Abstract
Background
Rapeseed is the third largest oil seed crop in the world. The seeds of this plant store lipids in oil bodies, and oleosin is the most important structural protein in oil bodies. However, the function of oleosin in oil crops has received little attention.
Results
In the present study, 48 oleosin sequences from the Brassica napus genome were identified and divided into four lineages (T, U, SH, SL). Synteny analysis revealed that most of the oleosin genes were conserved, and all of these genes experienced purifying selection during evolution. Three and four important oleosin genes from Arabidopsis and B. napus, respectively, were cloned and analyzed for function in Arabidopsis. Overexpression of these oleosin genes in Arabidopsis increased the seed oil content slightly, except for BnaOLE3. Further analysis revealed that the average oil body size of the transgenic seeds was slightly larger than that of the wild type (WT), except for BnaOLE1. The fatty acid profiles showed that the linoleic acid content (13.3% at most) increased and the peanut acid content (11% at most) decreased in the transgenic lines. In addition, the seed size and thousand-seed weight (TSW) also increased in the transgenic lines, which could lead to increased total lipid production.
Conclusion
We identified oleosin genes in the B. napus genome, and overexpression of oleosin in Arabidopsis seeds increased the seed weight and linoleic acid content (13.3% at most).
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1891-y) contains supplementary material, which is available to authorized users.
Keywords: Brassica napus, Oleosin, Gene evolution, Fatty acid, Oil content
Background
Brassica napus, which is closely related to the model plant Arabidopsis, is one of the most important oil crops, providing approximately 15% of the vegetable oil worldwide [1, 2]. B. napus shares a highly conserved genome sequence with Arabidopsis, especially for genes associated with lipid biosynthesis, which has been extensively studied [3, 4]. However, detailed analysis of the lipid metabolism pathway is required to understand homology gene function differentiation in this complex genome. Identification and functional analysis of homologous genes in B. napus has become easier with the release of the Arabidopsis and B. napus genomes.
Lipids are the main reserves in B. napus seeds and account for 35–50% of the dry weight. Triacylglycerol (TAG) is the main component of lipids and is stored in oil bodies [1, 5]. The oil body, an organelle that is widely distributed in lipid storage cells, is approximately 0.5 to 2 μm in diameter and contains a liquid matrix of neutral lipids surrounded by a hemi-unit membrane with structural proteins. The oil body serves as a natural protective system against fatty acid oxidation and maintains lipid stability under extreme stress conditions [6]. A well-known hypothesis is that the oil body is synthesized in the ER and then released to the cytoplasm via a budding mechanism [7, 8]. Although oil bodies can remain stable and do not aggregate or coalesce even after long-term storage when the environment remains unchanged, such as in mature seed cells or in vitro [9], the structure of this organelle does not remain constant during seed development, and the size of the oil body changes during seed maturation. The size of the oil body first increases and then decreases during seed development [10].
Three main proteins (oleosin, caleosin and steroleosins) are inserted in the phospholipid membrane [11, 12]. In addition, several putative adipose-regulatory proteins (such as SEIPIN), oil body-associated protein 1 (OBAP1) and lipid droplet-associated proteins were also reported to be present in the oil body [5, 13, 14]. Oleosin, which is synthesized in the ER, accounts for 80–90% of the oil body structure proteins and plays an important role in lipid storage [11]. Vance et al. [15] first identified the oleosin sequence from maize and proposed that oleosin may serve as the recognition signal for the specific binding of lipase to lipid bodies in the lipid degradation pathway [15, 16]. The oleosins from sesame, Pinus and B. napus were also cloned [17–19]. The oleosin peptide can be divided into three parts: a short amphipathic N-terminal sequence, a central hydrophobic domain and an amphipathic C-terminal sequence [5, 16, 20]. Thirty amino acids of the C-terminal sequence near the central domain can form an α-helical structure, which interacts horizontally with the PL layer on the oil body surface. The N- and C-terminal peptides present outside the oil body might be the target of lipase for oil body degradation or other metabolic pathways [7, 21]. The central hydrophobic domain, composed of 72 of the most highly conserved amino acids, is possibly one of the hallmarks of the oil body protein because this region has not been identified in the structural proteins of other lipid storage organelles [5, 21]. The conserved 72 amino acids can form a hairpin structure that is inserted into the phospholipid membrane, in which a highly conserved Pro knot (PX5SPX3P) forms the loop of the hairpin structure. The three Pro residues and one Ser residue are present in all oleosins from diverse plant species and green algae without any substitution, and substitution of the conserved Pro residues leads to abnormal localization [22]. Oleosins from diverse species have been reported, all of which can be divided into six lineages (P, U, SL, SH, T and M) [5, 16]. Among the six lineages, the M lineage is present in Lauraceae, and the T lineage was detected in only the tapeta of Brassicaceae [16, 20, 23]. The P lineage was mainly distributed in green algae and might be the origin of U oleosin, which further gave rise to the SL and SH lineages [5].
Many studies have proven that adjustment of the oleosin protein level could prevent fusion of the oil body and maintain oil body size during seed development [24–26]. The size of the oil droplets in wild-type (WT) and AtOLE1-knockout Arabidopsis seeds was uniform in the early stages of seed development [10]. The lack of oleosins causes the oil body to compress and fuse, resulting in an enlarged oil body in the middle stage of seed development [11, 27, 28]. Several homologous oleosin proteins from Arabidopsis thaliana were studied, and OLE1 and OLE4 were found to negatively regulate oil body size, whereas OLE2 might increase the oil body size [10]. In a OLE-knockout Arabidopsis mutant, although the oil body shape was irregular, the total fatty acid content did not change, except for the levels of eicosenoic acid (C20:1) and oleic acid (C18:1). The plants were also sensitive to low temperature during germination [6]. Similar results could be proved by heterologous expression of castor bean oleosin in Arabidopsis, which led to a 20% increase in the ricinoleic acid content in TAGs [29]. On the other hand, coexpression of oleosins with other genes in the TAG biosynthesis pathway could enhance the oil content [12], which might represent a new role of oleosins in the development of genetically modified crops. OLE also plays a role in Arabidopsis tapetosome formation and pollen development [30]. As a lipid storage structure, the quantity and size of oil bodies can reflect the oil content of seeds to some extent. Although there is some controversy regarding the relationship of oil body number and size with oil content [31, 32], high oil levels tend to lead to higher oil body production or sizes than low oil level [31], and the high oil content might be associated with increased oleosin gene copy numbers and increased gene expression levels.
However, the function of oleosins from important oil corps has received little attention. In the present study, all of the oleosin sequences from the B. napus genome were identified, and their structures, evolution and synteny relationship with oleosins from other Cruciferae plants were analyzed. In addition, 7 important oleosin genes from Arabidopsis and B. napus were cloned and transformed into Arabidopsis for functional analysis. The present results revealed that overexpression of oleosin in Arabidopsis could affect oil body size as well as seed size and seed weight, so oil production could be increased. In addition, transgenic seeds also show decreased freezing tolerance. This study provides a foundation for future studies regarding the oleosin superfamily in B. napus.
Results
Genome-wide identification of oleosin family genes in the genome of B. napus
Based on the gene sequences of 17 oleosin genes obtained from the Arabidopsis Information Resource (TAIR) using oleosin as the query, the oleosin genes from the B. napus genome were searched using BLAST in the CNS-Genoscope database. In total, 65 genes that share high similarity with Arabidopsis oleosin genes were identified, and 48 genes containing the oleosin family domain PF01277 in the Pfam database were selected for further synteny analysis (Table 1). In addition, 30 and 28 oleosin genes were identified from Brassica rapa and Brassica oleracea, respectively (Additional file 5: Table S1). The physicochemical parameters of each oleosin protein were calculated by ExPASy, and a considerable difference in these parameters was observed, which might result from the highly variable terminal sequences in these oleosin genes. The results revealed that most of the oleosin proteins had molecular masses < 25 kDa, except BnaOLE22, BnaOLE23, BnaOLE32, BnaOLE35 and BnaOLE37. Forty-three of these proteins have relatively high isoelectric points (pI> 7). Nearly all of these proteins have a high aliphatic index, and the GRAVY value of these proteins is close to zero, which shows that these oleosin proteins exhibit amphipathy.
