase activating protein (GAP) activity (6,8,9), regulation of cyclic AMP levels (10–12), and microtubule binding (13). Among these roles, the Ras GAP activity is thought to be most pertinent to NF-1 associated neoplasia. NF1 promotes the hydrolysis of Ras-bound GTP to GDP (8,9), thus transitioning Ras to its inactive state. Consequently, loss of function mutations in NF1 result in hyperactive Ras signaling, promoting aberrant cellular proliferation.
All NF-1 patients are either NF1 heterozygous or mosaic for an NF1 mutation, since homozygous germline mutations are embryonically lethal (14,15). It is unclear to what degree NF1 heterozygosity itself drives aspects of this disorder. However, germline mutations in the NF1 gene predispose patients to neoplasia in accordance with the Knudson two-hit hypothesis (16). In this regard, all neurofibromas that typify NF-1 result from loss of heterozygosity of NF1.
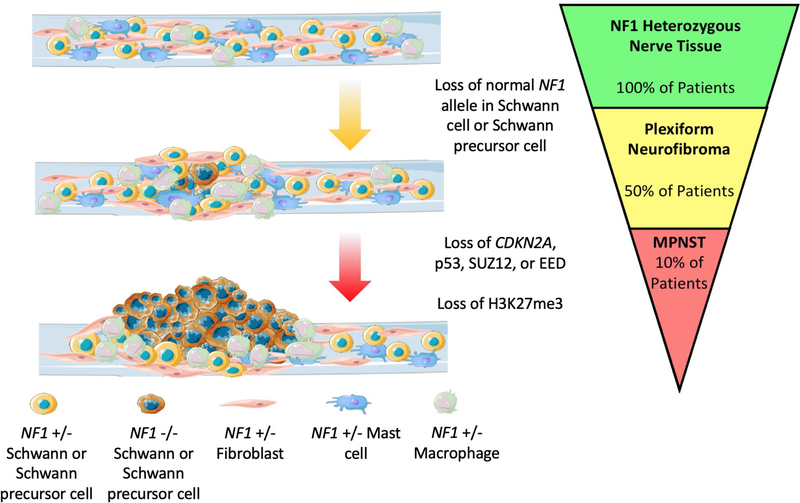
The most common NF-1-associated neoplasm is the café-au-lait macule. These are regions of hyperproliferative melanocytes that manifest clinically as areas of increased skin pigmentation (17). NF-1 patients also frequently develop dermal and cutaneous benign neurofibromas that arise from NF1 nullizygous progenitors termed skin derived precursor cells (18). Larger, more aggressive tumors, called plexiform neurofibromas (PNs) and MPNSTs, initially develop from NF1 loss of heterozygosity in Schwann precursor cells (SPCs) (19). Even the more severe skeletal malformities, like tibial dysplasia and pseudarthrosis, are associated with biallelic NF1 inactivation and aberrant osteoclast bone resorption (2,20).
Genotype-Phenotype Correlations in NF-1
While all NF1 germline loss of function mutations are fully penetrant (21,22) and result in NF-1, disease presentation is highly variable (23). Symptomatology and disease severity do not seem to correlate with any specific mutations, except in a few specific examples. The in-frame deletion c.2970–2972delAAT and missense mutations at this codon result in a relatively attenuated NF-1 phenotype (24). Patients with mutations affecting p.Arg1809 show a similar mild phenotype (25). Recently, Koczkowska et al. identified a set of missense mutations in NF1 codons 844–848 that correlate with more severe NF-1 manifestations. These patients present with a higher incidence of PNs as well as other NF-1-associated malignancies (26). NF1 microdeletion syndrome, in which the chromosomal locus 17q11.2 shows a 1.0–1.4 Mb deletion, is rare, but consistently severe clinically. These patients exhibit facial dysmorphism, scoliosis, and ADHD. They also suffer a higher risk of developing MPNSTs and other NF-1 associated neoplasms (27,28). Determining the precise genetic driver of each symptom in NF1 microdeletion syndrome is complicated by the fact that 14 protein coding genes and 4 microRNA genes are contained within the most common, 1.4 Mb deletion. Three of these genes are contained within an intron of NF1 on the antisense strand: EVI2A, EVI2B, and OMGP (7,29–31).
The difficulty in establishing genotype-phenotype correlations is in part attributable to NF1’s large size. The gene contains 60 exons and encodes a protein of 2,818 amino acids with multiple distinct functional domains (5,7). Several different NF1 splice variants are found in different tissues, and some of these variants have differential localization and function (32). The impact of various NF1 mutations on different isoforms and their respective functions is poorly understood.
Beyond its canonical RAS GAP activity, some isoforms of NF1 contain a tubulin binding domain and a nuclear localization signal. NF1 has been shown to associate with the microtubule-chromosome junction during cell division (33–36). Consistent with these observations, Koliou et al. showed that NF1 depletion in glioblastoma cells by siRNA disrupted proper chromosome congression (chromosomal alignment during metaphase) (33). This NF1 function may help to explain the frequent aneuploidy observed in NF-1-associated neoplasias (33,37–42). Interestingly, the tissues most affected in NF-1 are those which express the NF1 isoform that contains a nuclear localization signal, suggesting that nuclear NF1 functions may be particularly relevant for NF-1 associated tumorigenesis (32).
Some NF-1 patients develop symptoms in only one portion of the body, a condition termed segmental NF-1 (23,43,44). This subset of disease is caused by a de novo somatic NF1 mutation occurring early in embryonic development, rather than germline mutation. The resulting mosaicism leads to a phenotype in which only cells and tissues in the affected lineage manifest NF-1 phenotypes (44). These individuals will not pass on the mutation to their offspring unless it is present in the germline. This occurs in a small minority of patients with segmental disease.
NF-1 Associated Neoplasia
Individuals with NF-1 have a 60% lifetime risk of developing cancer (45) and are 4 times more likely to develop cancer compared to the general population (46,47) Glioblastoma, paraganglioma and pheochromocytoma, breast cancer, gastrointestinal stromal tumors, and MPNSTs all develop frequently in the context of NF-1 (46,48–51). While NF-1 patients exhibit a moderate predisposition to cancer generally, their likelihood of developing malignant neoplasms of the nervous system, such as malignant glioblastoma and MPNST, are 40-fold and 1000-fold higher, respectively than that of the general population (46). Indeed, MPNSTs develop in 8–13% of NF-1 patients (52) and represent the leading cause of death in NF-1. 50% of all MPNSTs develop in the context of NF-1, and MPNSTs constitute 10% of all malignant sarcomas overall (53). NF-1 patients are also highly predisposed to develop non-malignant tumors, such as dermal neurofibromas, PNs, and atypical neurofibromas (ANFs) (46); the latter two are precursor lesions to MPNST.
Dermal neurofibromas are benign growths that can develop in a cutaneous or subcutaneous setting. Each develops from a skin derived precursor cell that has somatically lost its functional allele of NF1 (17,18). These neurofibromas tend to be numerous and can cause itching and pain for patients. Treatment of these growths involves surgical removal or other local treatments (54). Dermal neurofibromas can also cause significant cosmetic concerns for NF-1 patients because these growths can develop on the face or on other exposed skin. Dermal neurofibromas typically develop during adolescence, exacerbating their psychological impact.
PNs, while still benign, are much larger tumors of the peripheral nerves that have the potential to cause disfigurement and can impose pressure on and disrupt the function of surrounding tissues and organs. These growths develop after loss of NF1 heterozygosity in an SPC (19) (Figure 1). Paracrine signaling within the tumor then occurs, recruiting fibroblasts (55), mast cells (55–58), macrophages (59), and Schwann cells (SCs) (58), thus creating a heterogenous mass of cells. Proliferation of neoplastic SCs is dependent upon growth factor signaling from surrounding cells (55–59); the relative contributions of cell autonomous versus paracrine signaling factors in these tumors have yet to be fully elucidated.
Figure 1. MPNST Pathogenesis.
Neoplastic growth in the nerves of an NF-1 patient is initiated after loss of the normal NF1 allele in an SPC. The ensuing paracrine signaling recruits fibroblasts, macrophages, and mast cells to the growing tumor. These tumors are termed plexiform neurofibromas (PNs), due to their heterogeneous composition. They develop in 50% of NF-1 patients. Typically, PNs only grow through adolescence. Further tumor growth in adulthood is observed to the context of ANF and MPNSTs. Upon loss of additional tumor suppressors (p53, CDKN2A, SUZ12, EED) in an NF1 null SC or SPC, SCs can transform into MPNST. This occurs in about 10% of NF-1 patients. These tumors metastasize readily and recur frequently after removal. High grade tumors are characterized by loss of H3K27me3. This figure was created in part from Servier Medical Art.
