Abstract
The outcome of chronic infections is highly variable. The heterogeneous disease outcomes in natural populations differ from genetically homogeneous infection models. Here, we use tuberculosis as a “case study” to contrast the genetic landscape in natural populations with standard infection models, discussing new strategies to bridge this gap.
Keywords: Infectious diseases, tuberculosis, host genetics, mouse models, genetic diversity, Collaborative Cross
Mouse models of tuberculosis
Much of what we know of protective immunity to Mycobacterium tuberculosis (Mtb) was discovered in the mouse model. The standard mouse model of tuberculosis (TB) consists of infecting an inbred line of mouse, often C57BL/6, with a lab-adapted strain of the pathogen. Given the lack of genetic or environmental diversity, this interaction produces a homogenous product; every animal becomes infected, mounts a Th1 biased T cell response, and eventually succumbs to disease. By deleting individual arms of immunity, it is clear that protection from TB in this model requires a robust T cell response and the production of interferon γ (IFNγ), since mice lacking this capacity are highly susceptible. Similarly, IFNγ production is an effective clinical diagnostic for TB and human patients lacking genes necessary for this immune axis are highly susceptible to mycobacterial disease [1]. Thus, mice and humans share a fundamental dependence on certain immune mechanisms for immunity to TB. In fact, if only Mendelian traits influenced TB susceptibility, one could view the human and mouse systems to be highly concordant, and it would be completely appropriate to base therapeutic development on the C57BL/6 model. But how well does this genetically and phenotypically homogenous system model TB in a naturally diverse population?
A short natural history of tuberculosis
The fact that only 5–10% of Mtb-infected individuals develop disease is often cited as evidence that the risk of developing TB differs quantitatively between individuals. More fundamentally, pathological and immunological metrics also suggest qualitative differences in the underlying biology of disease. For example, a rich literature describes the diverse histopathological manifestations of disease, which range from the canonical necrotic granuloma, to suppurative lesions or pneumonia. Similarly, while most individuals mount a Th1-biased cellular response when exposed to Mtb, the sensitivity of IFNγ-based diagnostics rarely surpass 80%, and increasing evidence indicates that this canonical response is not detectable in a substantial fraction of exposed individuals [2]. Together, these observations support the emerging view that human immunity to Mtb may be more diverse that previously appreciated and the interaction between Mtb and the human host in the natural setting produces a wide spectrum of biologically-diverse disease states.
While environmental factors such as nutrition and coinfection can influence disease risk, the development of TB is also influenced by the genetic composition of the host. Mendelian inheritance of rare loss of function alleles that disrupt Th1 immunity can produce severe susceptibility to mycobacterial infections. However, Mendelian inheritance does not explain the genetic influence on adult pulmonary TB, which appears to depend on a combination of more common alleles. A variety of association studies have identified host loci that influence TB risk [1]. Our understanding of these allelic variants remains incomplete, but the genes identified in these cohorts appear to function in a wider variety of pathways than those known to be involved in Medelian immunodeficiencies. While the establishment of a Th1-biased immunity remains prominent, regulation of disease-promoting inflammation, and alteration of the pathogen’s intracellular environment also contribute. The variety of implicated functions and preferential association between these variants with specific disease presentations, such as extrapulmonary TB, indicate that distinct biological processes control different manifestations of TB disease.
The human genetic variation described above only accounts for a portion of the complexity of the interaction between host and pathogen, as pathogen genotype accounts for some of the phenotypic diversity that remains unexplained by host genetics. Certainly, Mtb clinical isolates are phenotypically different, and display different degrees of virulence in animal models [4]. However, these types of studies oversimplify the contribution of pathogen genotype, which likely depends on interactions with host variants. Genome-to-genome analyses of other infection models often reveal that the effect of host polymorphisms depend on pathogen genotype [5]. Similarly, the long-term coevolution between Mtb and different human populations in distinct geographical locations has provided an opportunity for adaptation to geographically-restricted host populations, which has been proposed to produce a “sympatric” relationship between specific host and pathogen lineages [6]. While direct experimental evidence for this hypothesis is difficult to obtain, existing human genetic association studies that account for Mtb genotype discovered a differential association between human polymorphism and bacterial lineage [7]. Thus, in a natural population the risk of developing TB, the specific manifestation of disease, and the underlying pathophysiology of the infection are influenced by interactions between host and pathogen genotype (Figure 1).
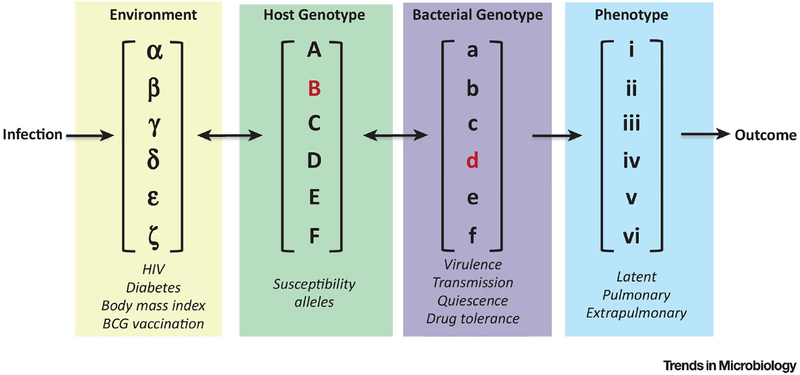
Figure 1. TB susceptibility is a complex trait.