Table 1.
OLE genes in B. napus genome and their sequence characteristics and physicochemical parameters prediction
| Gene ID | Name | family | chr. | gene position | gene length | exon | intron | Number of aa | Mol.Wt. | Theoretical pI | Aliphatic index | GRAVY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| start | end | ||||||||||||
| BnaA01g14480D | BnaOLE1A01 | SH | A01 | 7,305,082 | 7,306,307 | 1226 | 3 | 2 | 170 | 18,556.14 | 9.52 | 86.65 | −0.194 |
| BnaA01g26680D | BnaOLE8A01 | U | A01 | 18,669,237 | 18,669,731 | 495 | 1 | 0 | 164 | 18,141.98 | 9.39 | 96.4 | 0.232 |
| BnaA02g29000D | BnaOLE3 | SL | A02 | 21,253,735 | 21,254,824 | 1090 | 2 | 1 | 188 | 20,001.84 | 9.15 | 97.02 | −0.005 |
| BnaA02g35480D | BnaOLE18 | U | A02_R | 273,275 | 273,804 | 530 | 1 | 0 | 131 | 13,569.19 | 8.8 | 121.15 | 0.926 |
| BnaA03g02100D | BnaOLE19 | T | A03 | 962,155 | 963,313 | 1159 | 2 | 1 | 272 | 27,116.61 | 10.21 | 77.43 | 0.057 |
| BnaA03g02110D | BnaOLE20 | T | A03 | 964,861 | 966,041 | 1181 | 2 | 1 | 207 | 21,751.38 | 10 | 82.46 | −0.189 |
| BnaA03g02120D | BnaOLE13A03 | T | A03 | 967,459 | 968,374 | 916 | 1 | 0 | 79 | 7931.6 | 5.75 | 136.08 | 1.597 |
| BnaA03g02140D | BnaOLE21 | T | A03 | 975,911 | 977,658 | 1748 | 3 | 2 | 119 | 12,160.68 | 10.3 | 115.63 | 0.759 |
| BnaA03g27410D | BnaOLE22 | SL | A03 | 13,512,809 | 13,516,176 | 3368 | 5 | 4 | 778 | 86,260.73 | 7.3 | 93.07 | −0.03 |
| BnaA03g34830D | BnaOLE8A03 | U | A03 | 17,011,078 | 17,011,575 | 498 | 1 | 0 | 165 | 17,821.37 | 6.38 | 95.76 | 0.421 |
| BnaA03g40390D | BnaOLE17A03 | T | A03 | 20,190,729 | 20,192,198 | 1470 | 3 | 2 | 251 | 27,929.33 | 9.08 | 92.11 | −0.162 |
| BnaA03g47170D | BnaOLE1A03 | SH | A03 | 24,188,857 | 24,190,194 | 1338 | 2 | 1 | 183 | 20,001.69 | 9.3 | 84.81 | −0.234 |
| BnaA04g15210D | BnaOLE6A04 | SH | A04 | 12,660,472 | 12,660,912 | 441 | 1 | 0 | 146 | 15,657.25 | 8.66 | 92.88 | 0.277 |
| BnaOLE2 | BnaOLE2 | U | A04 | 9,050,747 | 9,051,738 | 992 | 1 | 0 | 220 | 23,048.11 | 9.1 | 77.59 | −0.083 |
| BnaA05g21590D | BnaOLE8A05 | U | A05 | 16,610,133 | 16,610,630 | 498 | 1 | 0 | 165 | 17,932.6 | 9.52 | 97.52 | 0.339 |
| BnaA06g03370D | BnaOLE23 | SL | A06 | 2,059,111 | 2,061,705 | 2595 | 7 | 6 | 517 | 57,648.71 | 8.59 | 92.75 | −0.032 |
| BnaA07g14960D | BnaOLE2A07 | SH | A07 | 13,061,213 | 13,062,594 | 1382 | 3 | 2 | 200 | 21,284.38 | 9.3 | 88.3 | −0.046 |
| BnaA08g14540D | BnaOLE1 | T | A08 | 12,282,608 | 12,283,829 | 1222 | 2 | 1 | 180 | 19,514.98 | 9.15 | 84.61 | −0.248 |
| BnaA08g22860D | BnaOLE24 | SL | A08 | 16,473,505 | 16,474,574 | 1070 | 2 | 1 | 209 | 20,340.83 | 9.7 | 99.43 | 0.75 |
| BnaA09g02110D | BnaOLE4A09 | U | A09 | 1,040,084 | 1,041,270 | 1187 | 2 | 1 | 188 | 19,879.71 | 6.91 | 96.97 | 0.045 |
| BnaA10g05240D | BnaOLE25 | SH | A10 | 2,963,139 | 2,963,633 | 495 | 1 | 0 | 164 | 17,864.6 | 9.1 | 98.17 | 0.347 |
| BnaA10g05760D | BnaOLE3A10 | T | A10 | 3,966,784 | 3,967,565 | 782 | 2 | 1 | 149 | 15,574.98 | 10.1 | 104.09 | 0.286 |
| BnaA10g23950D | BnaOLE26 | T | A10 | 15,705,746 | 15,706,808 | 1063 | 2 | 1 | 175 | 17,006.96 | 10.12 | 110.51 | 0.852 |
| BnaA10g23960D | BnaOLE27 | T | A10 | 15,711,917 | 15,713,045 | 1129 | 2 | 1 | 195 | 19,523.32 | 10.91 | 119.33 | 0.841 |
| BnaA10g30180D | BnaOLE28 | T | A10_R | 2,101,896 | 2,103,711 | 1816 | 2 | 1 | 204 | 20,585.57 | 9.4 | 78.04 | −0.185 |
| BnaA10g30190D | BnaOLE29 | SH | A10_R | 2,105,832 | 2,106,825 | 994 | 2 | 1 | 108 | 11,032.19 | 11.17 | 116.57 | 0.807 |
| BnaC01g17050D | BnaOLE1C01 | U | C01 | 11,676,131 | 11,677,343 | 1213 | 3 | 2 | 170 | 18,571.08 | 9.39 | 82.65 | −0.224 |
| BnaC01g34070D | BnaOLE8C01 | T | C01 | 33,257,398 | 33,257,892 | 495 | 1 | 0 | 164 | 17,864.6 | 9.1 | 98.17 | 0.347 |
| BnaC02g01550D | BnaOLE30 | T | C02 | 627,160 | 628,665 | 1506 | 2 | 1 | 149 | 14,826.83 | 11.28 | 107.65 | 0.682 |
| BnaC02g01560D | BnaOLE31 | T | C02 | 629,668 | 630,975 | 1308 | 2 | 1 | 157 | 15,236.32 | 10.72 | 109.87 | 0.73 |