The presence of non-neoplastic cells promotes neoplastic SC growth in PN through the generation of extracellular matrix components by fibroblasts (55,60) and provision of growth factors by mast cells (55). Secretion of KIT ligand combined with aberrant expression of KIT by NF1-null SCs may create an autocrine loop promoting SC proliferation (58). SC secretion of KIT ligand also recruits mast cells to the growing tumor (57). In turn, mast cells stimulate fibroblast recruitment, proliferation, and extracellular matrix production through TGF beta secretion (55). The increasingly fibrotic environment is conducive to further tumor growth (55,60). Thus, cell-cell interactions in PN create a self-perpetuating cycle fueling tumor expansion. Notably, accumulation of mast cells in peripheral nerves occurs after nerve injury, and PN formation is anecdotally associated with prior nerve injuries to the area in which PN arise (59,61–63).
Macrophages may also play a dual role in both inhibiting and promoting neurofibroma formation. Prior to neurofibroma development, macrophages suppress tumorigenesis through secretion of tumor necrosis factor. Conversely, in established neurofibromas, depletion of macrophages by dual c-KIT and FMS kinase inhibition induced tumor regression in a genetically engineered mouse model of neurofibroma formation (59). These results, however, are difficult to disentangle from other cell-cell or autocrine interactions within neurofibromas, since many of these interactions are mediated by KIT (56–59).
PN typically arise in early childhood and grow throughout adolescence. PN growth does not generally continue into adulthood, except in the context of malignant progression. Circulating steroid hormones have been implicated in this phenomenon. There have been reports of PN growth during pregnancy followed by postpartum PN regression. These observations prompted studies demonstrating that circulating progesterone and estrogen may stimulate PN growth. While there are conflicting and inconsistent results regarding the PN cell types targeted by these hormones, a unifying finding in the literature is that high doses of progesterone may stimulate neurofibroma growth, and thus caution should be taken when administering this hormone to NF-1 patients (64–67).
In spite of the aforementioned findings, the discontinuation of PN growth in adulthood is still not completely understood. Some PNs continue to grow into adulthood, developing distinct nodular sub-lesions (68), which protrude from, or are found adjacent to PN. Such lesions are histologically dissimilar to the PN (69,70). These nodular lesions display regions of hypercellularity, possess hyperchromatic nuclei relative to the associated PN (69,70) and manifest increased FDG uptake (68). Given their atypical histology, such growths are termed atypical neurofibromas. ANF is thought to represent a premalignant stage of MPNST, through loss of the tumor suppressor p16INK4A that occurs in ANF but not PN (70,71). Among patients with ANFs, surgical resection is largely successful when possible, and most patients do not develop recurrent disease (69). However, a subset of patients with ANF experience local recurrence and/or development of MPNST following ANF resection, lending further support to the hypothesis that ANFs represent precursor lesions of MPNST.
ANF and MPNST both exhibit loss of additional tumor suppressors beyond NF1 alone. This similarity, along with their histologic likeness, can render MPNST difficult to differentiate from ANF based on histologic criteria. MPNSTs, however, require wide margins of surgical resection (72,73), readily metastasize, and exhibit a significantly higher rate of recurrence (74,75). They also impart worse prognosis, with five-year survival rates of around 45%, based on a meta-analysis of over 1,800 patients (76). In a recent study, MPNST local recurrence rates ranged from 25% to 37%, depending on tumor grade (75). Of 9 patients who were treated with amputation for MPNST, 3 developed recurrent tumors at the amputation site (75). This finding is in stark contrast to a study that showed 100% disease specific survival at 200 months after surgical resection in patients with ANF (74). Moreover, even though the majority of the tumors in this study had positive resection margin, recurrent disease was still rare (74).
MPNST is typified by the loss of additional tumor suppressor loci in an NF1 null SC (71,77,78). Deletion of p16INK4A is the most common cooperating mutation with NF1 loss, occurring in about 75% of cases (71,77,79), though TP53 mutations are also common, occurring in 40% of MPNSTs (79). Genetically engineered mouse models with mutations in NF1 and either TP53 (80,81) or CDKN2A (82,83) generate tumors resembling human MPNSTs.
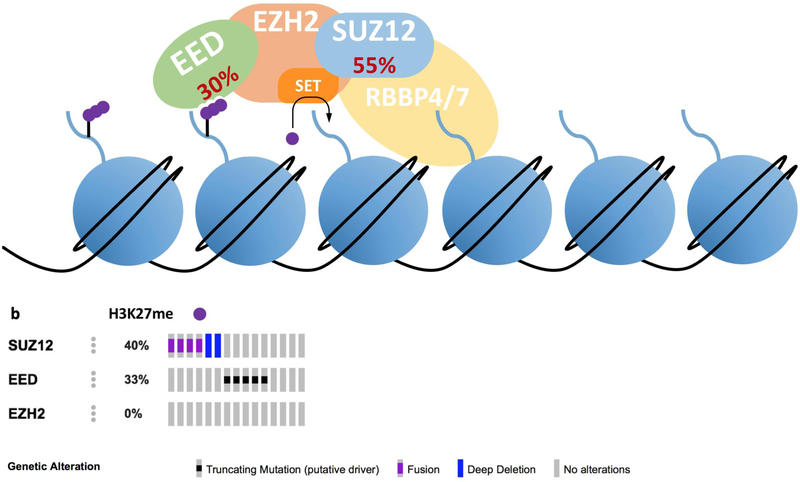
Despite compelling evidence that loss of function mutations in TP53 and CDKN2A contribute to MPNST tumorigenesis, the fact that these mutations occur in ANF and low grade MPNST means that defects in these genes cannot differentiate between non-malignant and malignant tumors. Recently, recurrent mutations in PRC2 components SUZ12 and EED have been identified in MPNST, and loss of the PRC2 product, histone H3 lysine 27 trimethylation (H3K27me3), is associated with progression from PN to MPNST (Figure 1). These mutations occur in 55% and 30% of MPNSTs respectively, and the mutations are typically mutually exclusive (p = 0.042) (Figure 2). These mutation frequencies were established in a meta-analysis of next generation sequencing studies of MPNST (79,84–87). The frequency of PRC2 mutations in MPNST has established H3K27me3 as a potentially useful biomarker to diagnose MPNST, and to distinguish this tumor from ANF and PN (84). The aforementioned clinical entities are summarized in Table 1.
Figure 2. PRC2 Function and Mutations in MPNST.
(a) PRC2 consists of four core components: EED, EZH2, SUZ12 and RBBP4/7 (158). There are also several PRC2 accessory polypeptides, not be discussed here. PRC2 trimethylates lysine 27 of histone H3 (158). EED (embryonic ectoderm development) recognizes trimethylated histone H3 lysine 27 (H3K27me3), allosterically activating the enzymatic activity of the entire complex (159,160). EZH2 is the catalytic component of PRC2, trimethylating H3K27 via its SET domain (158). SUZ12 is necessary for the catalytic activity of PRC2 and may regulate PRC2 activity through interactions with noncoding RNAs (161,162). RBBP4/7 recognizes unmodified histone H3, while active chromatin marks like H3K4me3 and H3K36me3 allosterically inhibit PRC2 activity (163). Loss of function mutation frequencies of PRC2 components in MPNST are denoted in red. (b) Publicly available data on PRC2 mutations in MPNSTs (cbioportal.org). Mutations in SUZ12 and EED occurred in 11 of 15 samples. Mutations in these two PRC2 core components are mutually exclusive (p = 0.042).
Table 1:
Clinical entities discussed in this review.
| Abbreviation | Term and Description |
|---|---|
| NF-1 | • Neurofibromatosis Type 1 • Disease results from heterozygous germline loss of function mutations in NF1 |
| NF1 | • Neurofibromin 1 • 2,818 amino acid protein • Functions include RAS GAP, cyclic AMP regulation, and microtubule binding • Tumor suppressor |
| SPC | • Schwann precursor cell • Cell type in the developmental lineage of Schwann cells • Plexiform neurofibromas, atypical neurofibromas, and malignant peripheral nerve sheath tumors arise from this lineage |
| SC | • Schwann cell • Supports peripheral nerves • NF-1 associated neurofibromas can rise from this cell type |
| PN | • Plexiform neurofibroma • Large benign neurofibroma variant • Heterogenous cellular composition • Can cause pain, disfigurement, local tissue dysfunction |
| ANF | • Atypical neurofibroma • Nonmalignant tumor • Thought to develop from plexiform neurofibromas • Has mutations in tumor suppressors in addition to NF1 |
| MPNST | • Malignant peripheral nerve sheath tumor • Malignant sarcoma with metastatic potential • Develop from nonmalignant neurofibromas in some NF-1 patients • Can arise sporadically in individuals without NF-1 |
| NF-1 Microdeletion Syndrome | • NF-1 variant • Entire NF1 gene deleted • Other genes in region are lost, including SUZ12 • More frequent and numerous plexiform neurofibromas • Higher MPNST risk |
Loss of Polycomb Repressive Complex 2 in MPNST
Gene expression is regulated in part regulated through post-translational modifications at the lysine 27 residue of histone H3 (H3K27). Acetylation at this residue (H3K27Ac) and consequent localization of bromodomain and extra-terminal domain (BET) proteins is associated with active transcription (88–91). Conversely, trimethylation at this residue (H3K27me3) compacts chromatin and represses transcription (92,93). PRC2 and KDM6A/KDM6B are respectively responsible for depositing and removing H3K27me3 (92–96). P300/CBP and the NuRD complex are responsible, respectively, for depositing and removing the acetyl mark (97,98). Together, these enzymes help to regulate transcription (99).