The variability of TB disease is determined by interactions between environment, host genotype and bacterial genotypes. In the lab we simplify these factors by controlling environment, infecting a single inbred mouse strain with a lab adapted Mtb strain and measure a small number of traits that we hypothesize are important for driving overall disease (highlighted in red). While classical inbred lines of mice represent tractable experimental systems, the lack of genetic diversity and homogenous disease states means we are only modeling a small component of the disease that is observed in humans.
If the biology of the infection varies, shouldn’t we expect that the response to an intervention would be equally variable? Indeed, our experience from TB control confirms this expectation. Diagnostics, vaccines, and antibiotics perform reliably in the mouse model. However, in humans, immunodiagnostics suffer from ~20% false negative rates, the efficacy of vaccination varies from 80% to zero in different populations, and even antibiotic therapy fails in up to 10% of patients even in well-controlled clinical trial settings [2,8]. This experience argues for more diverse model systems to understand how the heterogenous biology of disease impacts our interventions.
Modeling complexity
Clearly, the biological diversity of natural populations is not adequately modeled in a genetically homogenous animal model. One solution to this problem has been the use of alternative species, such as guinea pigs, rabbits, and nonhuman primates. All of these systems have advantages and are reasonably used to reproduce disease states not found in the standard mouse model. However, while these pathologies can be modeled they remain difficult to mechanistically characterize due to the relative intractability of these systems. Furthermore, the increased expense of these systems generally precludes the interrogation of different Mtb genotypes. A possible solution would be the introduction of controlled genetic diversity into the more tractable mouse model. Historically, this has been achieved through the use of classic substrains, however these strains were not generated for diversity and most are highly related. To overcome this lack of diversity, the mouse genetics field has invested in new model populations that have recently become available. The Collaborative Cross (CC) and Diversity Outbred (DO) panels are derived from the same eight parental mouse strains, which include wild-derived animals that were chosen for their genetic diversity. The DO’s are randomized outbred animals whose small recombination intervals make them ideal for high resolution for genetic mapping [8]. The CC represent >100 recombinant inbred lines and represent homozygous mosaics of the eight parent genomes. Unlike outbred populations, the structure of the CC allows the serial evaluation of genetically identical individuals and quantification of genotype specific effects of interventions. The DO and CC populations are beginning to be explored as infectious disease models, and have been found to recapitulate many of extreme disease traits previously undetected in traditional mouse models for a variety of pathogens and immune traits [9]. Similarly, the susceptibility to TB in the DO panel varies dramatically and produces immunological variation correlating with lung damage and increased inflammation in human TB patients [10]. These diverse traits are recapitulated in the reproducible CC panel, allowing more in-depth studies that verify that immune responses differ in these animals, and these immune biases correlate with vaccine efficacy [11]. Together these resources represent a new strategy to introduce known and controlled genetic variation into our most tractable mammalian model of infectious disease.
Concluding remarks and future perspectives
The goal of TB research is more effective interventions, and understanding the mechanisms by which genetic variation alters disease progression and treatment efficacy could play a key role in this process. In the future, additional non-genetic sources of variation, such as the microbiome composition that has strong immunomodulatory effects [12], need to be accounted for in these models. While it is unlikely that this information can be used to “personalize” treatment in the global health context, understanding why treatments fail in some individuals could still have a transformative effect, both in designing more powerful clinical trials and ultimately introducing an intervention, or combination of interventions, that is robust to genetic variation.
Acknowledgements
We thank Sarah Dunstan, Aditya Bandekar, Michael Kiritsy and Micah Belew for pivotal intellectual contributions. The authors wish to note that due to the format of this article, not all pertinent references could be included. This work was supported by grants to C.M. Smith from the Charles King Foundation and C.M. Sassetti from the NIH (AI132130).
References:
- 1.Abel L et al. (2018) Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives. The Lancet Infectious Diseases 18, e64–e75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazurek GH et al. Updated guidelines for using Interferon Gamma Release Assays to detect Mycobacterium tuberculosis infection - United States, 2010., MMWR. Recommendations and reports: Morbidity and mortality weekly report. Recommendations and reports, 59 25-June-(2010), 1–25 [PubMed] [Google Scholar]
- 3.Hernández-Pando R et al. (2012) Use of mouse models to study the variability in virulence associated with specific genotypic lineages of Mycobacterium tuberculosis. Infect. Genet. Evol 12, 725–731 [DOI] [PubMed] [Google Scholar]
- 4.Ansari MA et al. (2017) Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nat Genet 49, 666–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagneux S et al. (2006) Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl AcadSci USA 103, 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caws M et al. (2008) The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog 4, e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abubakar I et al. (2013) Systematic review and meta-analysis of the current evidence on the duration of protection by bacillus Calmette-Guérin vaccination against tuberculosis. Health Technol Assess 17, 1–372- v–vi [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogue MA et al. (2015) Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database. Mamm Genome 26, 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maurizio PL and Ferris MT (2016) The Collaborative Cross Resource for Systems Genetics Research of Infectious Diseases In Systems Genetics 1488pp. 579–596, Springer; New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramnik I and Beamer G (2016) Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin Immunopathol 38, 221–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CM et al. (2016) Tuberculosis Susceptibility and Vaccine Protection Are Independently Controlled by Host Genotype. mBio 7, e01516–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosshart SP et al. (2017) Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]