| BnaC02g01570D | BnaOLE32 | SL | C02 | 633,723 | 637,275 | 3553 | 3 | 2 | 870 | 88,165.02 | 10.4 | 51.98 | −0.891 |
| BnaC02g37030D | BnaOLE4C02 | T | C02 | 40,002,669 | 40,003,899 | 1231 | 2 | 1 | 191 | 20,248.17 | 7.89 | 101.1 | 0.058 |
| BnaC03g03140D | BnaOLE33 | T | C03 | 1,516,588 | 1,518,583 | 1996 | 3 | 2 | 214 | 22,584.38 | 10.17 | 81.59 | −0.196 |
| BnaC03g03150D | BnaOLE13C03 | T | C03 | 1,519,876 | 1,520,984 | 1109 | 2 | 1 | 116 | 12,072.39 | 9.3 | 106.98 | 0.456 |
| BnaC03g03170D | BnaOLE34 | SL | C03 | 1,525,165 | 1,526,940 | 1776 | 2 | 1 | 120 | 12,506.14 | 9.9 | 133.42 | 1.066 |
| BnaC03g32430D | BnOLE37 | U | C03 | 19,921,970 | 19,925,218 | 3249 | 4 | 3 | 777 | 86,175.33 | 6.29 | 93.56 | −0.004 |
| BnaC03g40520D | BnaOLE8C03 | SL | C03 | 25,478,827 | 25,479,324 | 498 | 1 | 0 | 165 | 17,778.43 | 6.9 | 101.7 | 0.474 |
| BnaC04g32530D | BnaOLE2C04 | SH | C04 | 34,250,286 | 34,251,730 | 1445 | 2 | 1 | 229 | 24,575.09 | 9.34 | 75.02 | −0.111 |
| BnaC04g38160D | BnaOLE6C04 | U | C04 | 39,433,420 | 39,433,857 | 438 | 1 | 0 | 145 | 15,567.15 | 7.85 | 98.21 | 0.334 |
| BnaC05g34600D | BnaOLE8C05 | U | C05 | 33,883,325 | 33,883,822 | 498 | 1 | 0 | 165 | 17,994.67 | 9.52 | 96.3 | 0.352 |
| BnaC06g02430D | BnaOLE35 | SL | C06 | 3,184,669 | 3,187,278 | 2610 | 7 | 6 | 517 | 57,886.95 | 8.78 | 92.17 | −0.05 |
| BnaC06g12930D | BnaOLE2C06 | T | C06 | 15,634,774 | 15,636,282 | 1509 | 3 | 2 | 200 | 21,275.37 | 9.3 | 88.3 | −0.048 |
| BnaC07g31360D | BnaOLE17C07 | SH | C07 | 35,479,311 | 35,480,772 | 1462 | 3 | 2 | 251 | 27,859.39 | 9.22 | 94.82 | −0.139 |
| BnaC07g39370D | BnaOLE1C07 | U | C07 | 40,278,874 | 40,279,959 | 1086 | 2 | 1 | 186 | 20,565.22 | 9.15 | 82.9 | −0.354 |
| BnaC08g03140D | BnaOLE5 | SH | C08 | 2,826,379 | 2,826,801 | 423 | 1 | 0 | 140 | 15,613.35 | 10.16 | 108.71 | 0.513 |
| BnaC08g11970D | BnaOLE1C08 | SH | C08 | 17,257,794 | 17,259,450 | 1657 | 3 | 2 | 179 | 19,424.86 | 9.15 | 85.08 | −0.288 |
| BnaC09g27370D | BnaOLE3C09 | T | C09 | 29,151,518 | 29,153,074 | 1557 | 2 | 1 | 149 | 15,711.18 | 9.99 | 104.09 | 0.311 |
| BnaC09g47930D | BnaOLE36 | SL | C09 | 47,002,663 | 47,003,720 | 1058 | 2 | 1 | 171 | 16,666.58 | 10.39 | 107.37 | 0.839 |
Sequence and phylogenetic analyses of the OLE genes
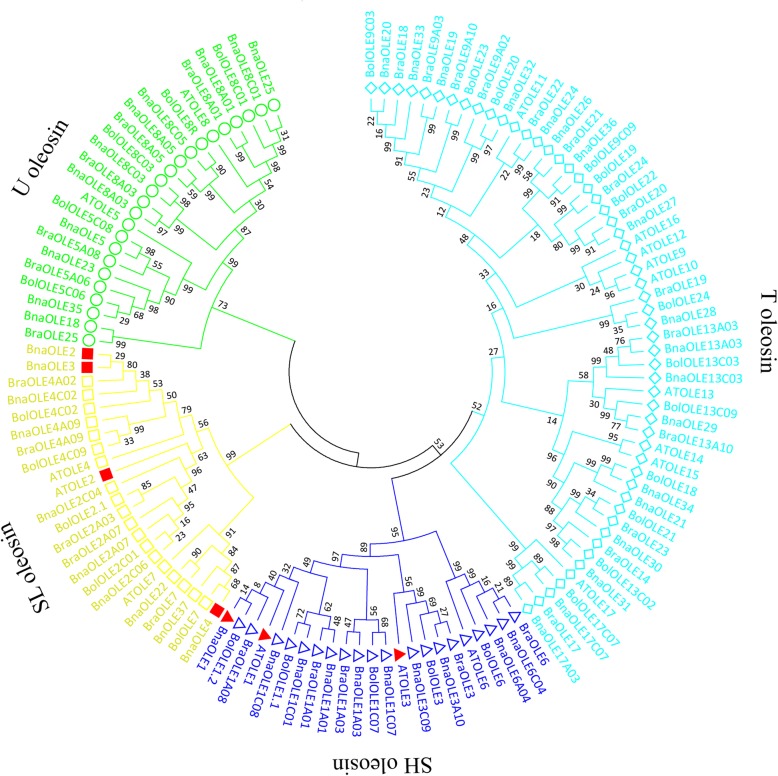
Sequence alignment was performed to analyze the similarity of oleosin genes between B. napus and other Brassica species using ClustalW. It was revealed that all of these oleosins contained a 72-amino-acid conserved sequence, exhibiting relatively high conservation. The conserved 72-residue sequence was submitted to Multiple Expectation Maximization for Motif Elicitation (MEME) for motif structure analysis, which indicated that the most well conserved sequence was the Pro loop (PX5SPX3P), and most of the X residues were the nonpolar Ile, Phe, Leu and Val in B. napus. BnaOLE17C07 and BnaOLE17A03 were different from the other oleosin genes; the third prolines of these proteins had been replaced by Thr (a neutral, nonpolar amino acid) (Additional file 1: Figure S1). The conserved 72-residue sequence was analyzed by phylogenetic analysis, revealing that the 48 oleosins of B. napus could be divided into the T, U, SL and SH lineages (Fig. 1, Additional file 2: Figure S2). Further analysis showed that T oleosin is the largest clade, including over 1/3 of the oleosins in B. napus, and that the other 3 lineages had similar members. The genes from the A genome and C genome of B. napus always appeared in pairs in most of the clades. The corresponding genes in their ancestors B. rapa and B. oleracea were also closely related, which indicated that these genes evolved from the same ancestor. Although syntenic genes were not identified for some BnaOLEs, we could speculate on the evolutionary relationships of these genes. For instance, BnaOLE5 was clustered in the same clade as BnaOLE8C01 and showed a close phylogenetic relationship with AtOLE5, which may indicate the evolutionary history of these genes.
Fig. 1.