PRC2 consists of the core components embryonic ectoderm development protein (EED), enhancer of zeste homologue 2 (EZH2), suppressor of zeste 12 homologue (SUZ12), retinoblastoma binding protein 4/7 (RBBP4/7), and several other accessory components. PRC2 mutations and aberrant H3K27me3 levels are characteristic of several different cancers (100). PRC2 was initially thought to play a general oncogenic role since many tumors exhibit copy number gains and gain of function mutations in the catalytic subunit EZH2 (101–106). However, PRC2 is frequently inactivated in MPNST, and loss of H3K27me3 is considered a predictor of poor outcome and an oncogenic driver in other cancers as well (Figure 3). For example, Wei et al. correlated loss of H3K27me3 with poor prognosis in breast, ovarian, and pancreatic cancers (107). PRC2 mutations and loss of H3K27me3 are also observed in acute lymphoblastic leukemias and myelodysplastic syndromes (108–111). Furthermore, mutations of lysine 27 to methionine in histone H3 lead to a global decrease in H3K27me3 through PRC2 sequestration and inhibition (112), and represent a key oncogenic driver in pediatric glioblastoma (112–117). Some of these mutations occur in conjunction with NF1 mutations (115).
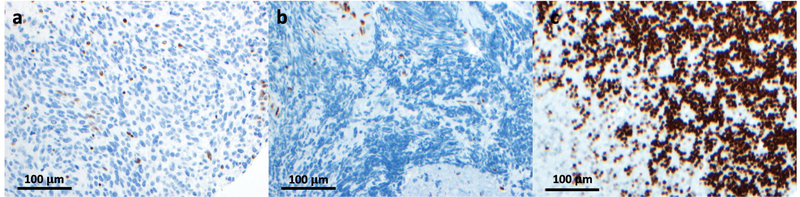
Figure 3. H3K27me3 Staining in MPNST.
MPNSTs frequently exhibit global loss of H3K27me3. (a,b) Immunohistochemical staining for H3K27me3 in MPNST tissue sections. Cells showing positive staining have been identified as inflammatory cells and endothelium (164). (c) H3K27me3 staining of granular neurons in human cerebellum, which exhibit high levels of H3K27me3, and serve as a positive control (117). Recent studies have highlighted the loss of this chromatin mark as an effective means of differentiating high grade MPNST from low grade MPNST and premalignant lesions. Images were provided by Drs. Sriram Venneti and Drew Pratt.
In MPNST, PRC2 is inactivated through loss of function mutations in SUZ12 and EED. Together, the PRC2 core components SUZ12 or EED are mutated in about 85% of MPNSTs, and these mutations are associated with more aggressive and more frequent tumors in the case of NF1 microdeletion syndrome (27,28,79). The result of these mutations is a global H3K27me3 loss (84–86). Lack of H3K27me3 allows for aberrant deposition of acetyl groups at loci normally silenced by PRC2 (84). Genetic and epigenetic aberrations in MPNST are summarized in Table 2. As previously mentioned, H3K27Ac recruits bromodomain extra-terminal domain (BET) proteins, specifically BRD4, to chromatin, which in turn promotes RNA polymerase II-mediated transcription (91). In this regard, De Raedt et al. demonstrated that SUZ12 null cell lines are more sensitive to the BRD4 inhibitor JQ1 (85). Moreover, reintroduction of ectopic SUZ12 into SUZ12 mutant MPNST cell lines was sufficient to reestablish H3K27me3 levels, deplete H3K27Ac, and reduce MPNST proliferation in cell culture (84). De Raedt et al. attributed these effects to down-regulation of a RAS transcriptional signature observed after SUZ12 reconstitution or JQ1 treatment. This paper identified a general inverse relationship between PRC2 activity and enrichment of RAS transcriptional signatures, though the precise mechanism of this interplay remains unclear (85). Patel et al. observed increased BRD4 protein level levels in mouse MPNSTs compared to their benign precursors, further supporting the notion that aberrant BRD4 expression is a pathogenic driver in MPNST (118). Alternatively, increased BRD4 protein levels in MPNST could represent a by-product of increased transcriptional demand imposed on rapidly proliferating cells.
Table 2:
Selected genes and chromatin elements discussed in this review.
| Abbreviation | Protein | Description |
|---|---|---|
| PRC2 | Polycomb repressive complex 2 | • Chromatin modifying complex responsible for laying down H3K27me3 • Contains SUZ12, EED, EZH2, and RBBP4/7 • Frequently disrupted in MPNST |
| SUZ12 | Suppressor of zeste 12 homologue | • Core component of PRC2 • Frequently mutated in MPNST • Deleted in NF-1 microdeletion syndrome |
| EED | Embryonic ectoderm development protein | • Core PRC2 component • Frequently mutated in MPNST |
| EZH2 | Enhancer of zeste homologue 2 | • Core, catalytic PRC2 component • Mutations not observed in MPNST • Increased expression in a subset of MPNSTs |
| CDKN2A | Cyclin dependent kinase inhibitor 2A | • Gene encoding tumor suppressors p16INK4A and p14ARF • p14ARF and p16INK4A transcripts transcribed in different reading frames • Often lost or silenced in MPNST |
| p16INK4A | Cyclin dependent kinase 4 inhibitor A | • A protein encoded by CDKN2A • Absent in approximately 75% of MPNSTs |
| H3K27me3 | Histone H3 lysine 27 trimethylation | • Epigenetic mark deposited by PRC2 • Typically lost along with mutations PRC2 core components |
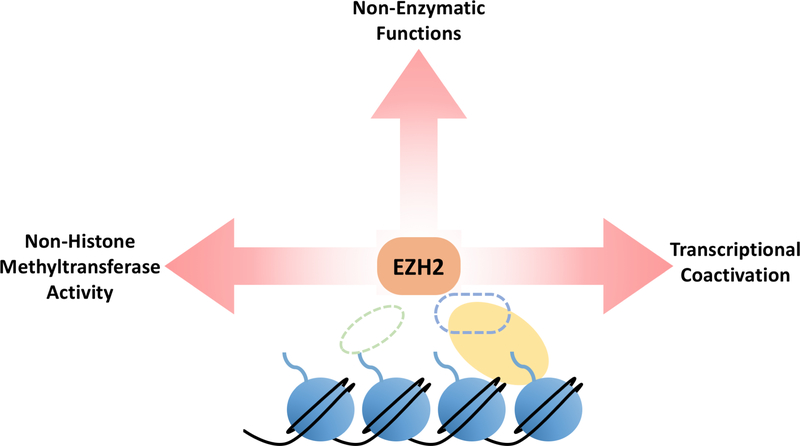
Despite the association of loss of PRC2 subunits with MPNST pathogenesis, loss of function mutations are not evenly distributed among PRC2 core components: nearly all PRC2 mutations in MPNST occur in SUZ12 and EED. By contrast, some MPNSTs exhibit overexpression of the PRC2 catalytic component, EZH2 (119). Moreover, EZH2 inhibition or depletion impair proliferation and promote apoptosis in cultured or xenografted MPNST cells (119,120). These data suggest that EZH2 may possess oncogenic functions in MPNST outside the context of PRC2, a phenomenon observed in other cancers. In many contexts, EZH2 expression negatively correlates with H3K27me3 levels but positively correlates with cancer cell proliferation and poor disease prognosis (121–123). There are also numerous examples of specific non-canonical EZH2 targets and interactions that drive tumor development and metastasis. For example, EZH2 functions as a transcriptional activator independent of its histone methyltransferase activity in breast cancer, and it can also hyperactivate Wnt signaling through interaction with PCNA associated factor and β-catenin (124–126). Yan et al. showed that overexpression of both EZH2 and catalytically inactive EZH2 conferred a growth advantage to Nasal-type Natural Killer/T-cell lymphoma cells in vitro (127). Phosphorylation of EZH2 at threonine 367 directs EZH2 to the cytoplasm and drives a metastatic phenotype in breast cancer mediated by EZH2 interactions with the cytoskeleton (128,129). In prostate cancer and glioblastoma, phosphorylation of EZH2 at serine 21 by AKT causes EZH2 to methylate the androgen receptor and STAT3 to drive disease progression. In prostate cancer, this occurs independently of other PRC2 components (130,131). Although the two aforementioned roles for EZH2 are not directly related to its canonical activity in PRC2, phosphorylation of EZH2 at this residue by AKT can also reduce H3K27me3 levels by diminishing EZH2’s affinity for histone H3 (132). Altogether, these data imply potential non-canonical roles for EZH2 in MPNST pathogenesis. Loss of other PRC2 core components could result in higher levels of unbound EZH2 that could promiscuously interact with other binding partners to potentiate disease progression.
To date, studies of PRC2 in MPNST have been unable to elucidate the precise mechanisms through which loss of SUZ12 and EED promotes MPNST malignancy. Two models through which PRC2 component loss contributes to MPNST pathogenesis emerge: PRC2 loss of function results in loss of the H3K27me3 mark and derepression of PRC2 target genes; and loss of SUZ12 or EED result in increased levels of unbound EZH2 that could participate in other, as-yet undefined oncogenic activities (Figure 4). These models are not mutually exclusive and could both function in MPNST. Additional work is needed to understand the precise mechanisms through which loss of SUZ12 and EED drive MPNST behavior.
Figure 4. Known Non-Canonical Roles of EZH2.
While SUZ12 or EED are mutated in approximately 85% of MPNSTs, EZH2 loss of function has not been observed. Overexpression of EZH2 has been reported in MPNSTs. EZH2 inhibition and depletion have also proven efficacious in slowing MPNST growth in preclinical models despite PRC2 already being genetically inactivated. This suggests that EZH2 may assume pathogenic functions in MPNST in the absence of other PRC2 core components. Non-canonical, pathogenic roles for EZH2 have been demonstrated in numerous other contexts. For example, overexpression of catalytically inactive EZH2 in natural killer/T-cell lymphoma conferred a growth advantage to these cells. Phosphorylated EZH2 has been shown to contribute to glioblastoma, breast cancer, and prostate cancer tumorigenicity through divergent mechanisms. The efficacy of EZH2 inhibition and depletion in slowing MPNST growth in preclinical models, as well as the clearly established non-canonical roles of EZH2 in other contexts represent an understudied avenue for potential MPNST therapies.
Current and Future Therapies for MPNST
Currently, the only effective therapy for MPNST is complete surgical resection to achieve negative margins (72,73). MPNSTs rapidly develop resistance to chemotherapy, and there is little data to indicate that such treatments improve patient outcome when employed in combination with surgery. Ill-defined margins in MPNST are a barrier to successful surgical resection (133,134), as tumor location and/or metastatic disease frequently are. There are currently no effective treatment options for patients with recurrent or metastatic disease or inoperable tumors. These individuals are encouraged to enroll in clinical trials (73).
Efforts to target MPNST pharmacologically have been met with little success to date. Two phase II clinical trials aimed at treating MPNST via tyrosine kinase (TK) inhibition have proven unsuccessful in recent years. In one trial, epidermal growth factor receptor inhibition with erlotinib failed to induce any clinical responses (135) despite encouraging preclinical data supporting this intervention. Another TK inhibitor, sorafenib, with activity against vascular endothelial growth factor receptor and platelet derived growth factor receptor, was tested in combination with the standard chemotherapy agent dacarbazine, also with limited success (136). A recent publication described a novel therapeutic strategy, in which YAP/TAZ signaling and PDGFR were targeted simultaneously to inhibit MPNST growth (137). The only clinical trial to date that has achieved promising results deployed doxorubicin, etoposide, and ifosfamide against chemotherapy-naive MPNSTs (138), with many patients exhibiting stable disease, and some even achieving partial responses.
Current clinical trials are largely focused on testing targeted therapies that have shown efficacy in other sarcomas. Many of these trials are evaluating mTOR inhibitors, TK inhibitors, or combination of these treatments. There is preclinical evidence suggesting that such interventions could be effective in MPNST (139–147), but such data has been poorly predictive of success against MPNST in the past. A candidate therapeutic approach that has been of great recent interest is the application of BET/BRD4 inhibitors. As previously noted, PRC2 core components are frequently mutated in MPNST, preventing PRC2-mediated deposition of the H3K27me3 repressive mark on chromatin, with a concomitant gain in acetylation at this site. Unfortunately, a phase II clinical trial involving the BET inhibitor CPI-0610 in MPNST was recently withdrawn due poor enrollment.
Encouragingly, there has been some success in targeted therapy for inoperable PN. A phase I clinical trial of the MEK inhibitor selumetinib achieved partial responses in 17/24 patients, and all patients experienced some decrease in tumor volume. Responses were maintained in 15/17 patients, with no patients exhibiting progressive disease (148). This trial is an important step forward in NF-1 treatment, since prevention of MPNST development may represent an effective means of reducing MPNST-associated mortality in NF-1. Selumetinib is currently in phase II clinical trials for PN.
Major therapeutic progress has been made in treatment of cutaneous melanoma and other cancers using immune checkpoint inhibitors. For example, interaction between programmed cell death protein 1 (PD-1) and its ligand, PD-L1, expressed by many neoplastic cells, leads to suppression of anti-tumoral T-cell immune responses. Blockade of these receptors or other immune checkpoint mediators (e.g. CTLA-4) can reactivate this response. Two recently published studies have attempted to characterize the MPNST immune microenvironment. One of these studies found that MPNSTs exhibit slightly increased PD-L1 expression compared to benign nerve tissue, significant CD8+ staining, and no PD-1 expression. It found no correlation between PD-L1 or CD8 staining and disease state or patient survival (149). It described the majority of tumor samples as non-inflamed, characterized by low neoantigen levels and limited response to PD pathway blockade (149,150). Haworth et al. report no difference in PD-L1 expression between benign and malignant NF-1 lesions (151). Both studies hypothesized that immunotherapy would only have limited utility in MPNST treatment, though inconsistencies between their findings underscores the need for further research in this area.
Considerations for Future Research
Many groups have developed genetically engineered mouse models of MPNST, which develop neoplasms that are histologically similar to human MPNST (152). Mo et al. (153) and Chau et al. (154) described a mouse model in which Nf1 null skin derived neurofibroma precursor cells identified by Le et al., 2009 (18) gave rise to PNs when orthotopically implanted in a nerve. When Nf1 and Tp53 null skin derived precursor cells were implanted into a nerve, they produced MPNSTs. Such models provide isogenic systems to examine the transition from benign PN to MPNST. Recently, Wu et al. developed a genetically engineered mouse model, in which Lats½ deficient mice rapidly develop tumors resembling MPNSTs (137). Additionally, Li et al. developed a series of immortalized SC lines from healthy individuals and from NF-1 patients. Furthermore, NF1 null and heterozygous immortalized cell lines were established from the same individual, providing isogenic cells for study (155). The availability of immortalized SC lines will expedite the process of understanding genetic and epigenetic aberrations that result from loss of NF1, and those that occur during MPNST evolution. This system will also aid in understanding MPNST pathogenesis by providing a platform on which to perform genetic manipulations of genes frequently mutated in MPNST. Crucially, potential interventions must be considered within the context of an NF1 heterozygous individual, since therapies that target hyperactive Ras signaling could exert deleterious effects in individuals with germline NF1 haploinsufficiency. NF1 heterozygous immortalized SC lines will be helpful in studying this narrowed therapeutic window (155).
Despite recent advances in understanding MPNSTs, progress is still hindered by lack of comprehensive genetic data on many MPNST cell lines and lack of robust, reliable NF1 antibodies. Additional steps should be taken toward establishing large databases of patient NF1 genotypes and outcomes in order to gain additional, detailed insights into genotype-phenotype correlations and how these might be leveraged to allow more effective MPNST treatment. Two patient registries have been established in recent years; more information is needed to understand the variability in NF-1 presentation (156,157).
While MPNST treatment is a critical area for study, the importance of potentially preventing MPNST altogether in NF-1 patients should not be overlooked. Currently available data indicate that premalignant NF-1 associated neoplasms likely respond better to pharmacological intervention than MPNSTs, and they more amenable to surgical cure. Drugs to treat PNs, and delay or even prevent progression to MPNST, may prove to be valuable therapies for individuals with NF-1. The MEK inhibitor selumetinib has showed promise in this context (148).
The outlook for patients with MPNST still remains guarded, but new findings regarding epigenetic aberrations in these tumors have provided a foundation on which new clinical trials may be built. Future studies should also elucidate in detail how loss of PRC2 components EED and SUZ12 contribute to malignancy, especially in the context of recent studies identifying EZH2 as a potential therapeutic target.
Implications.