Phylogenetic analysis of the A. thaliana, B. rapa, B. oleracea, and B. napus oleosin genes. The conserved central amino acid sequences of the OLE proteins were used to construct the cladogram using ClustalX and MEGA6. Different lineages of oleosins are colored differently, and the solid red graphics represent the gene that we selected for further functional analysis
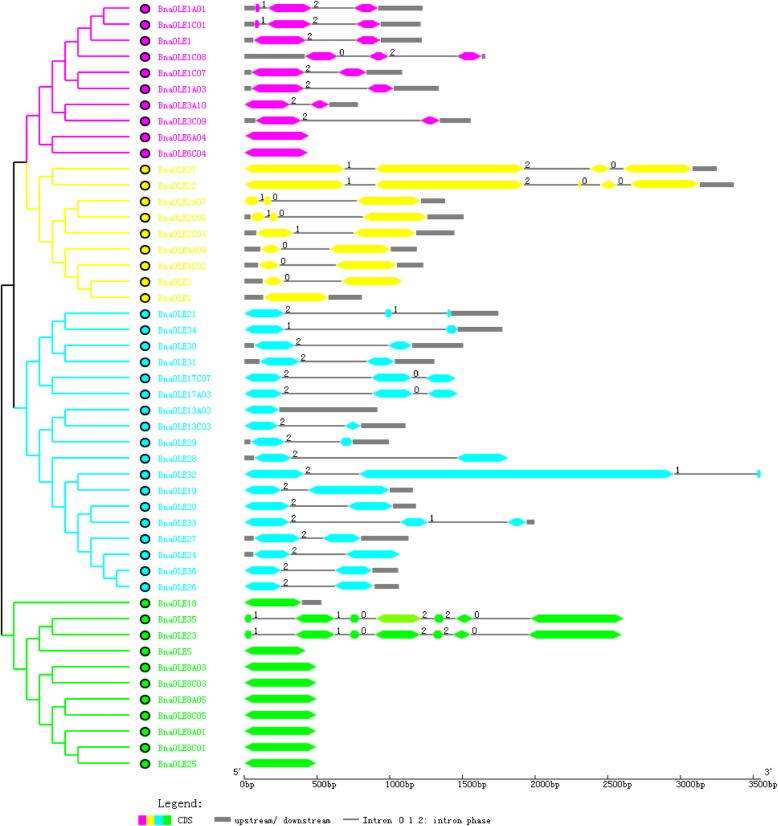
The lengths of most oleosin genes from B. napus were shorter than 2 kb, except for BnaOLE22, BnaOLE23, BnaOLE32, BnaOLE35 and BnaOLE37. These five genes did not belong to the same lineages and shared low sequence identity with each other in addition to the same lineages. Each clade of B. napus oleosins contains different intron-exon structural features (Fig. 2). In general, T oleosins possess more exons than the other three types of oleosins. Most U oleosins have no introns, except for BnaOLE23 and BnaOLE35, which are composed of many more amino acid residues than the other U oleosins. In addition to the other three long-chain oleosin genes, these genes also contain more exons than other genes from the same lineages. However, BnaOLE23 and BnaOLE35 show the same genetic structure as BnaOLE22 and BnaOLE37, so the gene pairs from the A genome and C genome have the same gene structure. Most of the oleosins from the other three clades contains two or three exons, with some exceptions that have no intron. For example, the SL oleosins BnaOLE22 and BnaOLE37 and the T oleosin BnaOLE32 possess more introns than the oleosins from the same lineages. Oleosins with a large number of introns often encode more amino acids than other oleosins. However, the SH oleosin genes BnaOLE6A04 and BnaOLE6C04, which have similar sequence lengths as other SH oleosin genes, have no introns. Despite the intron structure and sequence difference, all of the introns in oleosins are U2-type splice GT-AG introns [33, 34].
Fig. 2.
Genetic structure of B. napus oleosin genes. Different lineages of oleosins are colored differently. Green, yellow, dark blue and sky blue represent U, SL, SH and T oleosins, respectively. Round double-sided wedge boxes represent exons; black lines represent introns; gray boxes represent untranslated regions (UTRs); and the numbers near the introns indicate the intron phase. If an intron is between two complete codons, then the intron phase is defined as 0; if an intron is after the first or second nucleotide within the codon, the intron phase is defined as phase 1 or phase 2
Chromosomal location, duplication patterns and synteny analysis of the oleosin genes in B. napus
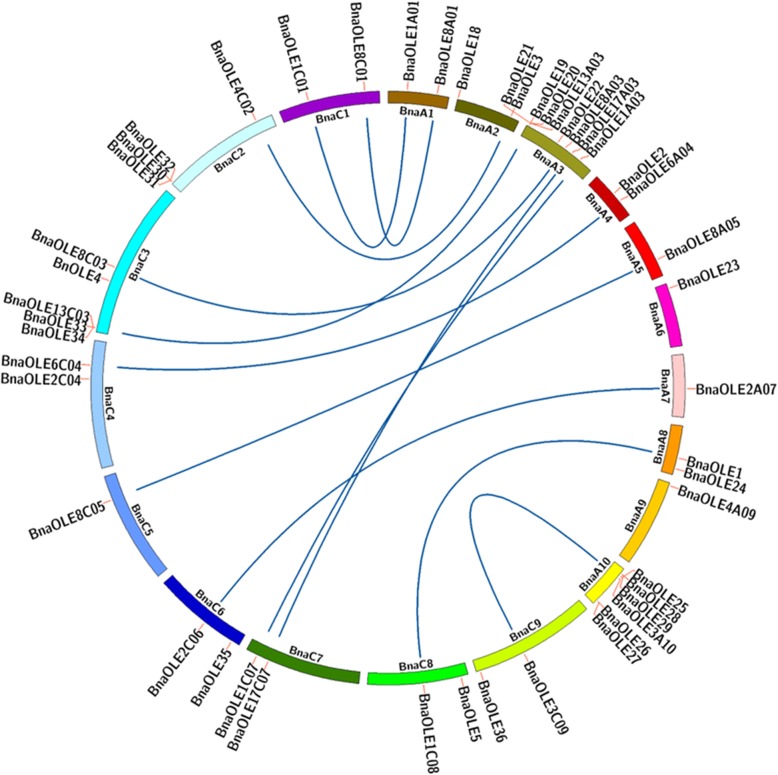
The chromosomal positions of the BnaOLE gene were analyzed, and most of the chromosomes possessed one or two oleosin genes except for the A03, A10, C02 and C03 chromosomes, and 8, 6, 4 and 5 oleosin genes were observed in these four chromosomes, respectively (Fig. 3). In addition, gene clusters mainly composed of T oleosins were found in some chromosomes; for instance, four genes encoding T oleosins, namely, BnaOLE19, BnaOLE20, BnaOLE13A03 and BnaOLE21, were located in clusters on the A03 chromosome, one of the reasons for which might be that nine T oleosins were clustered on A. thaliana chromosome 5. Most of the genes from the A genome appear in pairs with genes from the C genome. Intriguingly, two BnaOLE genes located on chromosome A3 were closely linked to paralogous genes located on chromosome C7, which might result from segmental duplication and chromosomal rearrangement during the long evolutionary history. The Brassica species experienced several whole-genome triplication (WGT) events, which contributed to gene-level evolution [35]. Gene duplication events were investigated to understand the genome expansion mechanism of the BnaOLE gene family in B. napus. Twelve tandemly duplicated genes (BnaOLE19/BnaOLE20/BnaOLE13A03/BnaOLE21, BnaOLE26/BnaOLE27, BnaOLE30/BnaOLE31/BnaOLE32 and BnaOLE33/BnaOLE13C03/BnaOLE34) located on chromosomes A03, A10, C02 and C03 were identified, and all twelve oleosins belonged to T oleosins.
Fig. 3.
Distribution of the oleosin gene in the B. napus genome and the syntenic relationship between oleosin genes. The chromosomal location information for the oleosin genes was obtained from the CNS-Genoscope database. The dark blue line indicates that these two genes located in the A genome and C genome are syntenic genes
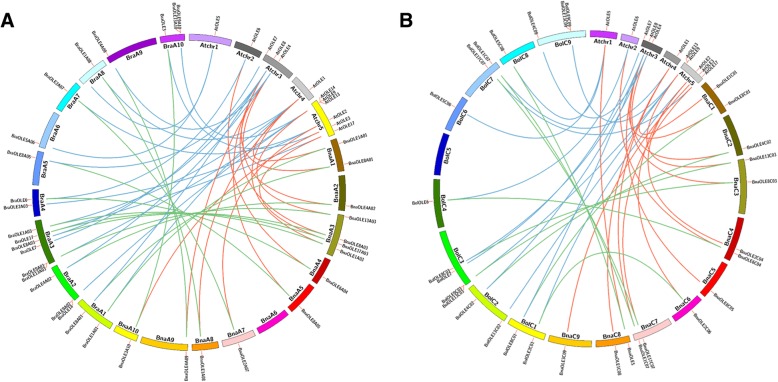
The Brassicaceae genome has experienced complex genome duplication and rearrangement [36]. We also analyzed the synteny maps of the oleosin genes in the B. napus genome and the homologous genes in A. thaliana, B. rapa and B. oleracea (Fig. 4). Gene duplication and gene loss were also observed in the OLE gene family. Except for some genes, such as AtOLE7, we did not identify syntenic genes. We found that most AtOLE genes had syntenic relationships with two or more BraOLE and BolOLE genes as a result of gene duplication, and these duplicated OLEs originating from the same AtOLEs in B. rapa and B. oleracea were distributed on different chromosomes. In addition, BraOLEs always appear in pairs with BolOLEs, and nearly all of these homologous BraOLE or BolOLE genes maintained a syntenic relationship with BnaOLEs. Given the close genetic relationship between B. rapa, B. oleracea and B. napus, a low level of gene loss was also observed in B. napus; for example, AtOLE1 had syntenic relationships with BraOLE1A01, BraOLE1A03 and BraOLE1A08, and syntenic genes of these three genes could be detected in the B. napus genome. In addition, we did not detect syntenic relationships of AtOLE10 in the A genome.
Fig. 4.
Synteny analysis map of oleosin genes. a Synteny analysis map of oleosin genes in the A genome in A. thaliana, B. rapa and B. napus chromosomes. The red lines indicate that these two genes from A. thaliana and B. napus are syntenic genes. The blue lines indicate that these two genes from A. thaliana and B. rapa are syntenic genes. The green lines indicate that these two genes from B. rapa and B. napus are syntenic genes. b Synteny analysis map of oleosin genes in the C genome in A. thaliana, B. oleracea and B. napus chromosomes. The red lines indicate that these two genes from A. thaliana and B. napus are syntenic genes. The blue lines indicate that these two genes from A. thaliana and B. oleracea are syntenic genes. The green lines indicate that these two genes from B. oleracea and B. napus are syntenic genes
Synonymous and nonsynonymous values were examined to assess the selective pressure on duplicated BnaOLE genes (Table 2). The largest Ka/Ks ratio of the gene pairs between B. napus and A. thaliana was 0.53, and the two largest Ka/Ks ratios were associated with AtOLE13. In contrast, the Ka/Ks ratio of AtOLE1 and the related orthologous OLE gene was the lowest. Different genes experience various degrees of selective pressure, and OLE1 is more highly conserved than other OLE genes, implying that OLE1 might perform more important functions than the other genes. In addition, the Ka/Ks ratio of T oleosins, such as AtOLE13 and AtOLE17, was relatively higher than that of the other oleosin lineages, which might be due to the high numbers of T oleosins.
Table 2.
Non-synonymous (Ka) and synonymous (Ks) nucleotide substitution rates for A. thaliana and B. napusole gene
| A. thaliana ID | A. thaliana gene | B. napus ID | B. napus gene | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|---|
| AT1G48990 | AtOLE5 | BnaC08g03140D | BnaOLE5 | 0.1408 | 0.61476 | 0.229032 |
| AT2G25890 | AtOLE6 | BnaA04g15210D | BnaOLE6A04 | 0.08926 | 0.53933 | 0.165502 |
| BnaC04g38160D | BnaOLE6C04 | 0.10466 | 0.54036 | 0.193686 | ||
| AT3G18570 | AtOLE8 | BnaA05g21590D | BnaOLE8A05 | 0.10166 | 0.46093 | 0.220554 |
| BnaA01g26680D | BnaOLE8A01 | 0.15212 | 0.41 | 0.371024 | ||
| BnaA03g34830D | BnaOLE8A03 | 0.11772 | 0.4827 | 0.243878 | ||
| BnaC05g34600D | BnaOLE8C05 | 0.09823 | 0.40958 | 0.239831 | ||
| BnaC01g34070D | BnaOLE8C01 | 0.13442 | 0.37374 | 0.359662 | ||
| BnaC03g40520D | BnaOLE8C03 | 0.12162 | 0.49017 | 0.248118 | ||
| AT3G27660 | AtOLE4 | BnaA02g29000D | BnaOLE4A02 | 0.08985 | 0.43793 | 0.20517 |
| BnaA09g02110D | BnaOLE4A09 | 0.10473 | 0.3736 | 0.280327 | ||
| BnaC02g37030D | BnaOLE4C02 | 0.10194 | 0.43748 | 0.233016 | ||
| AT4G25140 | AtOLE1 | BnaA01g14480D | BnaOLE1A01 | 0.05086 | 0.53381 | 0.095277 |
| BnaA03g47170D | BnaOLE1A03 | 0.06531 | 0.53037 | 0.12314 | ||
| BnaA08g14540D | BnaOLE1A08 | 0.05022 | 0.48812 | 0.102885 | ||
| BnaC01g17050D | BnaOLE1C01 | 0.04063 | 0.57389 | 0.070798 | ||
| BnaC07g39370D | BnaOLE1C07 | 0.08101 | 0.5862 | 0.138195 | ||
| BnaC08g11970D | BnaOLE1C08 | 0.05527 | 0.47217 | 0.117055 | ||
| AT5G07550 | AtOLE13 | BnaA03g02120D | BnaOLE13A03 | 0.18617 | 0.35046 | 0.531216 |
| BnaC03g03150D | BnaOLE13C03 | 0.17112 | 0.37186 | 0.460173 | ||
| AT5G40420 | AtOLE2 | BnaA07g14960D | BnaOLE2A07 | 0.08676 | 0.46651 | 0.185977 |
| BnaC04g32530D | BnaOLE2C04 | 0.1523 | 0.56784 | 0.268209 | ||
| BnaC06g12930D | BnaOLE2C06 | 0.08442 | 0.4767 | 0.177093 | ||
| AT5G51210 | AtOLE3 | BnaA10g05760D | BnaOLE3A10 | 0.08088 | 0.43697 | 0.185093 |
| BnaC09g27370D | BnaOLE3C09 | 0.08129 | 0.47551 | 0.170953 | ||
| AT5G61610 | AtOLE17 | BnaA03g40390D | BnaOLE17A03 | 0.24568 | 0.43828 | 0.560555 |
Gene cloning of AtOLEs and subcellular localization of oleosin proteins
Previous research has shown that the ortholog of OLE1 and OLE2 is located in the region of the quantitative trait locus (QTL) of seed oil content [37], which is evidence that oleosin might contribute to TAG accumulation. Therefore, we cloned three oleosin genes from A. thaliana and four of the homologous genes from B. napus to verify the effect of these genes on seed oil content in Arabidopsis. AtOLE1, AtOLE2 and AtOLE3 possess 173, 199 and 141 amino acids, respectively, and BnaOLEs possess relatively high numbers of amino acids. BnaOLE1, BnaOLE2, BnaOLE3 and BnaOLE4 possess 180, 220, 188 and 193 amino acids, respectively.
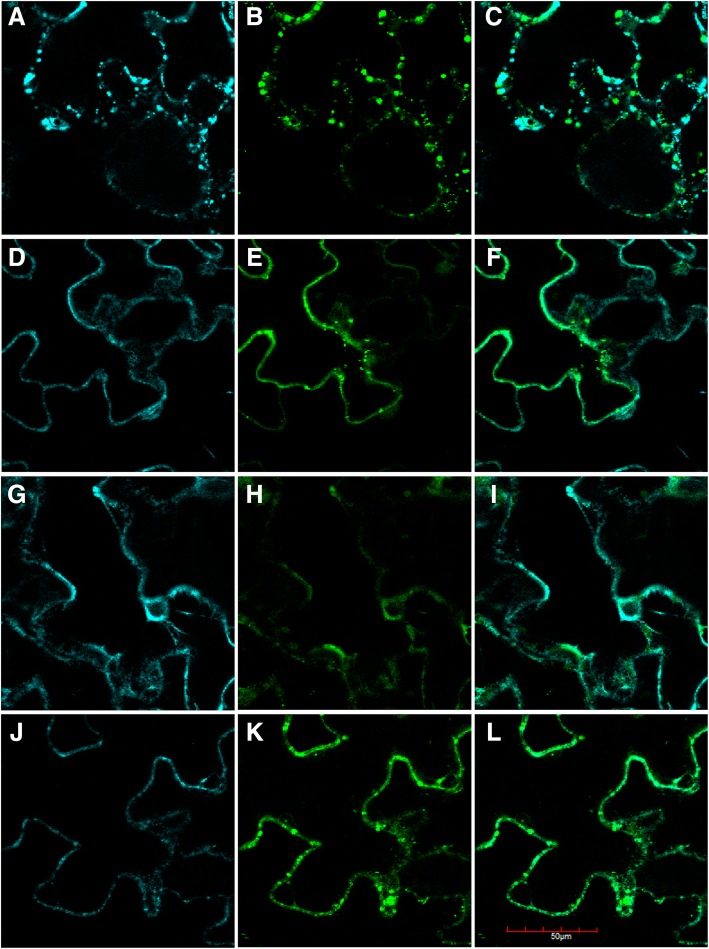
The subcellular localization of oleosins from B. napus was analyzed, and the vector pCambia1303-EGFP-DsRed3 containing the target protein was constructed, in which the fluorescent protein was attached to the oleosin protein. Therefore, we could check the oleosin distribution by monitoring the fluorescence signal in cells. The constructed plasmids were transformed into tobacco leaves for transient expression with Agrobacterium GV3101. The results revealed that the cellular localization of BnaOLEs partially overlapped with the cell membrane marker (Fig. 5). The results for AtOLEs were similar, and the fluorescence signals for these proteins coincided with the cell membrane and partially overlapped with the chloroplast (Additional file 3: Figure S3). This result indicated that the oleosin protein may be located in the cell membrane before oil body formation.