Identification of mutations in the PRC2 components EED and SUZ12 in the majority of MPNSTs may imply noncanonical oncogenic activities of the intact component, EZH2, and provide new opportunities for therapeutic intervention.
Acknowledgements
The authors apologize to investigators whose work was not cited due to space constraints. Supported by awards from DoD (NF170044) and NIH (R01GM101171 and R01HL114858). Figure 1 was produced in part using Servier Medical Art and Figure 2 was produced in part from data and images available through cbioportal.org. The authors are grateful to Drs. Rajesh Rao and Sriram Venneti for helpful comments on the manuscript, and to Drs. Venneti and Drew Pratt for the images in Figure 3.
Footnotes
Conflict of interest statement: D.B.L. has stock interest in ABBV, GILD, ILMN, INFI, JNJ, LCI, and TGTX. J.L.K. has no relevant financial interests.
References
- 1.Uusitalo E, Leppävirta J, Koffert A, Suominen S, Vahtera J, Vahlberg T, et al. Incidence and Mortality of Neurofibromatosis: A Total Population Study in Finland. Journal of Investigative Dermatology 2015;135(3):904–6 doi 10.1038/jid.2014.465. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Nyman JS, Ono K, Stevenson DA, Yang X, Elefteriou F. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Human molecular genetics 2011;20(20):3910–24 doi 10.1093/hmg/ddr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson DA, Birch PH, Friedman JM, Viskochil DH, Balestrazzi P, Boni S, et al. Descriptive analysis of tibial pseudarthrosis in patients with neurofibromatosis 1. American Journal of Medical Genetics 1999;84(5):413–9 doi . [DOI] [PubMed] [Google Scholar]
- 4.Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology 2005;65(7):1037 doi 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- 5.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 1990;249(4965):181 doi 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 6.Gutmann DH, Wood DL, Collins FS. Identification of the neurofibromatosis type 1 gene product. Proceedings of the National Academy of Sciences 1991;88(21):9658 doi 10.1073/pnas.88.21.9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cawthon RM, Weiss R, Xu G, Viskochil D, Culver M, Stevens J, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 1990;62(1):193–201 doi 10.1016/0092-8674(90)90253-B. [DOI] [PubMed] [Google Scholar]
- 8.Martin GA, Viskoohil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 1990;63(4):843–9 doi 10.1016/0092-8674(90)90150-D. [DOI] [PubMed] [Google Scholar]
- 9.Xu G, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, et al. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 1990;63(4):835–41 doi 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 10.Anastasaki C, Gutmann DH. Neuronal NF1/RAS regulation of cyclic AMP requires atypical PKC activation. Human Molecular Genetics 2014;23(25):6712–21 doi 10.1093/hmg/ddu389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein–stimulated adenylyl cyclase activity. Nature Neuroscience 2002;5:95 doi 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- 12.Guo H-F, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 2000;403:895 doi 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- 13.Bollag G, McCormick F, Clark R. Characterization of full-length neurofibromin: tubulin inhibits Ras GAP activity. The EMBO journal 1993;12(5):1923–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes & Development 1994;8(9):1019–29 doi 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 15.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nature Genetics 1994;7(3):353–61 doi 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 16.Knudson AG Jr., Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America 1971;68(4):820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Schepper S, Maertens O, Callens T, Naeyaert J-M, Lambert J, Messiaen L. Somatic Mutation Analysis in NF1 Café au lait Spots Reveals Two NF1 Hits in the Melanocytes. Journal of Investigative Dermatology 2008;128(4):1050–3 doi 10.1038/sj.jid.5701095. [DOI] [PubMed] [Google Scholar]
- 18.Le LQ, Shipman T, Burns DK, Parada LF. Cell of origin and microenvironment contribution for NF1-associated dermal neurofibromas. Cell stem cell 2009;4(5):453–63 doi 10.1016/j.stem.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Liu C, Patel AJ, Liao C-P, Wang Y, Le LQ. Cells of origin in the embryonic nerve roots for NF1-associated plexiform neurofibroma. Cancer cell 2014;26(5):695–706 doi 10.1016/j.ccell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson DA, Zhou H, Ashrafi S, Messiaen LM, Carey JC, D’Astous JL, et al. Double inactivation of NF1 in tibial pseudarthrosis. American journal of human genetics 2006;79(1):143–8 doi 10.1086/504441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riccardi VM, Lewis RA. Penetrance of von Recklinghausen neurofibromatosis: a distinction between predecessors and descendants. American journal of human genetics 1988;42(2):284–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Huson SM, Compston DA, Clark P, Harper PS. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. I. Prevalence, fitness, mutation rate, and effect of parental transmission on severity. Journal of medical genetics 1989;26(11):704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riccardi VM. Neurofibromatosis: Clinical heterogeneity. Current Problems in Cancer 1982;7(2):1–34 doi 10.1016/S0147-0272(82)80016-0. [DOI] [PubMed] [Google Scholar]
- 24.Upadhyaya M, Huson SM, Davies M, Thomas N, Chuzhanova N, Giovannini S, et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970–2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. American journal of human genetics 2007;80(1):140–51 doi 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojnueangnit K, Xie J, Gomes A, Sharp A, Callens T, Chen Y, et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype-Phenotype Correlation. Human mutation 2015;36(11):1052–63 doi 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koczkowska M, Chen Y, Callens T, Gomes A, Sharp A, Johnson S, et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844–848. American journal of human genetics 2018;102(1):69–87 doi 10.1016/j.ajhg.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kayes LM, Burke W, Riccardi VM, Bennett R, Ehrlich P, Rubenstein A, et al. Deletions spanning the neurofibromatosis 1 gene: identification and phenotype of five patients. American journal of human genetics 1994;54(3):424–36. [PMC free article] [PubMed] [Google Scholar]
- 28.Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, et al. Deletions spanning the neurofibromatosis type 1 gene: Implications for genotype-phenotype correlations in neurofibromatosis type 1? Human Mutation 1997;9(5):458–64 doi . [DOI] [PubMed] [Google Scholar]
- 29.O’Connell P, Viskochil D, Buchberg AM, Fountain J, Cawthon RM, Culver M, et al. The human homolog of murine Evi-2 lies between two von Recklinghausen neurofibromatosis translocations. Genomics 1990;7(4):547–54. [DOI] [PubMed] [Google Scholar]
- 30.Viskochil D, Cawthon R, O’Connell P, Xu GF, Stevens J, Culver M, et al. The gene encoding the oligodendrocyte-myelin glycoprotein is embedded within the neurofibromatosis type 1 gene. Molecular and cellular biology 1991;11(2):906–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cawthon RM, Andersen LB, Buchberg AM, Xu GF, O’Connell P, Viskochil D, et al. cDNA sequence and genomic structure of EV12B, a gene lying within an intron of the neurofibromatosis type 1 gene. Genomics 1991;9(3):446–60. [DOI] [PubMed] [Google Scholar]
- 32.Vandenbroucke I, Vandesompele J, De Paepe A, Messiaen L. Quantification of NF1 transcripts reveals novel highly expressed splice variants. FEBS Letters 2002;522(1–3):71–6 doi 10.1016/S0014-5793(02)02887-9. [DOI] [PubMed] [Google Scholar]
- 33.Koliou X, Fedonidis C, Kalpachidou T, Mangoura D. Nuclear import mechanism of neurofibromin for localization on the spindle and function in chromosome congression. Journal of Neurochemistry 2016;136(1):78–91 doi 10.1111/jnc.13401. [DOI] [PubMed] [Google Scholar]
- 34.Gregory PE, Gutmann DH, Mitchell A, Park S, Boguski M, Jacks T, et al. Neurofibromatosis type 1 gene product (neurofibromin) associates with microtubules. Somat Cell Mol Genet 1993;19(3):265–74. [DOI] [PubMed] [Google Scholar]
- 35.Li C, Cheng Y, Gutmann DA, Mangoura D. Differential localization of the neurofibromatosis 1 (NF1) gene product, neurofibromin, with the F-actin or microtubule cytoskeleton during differentiation of telencephalic neurons. Developmental Brain Research 2001;130(2):231–48 doi 10.1016/S0165-3806(01)00190-0. [DOI] [PubMed] [Google Scholar]
- 36.Ozawa T, Araki N, Yunoue S, Tokuo H, Feng L, Patrakitkomjorn S, et al. The Neurofibromatosis Type 1 Gene Product Neurofibromin Enhances Cell Motility by Regulating Actin Filament Dynamics via the Rho-ROCK-LIMK2-Cofilin Pathway. Journal of Biological Chemistry 2005;280(47):39524–33 doi 10.1074/jbc.M503707200. [DOI] [PubMed] [Google Scholar]
- 37.Fang Y, Elahi A, Denley RC, Rao PH, Brennan MF, Jhanwar SC. Molecular Characterization of Permanent Cell Lines from Primary, Metastatic and Recurrent Malignant Peripheral Nerve Sheath Tumors (MPNST) with Underlying Neurofibromatosis-1. Anticancer Research 2009;29(4):1255–62. [PubMed] [Google Scholar]