Fig. 5.
Subcellular localization of BnaOLEs. a, b, and c represent the subcellular location of BnaOLE1; d, e and f represent the subcellular location of BnaOLE2; g, h and i represent the subcellular location of BnaOLE3; and j, k and l represent the subcellular location of BnaOLE4. a, d, g, j: plasma membrane-CFP marker; b, e, h, k: EGFP fluorescence; c, f, i, l: merged fluorescence of EGFP and plasma membrane-CFP marker. Scale bar: 50 μm
Overexpression of OLEs in Arabidopsis could affect the fatty acid content
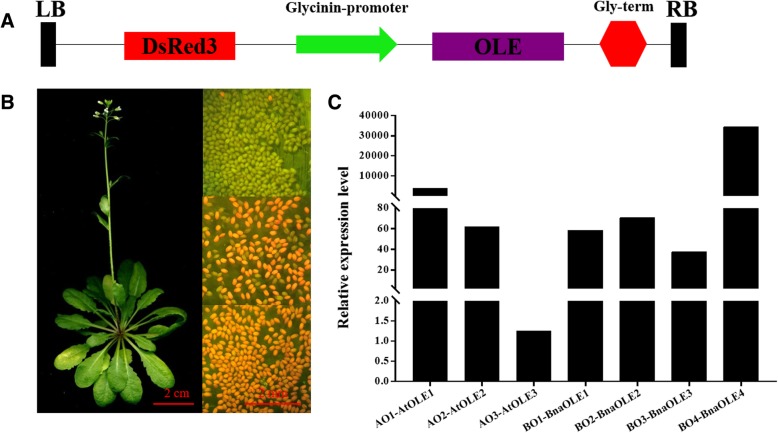
The plant expression vector with the glycinin promoter was constructed and then transferred into Arabidopsis to generate the OLE overexpression lines. DsRED, a type of fluorescent protein, was also added to the vector (Fig. 6). It was easy to identify the transgenic seeds by observing the red fluorescence through a filter. This approach is more convenient than the selection of transgenic plants based on antibiotic resistance. The transgenic lines overexpressing OLEs showed no visible morphological variation compared with the WT. To investigate the influence of overexpression of these genes on lipid metabolism, homozygous transgenic T3 lines were used to analyze the oil content and fatty acid profiles using gas chromatography.
Fig. 6.
Plant transformation and selection of transgenic seeds. a Schematic representation of the plant expression vector used for plant transformation. b Different generations of transgenic plants under green light. The red seeds and siliques represent the transgenic lines, while the green represents the WT. c Relative expression of target genes in the siliques of the T3 generation of transgenic plants. The expression of AtOLE3 in WT was assigned as 1
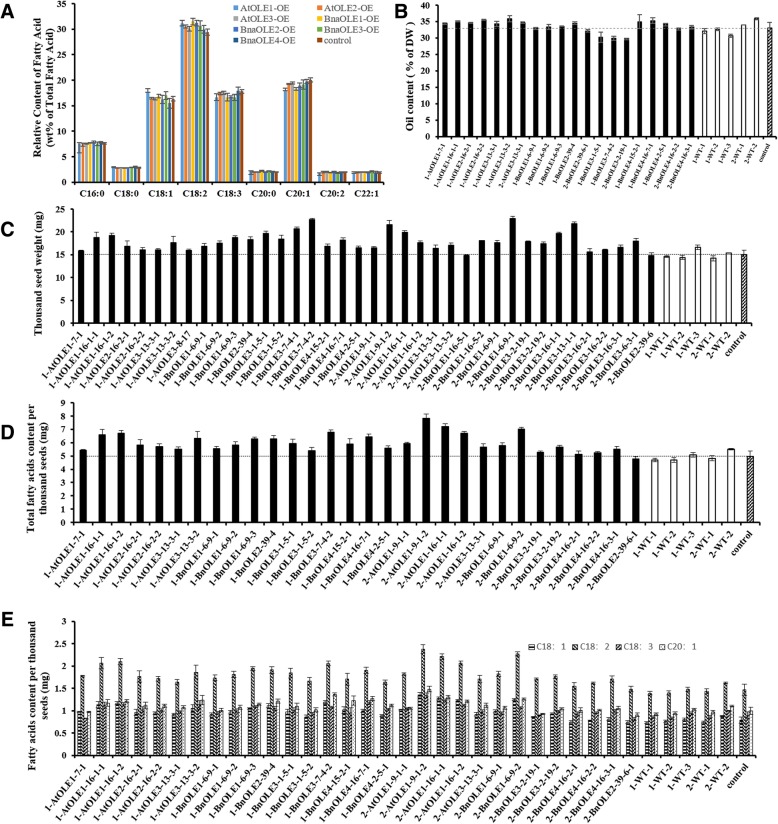
It was revealed that the oil content of transgenic lines showed different variation compared with the WT (Fig. 7a, b, Table 3). The seed oil content of OLE-OE in different lines varied from 29.2 to 35.6%. Compared with the WT (with an oil content of 32.3%), the oil content in most of the transgenic lines increased slightly, except in BnaOLE3-OE. Further analysis showed that the oil content of the AtOLE3-OE seeds was the highest, reaching 35.8%, an increase of 10.8% compared with the WT. The average oil content also increased to a similar extent in AtOLE1-OE (34.7%), AtOLE3-OE (35.0%), BnaOLE1-OE (33.3%), BnaOLE2-OE (33.3%) and BnaOLE4-OE (34.8%). On the other hand, the oil content in the BnaOLE3-OE seeds decreased to a value of only 29.9%.
Fig. 7.
Oil content and fatty acids profiles in Arabidopsis seeds. a Relative levels of fatty acids in transgenic seeds and the WT; (b) oil content in transgenic seeds and the WT; (c) thousand-seed weight of transgenic plants and the WT; (d) total fatty acid weight per thousand seeds of transgenic plants and the WT; (e) levels of different types of fatty acids per thousand seeds of transgenic plants and the WT. At least three replicates were used for each sample. T-test was used to measure significance differences between lines and the transgenic lines showed significant difference with the control
Table 3.