- 38.Krone W, Högemann I. Cell culture studies on neurofibromatosis (von Recklinghausen). Human Genetics 1986;74(4):453–5 doi 10.1007/BF00280506. [DOI] [PubMed] [Google Scholar]
- 39.Mechtersheimer G, Otaño-Joos M, Ohl S, Benner A, Lehnert T, Willeke F, et al. Analysis of chromosomal imbalances in sporadic and NF1-associated peripheral nerve sheath tumors by comparative genomic hybridization. Genes, Chromosomes and Cancer 1999;25(4):362–9 doi . [DOI] [PubMed] [Google Scholar]
- 40.Plaat BEC, Molenaar WM, Mastik MF, Hoekstra HJ, te Meerman GJ, van den Berg E. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: Sporadic Vs. neurofibromatosis-type-1-associated malignant schwannomas. International Journal of Cancer 1999;83(2):171–8 doi . [DOI] [PubMed] [Google Scholar]
- 41.Schmidt H, Taubert H, Würl P, Bache M, Bartel F, Holzhausen H-J, et al. Cytogenetic characterization of six malignant peripheral nerve sheath tumors: comparison of karyotyping and comparative genomic hybridization. Cancer Genetics and Cytogenetics 2001;128(1):14–23 doi 10.1016/S0165-4608(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt H, Taubert H, Meye A, Würl P, Bache M, Bartel F, et al. Gains in chromosomes 7, 8q, 15q and 17q are characteristic changes in malignant but not in benign peripheral nerve sheath tumors from patients with Recklinghausen’s disease. Cancer Letters 2000;155(2):181–90 doi 10.1016/S0304-3835(00)00426-2. [DOI] [PubMed] [Google Scholar]
- 43.Miller RM, Sparkes RS. Segmental Neurofibromatosis. Archives of Dermatology 1977;113(6):837–8 doi 10.1001/archderm.1977.01640060133020. [DOI] [PubMed] [Google Scholar]
- 44.Paine RS. A clinical, pathological and genetic study of multiple neurofibromatosis. American Journal of Human Genetics 1956;8(3):190–1. [Google Scholar]
- 45.Uusitalo E, Rantanen M, Kallionpää RA, Pöyhönen M, Leppävirta J, Ylä-Outinen H, et al. Distinctive Cancer Associations in Patients With Neurofibromatosis Type 1. Journal of Clinical Oncology 2016;34(17):1978–86 doi 10.1200/JCO.2015.65.3576. [DOI] [PubMed] [Google Scholar]
- 46.Seminog OO, Goldacre MJ. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. British journal of cancer 2013;108(1):193–8 doi 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zöller MET, Rembeck B, Odén A, Samuelsson M, Angervall L. Malignant and benign tumors in patients with neurofibromatosis type 1 in a defined Swedish population. Cancer 1997;79(11):2125–31 doi . [DOI] [PubMed] [Google Scholar]
- 48.Zinnamosca L, Petramala L, Cotesta D, Marinelli C, Schina M, Cianci R, et al. Neurofibromatosis type 1 (NF1) and pheochromocytoma: prevalence, clinical and cardiovascular aspects. Archives of Dermatological Research 2011;303(5):317–25 doi 10.1007/s00403-010-1090-z. [DOI] [PubMed] [Google Scholar]
- 49.Stiller CA, Chessells JM, Fitchett M. Neurofibromatosis and childhood leukaemia/lymphoma: a population-based UKCCSG study. British journal of cancer 1994;70(5):969–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharif S, Moran A, Huson SM, Iddenden R, Shenton A, Howard E, et al. Women with neurofibromatosis 1 are at a moderately increased risk of developing breast cancer and should be considered for early screening. Journal of medical genetics 2007;44(8):481–4 doi 10.1136/jmg.2007.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlenterie M, Flucke U, Hofbauer LC, Timmers HJLM, Gastmeier J, Aust DE, et al. Pheochromocytoma and Gastrointestinal Stromal Tumors in Patients With Neurofibromatosis Type I. The American Journal of Medicine 2013;126(2):174–80 doi 10.1016/j.amjmed.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Evans DGR, Baser ME, McGaughran J, Sharif S, Howard E, Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. Journal of medical genetics 2002;39(5):311–4 doi 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss SW, Goldblum JR. Enzinger and Weiss’ soft tissue tumors [electronic resource] / Weiss Sharon W., Goldblum John R. St. Louis Mo.; London: :: Mosby Elsevier Science & Technology; 2007. [Google Scholar]
- 54.Ferner RE, Huson SM, Thomas N, Moss C, Willshaw H, Evans DG, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. Journal of medical genetics 2007;44(2):81–8 doi 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang F-C, Chen S, Clegg T, Li X, Morgan T, Estwick SA, et al. Nf1+/− mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. Human molecular genetics 2006;15(16):2421–37 doi 10.1093/hmg/ddl165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang F-C, Ingram DA, Chen S, Zhu Y, Yuan J, Li X, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell 2008;135(3):437–48 doi 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F-C, Ingram DA, Chen S, Hingtgen CM, Ratner N, Monk KR, et al. Neurofibromin-deficient Schwann cells secrete a potent migratory stimulus for Nf1+/− mast cells. The Journal of clinical investigation 2003;112(12):1851–61 doi 10.1172/JCI19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan JJ, Klein KA, Neuberger TJ, Leftwich JA, Westin EH, Kauma S, et al. Role for the stem cell factor/KIT complex in schwann cell neoplasia and mast cell proliferation associated with neurofibromatosis. Journal of Neuroscience Research 1994;37(3):415–32 doi 10.1002/jnr.490370314. [DOI] [PubMed] [Google Scholar]
- 59.Prada CE, Jousma E, Rizvi TA, Wu J, Dunn RS, Mayes DA, et al. Neurofibroma-associated macrophages play roles in tumor growth and response to pharmacological inhibition. Acta neuropathologica 2013;125(1):159–68 doi 10.1007/s00401-012-1056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaakkola S, Peltonen J, Riccardi V, Chu ML, Uitto J. Type 1 neurofibromatosis: selective expression of extracellular matrix genes by Schwann cells, perineurial cells, and fibroblasts in mixed cultures. The Journal of clinical investigation 1989;84(1):253–61 doi 10.1172/JCI114148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cichowski K, Jacks T. NF1 Tumor Suppressor Gene Function: Narrowing the GAP. Cell 2001;104(4):593–604 doi 10.1016/S0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 62.Olsson Y MAST CELLS IN HUMAN PERIPHERAL NERVE. Acta Neurologica Scandinavica 1971;47(3):357–68 doi 10.1111/j.1600-0404.1971.tb07490.x. [DOI] [PubMed] [Google Scholar]
- 63.Rizvi TA, Huang Y, Sidani A, Atit R, Largaespada DA, Boissy RE, et al. A novel cytokine pathway suppresses glial cell melanogenesis after injury to adult nerve. The Journal of neuroscience : the official journal of the Society for Neuroscience 2002;22(22):9831–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLaughlin ME, Jacks T. Progesterone Receptor Expression in Neurofibromas. Cancer Research 2003;63(4):752. [PubMed] [Google Scholar]
- 65.Fishbein L, Zhang X, Fisher LB, Li H, Campbell-Thompson M, Yachnis A, et al. In vitro studies of steroid hormones in neurofibromatosis 1 tumors and schwann cells. Molecular Carcinogenesis 2007;46(7):512–23 doi 10.1002/mc.20236. [DOI] [PubMed] [Google Scholar]
- 66.Overdiek A, Winner U, Mayatepek E, Rosenbaum T. Schwann Cells From Human Neurofibromas Show Increased Proliferation Rates Under the Influence of Progesterone. Pediatric Research 2008;64:40 doi 10.1203/PDR.0b013e31817445b8. [DOI] [PubMed] [Google Scholar]
- 67.Lammert M, Mautner V-F, Kluwe L. Do hormonal contraceptives stimulate growth of neurofibromas? A survey on 59 NF1 patients. BMC Cancer 2005;5(1):16 doi 10.1186/1471-2407-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meany H, Dombi E, Reynolds J, Whatley M, Kurwa A, Tsokos M, et al. 18-fluorodeoxyglucose-positron emission tomography (FDG-PET) evaluation of nodular lesions in patients with neurofibromatosis type 1 and plexiform neurofibromas (PN) or malignant peripheral nerve sheath tumors (MPNST). Pediatric Blood & Cancer 2013;60(1):59–64 doi 10.1002/pbc.24212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baldwin A, Dombi E, Widemann BC, Higham CS, Bhaumik S, Rogiers A, et al. The characteristics of 76 atypical neurofibromas as precursors to neurofibromatosis 1 associated malignant peripheral nerve sheath tumors. Neuro-Oncology 2018;20(6):818–25 doi 10.1093/neuonc/noy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beert E, Brems H, Daniëls B, De Wever I, Van Calenbergh F, Schoenaers J, et al. Atypical neurofibromas in neurofibromatosis type 1 are premalignant tumors. Genes, Chromosomes and Cancer 2011;50(12):1021–32 doi 10.1002/gcc.20921. [DOI] [PubMed] [Google Scholar]
- 71.Kourea HP, Orlow I, Scheithauer BW, Cordon-Cardo C, Woodruff JM. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. The American journal of pathology 1999;155(6):1855–60 doi 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferner RE, Gutmann DH. International Consensus Statement on Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis 1. Cancer Research 2002;62(5):1573. [PubMed] [Google Scholar]
- 73.Bradford D, Kim A. Current Treatment Options for Malignant Peripheral Nerve Sheath Tumors. Current Treatment Options in Oncology 2015;16(3):12 doi 10.1007/s11864-015-0328-6. [DOI] [PubMed] [Google Scholar]
- 74.Bernthal NM, Putnam A, Jones KB, Viskochil D, Randall RL. The effect of surgical margins on outcomes for low grade MPNSTs and atypical neurofibroma. Journal of Surgical Oncology 2014;110(7):813–6 doi 10.1002/jso.23736. [DOI] [PubMed] [Google Scholar]
- 75.Watson KL, Al Sannaa GA, Kivlin CM, Ingram DR, Landers SM, Roland CL, et al. Patterns of recurrence and survival in sporadic, neurofibromatosis Type 1-associated, and radiation-associated malignant peripheral nerve sheath tumors. Journal of neurosurgery 2017;126(1):319–29 doi 10.3171/2015.12.JNS152443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolberg M, Høland M, Agesen TH, Brekke HR, Liestøl K, Hall KS, et al. Survival meta-analyses for >1800 malignant peripheral nerve sheath tumor patients with and without neurofibromatosis type 1. Neuro-oncology 2013;15(2):135–47 doi 10.1093/neuonc/nos287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nielsen GP, Stemmer-Rachamimov AO, Ino Y, Moller MB, Rosenberg AE, Louis DN. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. The American journal of pathology 1999;155(6):1879–84 doi 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahrmann EP, Moriarity BS, Otto GM, Watson AL, Choi K, Collins MH, et al. Trp53 haploinsufficiency modifies EGFR-driven peripheral nerve sheath tumorigenesis. The American journal of pathology 2014;184(7):2082–98 doi 10.1016/j.ajpath.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brohl AS, Kahen E, Yoder SJ, Teer JK, Reed DR. The genomic landscape of malignant peripheral nerve sheath tumors: diverse drivers of Ras pathway activation. Scientific Reports 2017;7(1):14992 doi 10.1038/s41598-017-15183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse Models of Tumor Development in Neurofibromatosis Type 1. Science 1999;286(5447):2172 doi 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 81.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science (New York, NY) 1999;286(5447):2176–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joseph NM, Mosher JT, Buchstaller J, Snider P, McKeever PE, Lim M, et al. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer cell 2008;13(2):129–40 doi 10.1016/j.ccr.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dodd RD, Mito JK, Eward WC, Chitalia R, Sachdeva M, Ma Y, et al. NF1 deletion generates multiple subtypes of soft-tissue sarcoma that respond to MEK inhibition. Molecular cancer therapeutics 2013;12(9):1906–17 doi 10.1158/1535-7163.MCT-13-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nature Genetics 2014;46:1227 doi 10.1038/ng.309510.1038/ng.3095https://www.nature.com/articles/ng.3095#supplementary-informationhttps://www.nature.com/articles/ng.3095#supplementary-information . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014;514:247 doi 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 86.Zhang M, Wang Y, Jones S, Sausen M, McMahon K, Sharma R, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nature Genetics 2014;46:1170 doi 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sohier P, Luscan A, Lloyd A, Ashelford K, Laurendeau I, Briand-Suleau A, et al. Confirmation of mutation landscape of NF1-associated malignant peripheral nerve sheath tumors. Genes, Chromosomes and Cancer 2017;56(5):421–6 doi 10.1002/gcc.22446. [DOI] [PubMed] [Google Scholar]
- 88.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences 2003;100(15):8758 doi 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roe J-S, Mercan F, Rivera K, Pappin Darryl J, Vakoc Christopher R BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Molecular Cell 2015;58(6):1028–39 doi 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJT, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. The Journal of biological chemistry 2012;287(51):43137–55 doi 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Molecular Cell 2005;19(4):523–34 doi 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 92.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002;298(5595):1039 doi 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 93.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Reviews Genetics 2007;8:9 doi 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 94.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proceedings of the National Academy of Sciences of the United States of America 2007;104(47):18439–44 doi 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee MG, Villa R, Trojer P, Norman J, Yan K-P, Reinberg D, et al. Demethylation of H3K27 Regulates Polycomb Recruitment and H2A Ubiquitination. Science 2007;318(5849):447 doi 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 96.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449:731 doi 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 97.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development (Cambridge, England) 2009;136(18):3131–41 doi 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. The EMBO journal 2012;31(3):593–605 doi 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic acids research 2010;38(15):4958–69 doi 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Current Opinion in Cell Biology 2015;37:42–8 doi 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 101.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624 doi 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 102.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America 2003;100(20):11606–11 doi 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal 2003;22(20):5323–35 doi 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics 2010;42(2):181–5 doi 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proceedings of the National Academy of Sciences of the United States of America 2012;109(8):2989–94 doi 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Letters 2012;586(19):3448–51 doi 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 107.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Molecular carcinogenesis 2008;47(9):701–6 doi 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine 2012;18(2):298–301 doi 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481(7380):157–63 doi 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genetics 2010;42:722 doi 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 111.Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nature Communications 2014;5:4177 doi 10.1038/ncomms5177. [DOI] [PubMed] [Google Scholar]
- 112.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science (New York, NY) 2013;340(6134):857–61 doi 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones David TW, Konermann C, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 2012;22(4):425–37 doi 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 114.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics 2012;44(3):251–3 doi 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226 doi 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 116.Khuong-Quang D-A, Buczkowicz P, Rakopoulos P, Liu X-Y, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica 2012;124(3):439–47 doi 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, et al. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain pathology (Zurich, Switzerland) 2013;23(5):558–64 doi 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Patel AJ, Liao C-P, Chen Z, Liu C, Wang Y, Le LQ. BET bromodomain inhibition triggers apoptosis of NF1-associated malignant peripheral nerve sheath tumors through Bim induction. Cell reports 2014;6(1):81–92 doi 10.1016/j.celrep.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang P, Garnett J, Creighton CJ, Al Sannaa GA, Igram DR, Lazar A, et al. EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve sheath tumour cell survival and tumourigenesis. The Journal of pathology 2014;232(3):308–18 doi 10.1002/path.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang P, Yang X, Ma X, Ingram DR, Lazar AJ, Torres KE, et al. Antitumor effects of pharmacological EZH2 inhibition on malignant peripheral nerve sheath tumor through the miR-30a and KPNB1 pathway. Molecular cancer 2015;14:55- doi 10.1186/s12943-015-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rao RC, Chan MP, Andrews CA, Kahana A. Epigenetic markers in basal cell carcinoma: universal themes in oncogenesis and tumor stratification? - a short report. Cellular Oncology 2018;41(6):693–8 doi 10.1007/s13402-018-0402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rao RC, Chan MP, Andrews CA, Kahana A. EZH2, Proliferation Rate, and Aggressive Tumor Subtypes in Cutaneous Basal Cell Carcinoma. JAMA oncology 2016;2(7):962–3 doi 10.1001/jamaoncol.2016.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Holm K, Grabau D, Lövgren K, Aradottir S, Gruvberger-Saal S, Howlin J, et al. Global H3K27 trimethylation and EZH2 abundance in breast tumor subtypes. Molecular oncology 2012;6(5):494–506 doi 10.1016/j.molonc.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Molecular and cellular biology 2007;27(14):5105–19 doi 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee Shuet T, Li Z, Wu Z, Aau M, Guan P, Karuturi RKM, et al. Context-Specific Regulation of NF-κB Target Gene Expression by EZH2 in Breast Cancers. Molecular Cell 2011;43(5):798–810 doi 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 126.Jung H-Y, Jun S, Lee M, Kim H-C, Wang X, Ji H, et al. PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Molecular cell 2013;52(2):193–205 doi 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yan J, Li B, Lin B, Lee PT, Chung T-H, Tan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 2016;128(7):948 doi 10.1182/blood-2016-01-690701. [DOI] [PubMed] [Google Scholar]