Relative content of three C18 fatty acids of transgenic seeds
| AtOLE1-OE | AtOLE2-OE | AtOLE3-OE | BnOLE1-OE | BnOLE2-OE | BnOLE3-OE | BnOLE4-OE | WT | |
|---|---|---|---|---|---|---|---|---|
| C18:1 | 17.77124 | 16.30847 | 16.34432 | 16.85946 | 16.26213 | 16.99048 | 15.48557 | 16.36811 |
| C18:2 | 31.1659 | 30.47247 | 30.11299 | 31.50722 | 31.20111 | 30.65505 | 29.96506 | 29.40841 |
| C18:3 | 16.68444 | 17.51728 | 17.55592 | 16.67468 | 16.76694 | 16.61466 | 17.91633 | 17.8015 |
The fatty acid profile also changed, especially for the 18C unsaturated fatty acids. The levels of all three of the main 18C unsaturated fatty acids in the seeds of the OLE lines increased or decreased to different degrees. In addition to BnaOLE4, the transgenic lines showed increased oleic and linoleic acid levels; however, the relative level of linolenic acid decreased. In addition, the eicosaenoic acid content also showed a decreasing trend. Among all of the transgenic lines, the fatty acid profile of AtOLE1-OE seeds showed the maximum variation. The oleic acid content increased from 15.8 to 17.2%, and the linoleic acid content increased from 29.5 to 32.6%, accompanied by decreased linolenic acid (from 17.8 to 15.3%) and eicosaenoic acid (from 20.1 to 17.8%) levels. BnaOLE1 exhibited the largest increase in linoleic acid content (7.1%), followed by BnaOLE2 (6.1%) and BnaOLE3 (6.0%). As a consequence, the linolenic acid and eicosaenoic acid levels of these three transgenic lines decreased more than those of the other transgenic lines.
In addition to lipid metabolism, seed weight was also affected in the transgenic lines (Fig. 7c), and it was revealed that the thousand-seed weight (TSW) in all of the transgenic lines increased to different degrees. BnaOLE1-OEs had the largest average TSW, which was 24.4% higher than that of the WT. Considering the seed oil content and the TSWs of the transgenic lines, it was concluded that overexpression of oleosin could increase the total fatty acid production per seed. The linoleic acid content can reach 1.98 mg per thousand seeds in the AtOLE1-OE lines, while the level in the control is only 1.44 mg, representing a 34.7% increase in the AtOLE1-OE lines, more than that in the other transgenic lines. The oleic acid content per thousand seeds in the transgenic lines also showed a similar increase as the linoleic acid content. Moreover, the linolenic acid and eicosaenoic acid levels showed a slight increase compared with the levels in the WT. It was revealed that AtOLE1-OE contains 1.03 mg of linolenic acid and 1.12 mg of eicosaenoic acid per thousand seeds, while the WT seeds contain 0.9 mg linolenic acid and 0.99 mg eicosaenoic acid. In a word, overexpression of OLE might affect the relative content of fatty acids, but could enhancing the linoleic acid content without decreasing the yield of other fatty acids.
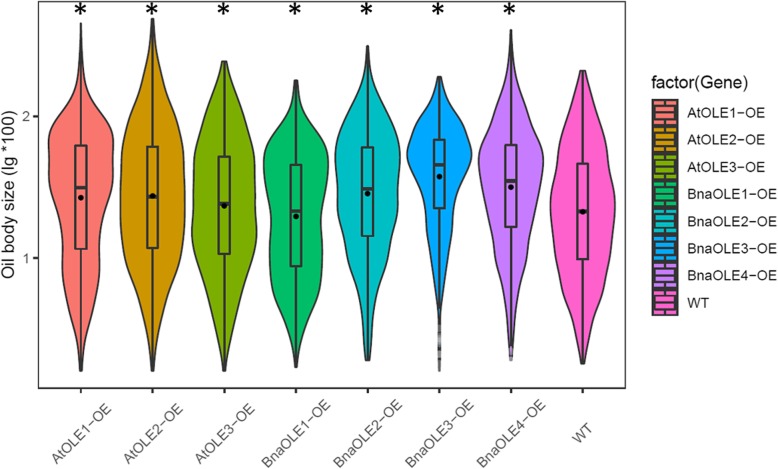
Overexpression of OLEs in Arabidopsis could affect oil body size
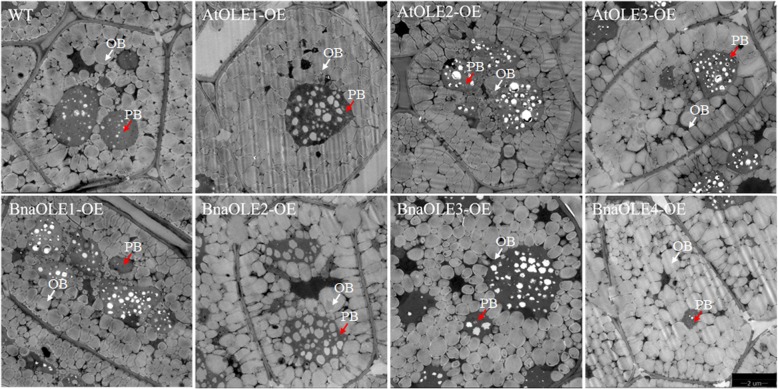
The oil body size in the seeds of transgenic lines was also analyzed. The average oil body size of most of the transgenic lines was significantly higher than that of the WT, except for BnaOLE1-OEs (P < 0.01) (Fig. 8, Fig. 9). The average oil body size of BnaOLE1-OEs was only 0.29 μm2, whereas the average oil body size of the WT was 0.33 μm2. The average oil body size of all six transgenic lines ranged from 0.41 to 0.48 μm2, representing a 24 to 45% increase compared to the control. Further analysis revealed that the largest oil body was observed in AtOLE2-OEs, reaching 4.842 μm2, more than two times larger than the largest oil body in WT seeds (2.08 μm2). Considering the different oil body size distributions, in cotyledons of BnaOLE1-OEs, the percentage of oil bodies with sizes less than 0.6 μm2 was similar to that in the WT, whereas the number of oil bodies with sizes less than 0.9 μm2 was greater than that in the WT. Taken together, these results showed that overexpression of BnaOLE1 resulted in relatively few large oil bodies, which led to a relatively small average oil body size (Additional file 4: Figure S4).
Fig. 8.
TEM images of oil bodies in cotyledons of transgenic seeds. (Bar = 2 μm). WT represents the cotyledons of wild-type seeds; AtOLE1-OE, AtOLE2-OE, AtOLE3-OE, BnOLE1-OE, BnOLE2-OE, BnOLE3-OE and BnOLE4-OE represent the cotyledons of transgenic seeds overexpressing AtOLE1, AtOLE2, AtOLE3, BnOLE1, BnOLE2, BnOLE3 and BnOLE4, respectively. OB indicates oil bodies, and PB indicates protein bodies
Fig. 9.
Distribution of different sizes of oil bodies in transgenic seeds. The Y-axis represents the logarithm of oil body area to the base 10 of the concentration. Different colors represent different transgenic lines. T-test was used to measure significance differences between lines and the transgenic lines showed significant difference with the control (* represent that P < 0.05)
Overexpression of OLE genes could affect freezing tolerance
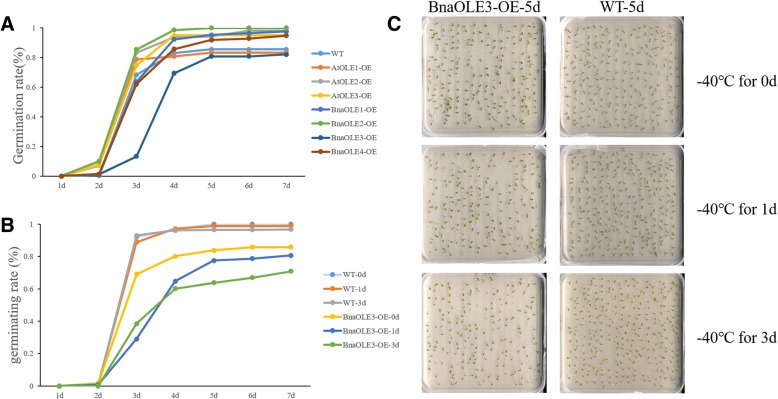
A previous study showed that oleosin might increase the resistance of seeds to freezing, so we asked whether the evaluated oleosin level could reduce the injuries caused by freezing treatment. The transgenic and WT seeds were treated by freezing at − 40 °C for 1 day, followed by sowing on MS medium containing 1% agar. Surprisingly, the freezing treatment did not affect seed germination, except for seeds expressing BnaOLE3 (Fig. 10). Three days after sowing, the germination rate of seeds expressing BnaOLE3 was lower than that of other seeds. Further analysis showed that prolonged freezing time could harm seed germination severely. After treatment with low temperature for 3 days, the germination rate decreased from 85.7 to 70%. These results show that heterologous expression of BnaOLE3 could reduce cold resistance.