- 128.Anwar T, Arellano-Garcia C, Ropa J, Chen Y-C, Kim HS, Yoon E, et al. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nature Communications 2018;9(1):2801 doi 10.1038/s41467-018-05078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su Ih, Dobenecker M-W, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb Group Protein Ezh2 Controls Actin Polymerization and Cell Signaling. Cell 2005;121(3):425–36 doi 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 130.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, NY) 2012;338(6113):1465–9 doi 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim E, Kim M, Woo D-H, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell 2013;23(6):839–52 doi 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cha T-L, Zhou BP, Xia W, Wu Y, Yang C-C, Chen C-T, et al. Akt-Mediated Phosphorylation of EZH2 Suppresses Methylation of Lysine 27 in Histone H3. Science 2005;310(5746):306 doi 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 133.Crim JR, Seeger LL, Yao L, Chandnani V, Eckardt JJ. Diagnosis of soft-tissue masses with MR imaging: can benign masses be differentiated from malignant ones? Radiology 1992;185(2):581–6 doi 10.1148/radiology.185.2.1410377. [DOI] [PubMed] [Google Scholar]
- 134.Levine E, Huntrakoon M, Wetzel LH. Malignant nerve-sheath neoplasms in neurofibromatosis: distinction from benign tumors by using imaging techniques. American Journal of Roentgenology 1987;149(5):1059–64 doi 10.2214/ajr.149.5.1059. [DOI] [PubMed] [Google Scholar]
- 135.Albritton KH, Rankin C, Coffin CM, Ratner N, Budd GT, Schuetze SM, et al. Phase II study of erlotinib in metastatic or unresectable malignant peripheral nerve sheath tumors (MPNST). Journal of Clinical Oncology 2006;24(18_suppl):9518- doi 10.1200/jco.2006.24.18_suppl.9518. [DOI] [Google Scholar]
- 136.D’Adamo DR, Dickson MA, Keohan ML, Carvajal RD, Hensley ML, Hirst CM, et al. A Phase II Trial of Sorafenib and Dacarbazine for Leiomyosarcoma, Synovial Sarcoma, and Malignant Peripheral Nerve Sheath Tumors. The Oncologist 2018. doi 10.1634/theoncologist.2018-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu LMN, Deng Y, Wang J, Zhao C, Wang J, Rao R, et al. Programming of Schwann Cells by Lats½-TAZ/YAP Signaling Drives Malignant Peripheral Nerve Sheath Tumorigenesis. Cancer Cell 2018;33(2):292–308.e7 doi 10.1016/j.ccell.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Higham CS, Steinberg SM, Dombi E, Perry A, Helman LJ, Schuetze SM, et al. SARC006: Phase II Trial of Chemotherapy in Sporadic and Neurofibromatosis Type 1 Associated Chemotherapy-Naive Malignant Peripheral Nerve Sheath Tumors. Sarcoma 2017;2017:8685638- doi 10.1155/2017/8685638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Patwardhan PP, Surriga O, Beckman MJ, de Stanchina E, Dematteo RP, Tap WD, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clinical cancer research : an official journal of the American Association for Cancer Research 2014;20(12):3146–58 doi 10.1158/1078-0432.CCR-13-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mahller YY, Vaikunth SS, Currier MA, Miller SJ, Ripberger MC, Hsu Y-H, et al. Oncolytic HSV and Erlotinib Inhibit Tumor Growth and Angiogenesis in a Novel Malignant Peripheral Nerve Sheath Tumor Xenograft Model. Molecular Therapy 2007;15(2):279–86 doi 10.1038/sj.mt.6300038. [DOI] [PubMed] [Google Scholar]
- 141.Du X, Yang J, Ylipää A, Zhu Z. Genomic amplification and high expression of EGFR are key targetable oncogenic events in malignant peripheral nerve sheath tumor. Journal of hematology & oncology 2013;6:93- doi 10.1186/1756-8722-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Ambrosini G, Cheema HS, Seelman S, Teed A, Sambol EB, Singer S, et al. Sorafenib inhibits growth and mitogen-activated protein kinase signaling in malignant peripheral nerve sheath cells. Molecular cancer therapeutics 2008;7(4):890–6 doi 10.1158/1535-7163.MCT-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proceedings of the National Academy of Sciences of the United States of America 2005;102(24):8573–8 doi 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zou CY, Smith KD, Zhu Q-S, Liu J, McCutcheon IE, Slopis JM, et al. Dual targeting of AKT and mammalian target of rapamycin: A potential therapeutic approach for malignant peripheral nerve sheath tumor. Molecular Cancer Therapeutics 2009;8(5):1157 doi 10.1158/1535-7163.MCT-08-1008. [DOI] [PubMed] [Google Scholar]
- 145.Johansson G, Mahller YY, Collins MH, Kim M-O, Nobukuni T, Perentesis J, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Molecular cancer therapeutics 2008;7(5):1237–45 doi 10.1158/1535-7163.MCT-07-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bhola P, Banerjee S, Mukherjee J, Balasubramanium A, Arun V, Karim Z, et al. Preclinical in vivo evaluation of rapamycin in human malignant peripheral nerve sheath explant xenograft. International Journal of Cancer 2010;126(2):563–71 doi 10.1002/ijc.24783. [DOI] [PubMed] [Google Scholar]
- 147.Johannessen CM, Johnson BW, Williams Sybil MG, Chan AW, Reczek EE, Lynch RC, et al. TORC1 Is Essential for NF1-Associated Malignancies. Current Biology 2008;18(1):56–62 doi 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 148.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of Selumetinib in Neurofibromatosis Type 1–Related Plexiform Neurofibromas. New England Journal of Medicine 2016;375(26):2550–60 doi 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shurell E, Singh AS, Crompton JG, Jensen S, Li Y, Dry S, et al. Characterizing the immune microenvironment of malignant peripheral nerve sheath tumor by PD-L1 expression and presence of CD8+ tumor infiltrating lymphocytes. Oncotarget 2016;7(39):64300–8 doi 10.18632/oncotarget.11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Science translational medicine 2016;8(328):328rv4–rv4 doi 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haworth KB, Arnold MA, Pierson CR, Choi K, Yeager ND, Ratner N, et al. Immune profiling of NF1-associated tumors reveals histologic subtype distinctions and heterogeneity: implications for immunotherapy. Oncotarget 2017;8(47):82037–48 doi 10.18632/oncotarget.18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim A, Stewart DR, Reilly KM, Viskochil D, Miettinen MM, Widemann BC. Malignant Peripheral Nerve Sheath Tumors State of the Science: Leveraging Clinical and Biological Insights into Effective Therapies. Sarcoma 2017;2017:7429697- doi 10.1155/2017/7429697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, et al. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell 2013;152(5):1077–90 doi 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chau V, Lim SK, Mo W, Liu C, Patel AJ, McKay RM, et al. Preclinical therapeutic efficacy of a novel pharmacologic inducer of apoptosis in malignant peripheral nerve sheath tumors. Cancer research 2014;74(2):586–97 doi 10.1158/0008-5472.CAN-13-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li H, Chang L-J, Neubauer DR, Muir DF, Wallace MR. Immortalization of human normal and NF1 neurofibroma Schwann cells. Laboratory Investigation 2016;96:1105 doi 10.1038/labinvest.2016.88. [DOI] [PubMed] [Google Scholar]
- 156.Johnson KJ, Hussain I, Williams K, Santens R, Mueller NL, Gutmann DH. Development of an international internet-based neurofibromatosis Type 1 Patient registry. Contemporary Clinical Trials 2013;34(2):305–11 doi 10.1016/j.cct.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 157.Seidlin M, Holzman R, Knight P, Korf B, Rangel Miller V, Viskochil D, et al. Characterization and utilization of an international neurofibromatosis web-based, patient-entered registry: An observational study. PloS one 2017;12(6):e0178639-e doi 10.1371/journal.pone.0178639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002;111(2):197–208 doi 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 159.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009;461(7265):762–7 doi 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nature Cell Biology 2008;10:1291 doi 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 161.Cao R, Zhang Y. SUZ12 Is Required for Both the Histone Methyltransferase Activity and the Silencing Function of the EED-EZH2 Complex. Molecular Cell 2004;15(1):57–67 doi 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 162.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA (New York, NY) 2015;21(12):2007–22 doi 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schmitges Frank W, Prusty Archana B, Faty M, Stützer A, Lingaraju Gondichatnahalli M, Aiwazian J, et al. Histone Methylation by PRC2 Is Inhibited by Active Chromatin Marks. Molecular Cell 2011;42(3):330–41 doi 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 164.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Science Translational Medicine 2016;8(366):366ra161 doi 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]