Fig. 10.
Germination rate of Arabidopsis seeds after freezing treatment. a Germination rate of Arabidopsis seeds after freezing treatment for 24 h. b Germination rate of Arabidopsis seeds overexpressing BnaOLE3 and WT seeds after freezing treatment for different durations. T-test was used to measure significance differences between lines and the transgenic lines showed significant difference with the control. c Germination rate of Arabidopsis seeds overexpressing BnaOLE3 and WT seeds after freezing treatment for different durations 5 days after sowing
Discussion
Plant seeds that store lipids in oil bodies are used for seed germination and other physiological and biochemical metabolic processes. The oil body consists of TAG, a phospholipid monolayer enclosing the neutral lipids and oleosins inserted into the half-unit membrane [5, 7]. From primitive green algae to higher plants, oleosin is widely distributed in different species [21], indicating the important roles of oleosin in oil body biogenesis and stabilization. Due to the high conservation of oleosin, many studies have focused on the evolutionary relationships of oleosins [21, 23]. It was revealed that there are six lineages of oleosins (P, U, SL, SH, T and M) in plants; some lineages exist in only a single family, such as T oleosin and M oleosin, which are found in only Lauraceae and Brassicaceae, respectively. The gene evolution of oleosins has been reviewed for some species; however, the identification and annotation of oleosins in B. napus, an important oil crop worldwide, has received little attention. In the present study, 48 oleosin genes from the B. napus genome were identified based on the conserved domain, and these genes were divided into four lineages of oleosins (U, SL, SH, and T). We did not detect P oleosins and M oleosins in B. napus; these lineages were only found in algae and Lauraceae, respectively. Nearly half of the identified oleosins were T oleosins, and the other three lineages were present at similar levels. A previous study has shown that most oleosins possess no introns [23]. Liu et al [38] hypothesized that introns of oleosins were gained early in evolution and then lost, so most of oleosins possess no introns. The present results revealed the presence of at least one intron in the oleosin sequences, except U oleosins, in B. napus. Which was similar with the previous report that there are few introns in the oleosin genes of other species except for Brassicaceae plants [16]. And in Brassicaceae plants, most of SL, SH and T oleosins contain introns and U oleosins contain few oleosins. Although SH oleosins and SL oleosins evolved from the U oleosins [5], there is a difference in intron number between these oleosins; SH oleosins have more introns than SL oleosins. Based on the genomic structures of oleosins, BnaOLE23 and BnaOLE35 were the two exceptions among U oleosins, each possessing six introns; however, the homologous genes in Arabidopsis AtOLE5 had no intronic structure. These two genes contain more nucleotides than other oleosin genes and it might indicate that the increased gene length during evolution lead to the more complex gene structures. Although the intron structures and sequences differed, all of the introns in the oleosins were U2-type splice GT-AG introns [33, 34].
Gene duplication increased the gene number and led to functional differentiation of genes for adaptation to the environment during evolution. Many studies have revealed that Brassica species have undergone whole-genome duplication (WGD) events during evolution. In addition, several independent lineage-specific WGD events have been identified in Brassicaceae [35, 39]. For the WGT event in Brassica species, the BnaOLE genes experienced gene duplication events. Genetic structure and phylogenetic analyses showed that BnaOLEs share a close phylogenetic relationship and similar structure with corresponding homologous genes in three other Brassicaceae plants. The results were also further confirmed by the Ka/Ks ratios (less than one), suggesting that the oleosin genes in Brassicaceae plants had experienced purifying selection (Additional file 6: Table S2). The tandemly duplicated genes were also detected in the BnaOLE gene family, and all twelve tandemly duplicated genes belonged to T oleosins. One underlying reason might be that T oleosins formed gene clusters in the Arabidopsis genome. Gene duplication played an important role in the genetic evolution of this locus; however, redundant gene numbers led to relatively low conservation because the Ka/Ks ratio of T oleosins was relatively higher than that of the other three lineages.
Oleosin was first identified in maize in 1990 [15], and previous studies have shown that oleosin, the main structural protein of the oil body, can keep oil droplets stable and prevent oil body aggregation during seed dehydration [40]. However, the role of oleosin in the storage and activation of lipids in seeds and other cellular activities remains unclear. Oleosins play an important role in the formation of oil droplets and accumulation of lipids. Reduction in oleosin content in rice seeds can lead to decreased oil content and increased oil body size in seeds [26]. However, few studies have investigated the relationship between the oleosin protein content and oil body size. In the present study, three Arabidopsis oleosin genes and their homologs from B. napus were selected for verification of their effect on seed oil content in Arabidopsis. Homozygous T3 transgenic seeds were used for all further analyses. The cotyledons of the seeds were used for transmission electron microscopy (TEM) analysis; the seed oil body sizes of the transgenic lines were significantly higher than those of the WT, except for BnaOLE1 (P < 0.01). BnaOLE1 overexpression resulted in a significant decrease in oil body size compared with that in WT seeds (P < 0.01), with an average size of 0.292 to 0.332 μm2, which is consistent with the previous hypothesis that AtOLE1 might restrict oil body size. Considering the different oil body size distributions, similar to the overall result, there were more small oil bodies in transgenic seeds than in WT seeds. In the BnaOLE1-OE lines, the percentage of oil bodies with sizes less than 0.9 μm2 was higher than that in the WT, leading to a small oil body size overall. In contrast, the percentage of oil bodies in other transgenic seeds was higher than that in the WT, and the largest oil body, which was two times larger than the largest oil body in the WT, was observed in AtOLE2 seeds. These results indicate that abnormal gene expression could disrupt the natural oil body formation process, which could result in the emergence of a relatively large oil body, even an unusually large oil body, instead of a simple negative correlation between oil body size and oleosin content.
After knocking out the Arabidopsis oil body protein gene, the oleic acid (C18:1) content in seeds decreased, and the eicosenoic acid (C20:1) content increased [29]. On the other hand, overexpression of castor oleosin in A. thaliana can greatly increase the accumulation of hydroxyl fatty acids in seeds [29], and coexpression of oleosin with other important lipid synthesis-related genes, such as DGAT, FADX and WRI1, could further improve target production [41–44]. For example, coexpression of ScDGA1 and AtOLE3 resulted in a nearly 50% increase in oil content compared with transformants that only expressed the single gene ScDGA1 [42]. In the present study, the linoleic acid (C18:2) content in transgenic seeds was higher than that in the control, and the eicosaenoic acid (C20:1) content was lower than that in the WT seeds, which was opposite trend compared to that in the ole mutant seeds reported previously [45]. This result could be partly explained by the fact that oleosin can be coexpressed with FAD2 and FAD3 in Arabidopsis [46, 47]; therefore, the expression of oleosin could affect the levels of the substrate and product of the two related enzymes. Previous research has shown that overexpression of oleosin from Carthamus tinctorius L. and soybean could alter the seed oil content [48, 49]. Although in the present study, the oil content of the transgenic lines showed different variation trends, the highest oil content reached 37.9%, which was 14.8% higher than that in the WT. Lipids are the main form of storage in Arabidopsis seeds, and changes in oil content may affect seed quality. Alteration of the expression of lipid metabolism-related genes such as DGAT, FAX1 and WRI1 affects oil content and seed weight [50–52]. The most important phenomenon observed in the present study was that the TSWs of the transgenic lines increased. Taken together, these results showed that overexpression of OLEs could increase oil production and the linoleic acid yield. For example, the linoleic acid content in AtOLE1-OE seeds reached 2.37 mg per thousand seeds, while the value was only 1.46 mg for the control. The highest oil output per thousand seeds of the transgenic lines reached 7.8 mg, which was 57% higher than the average yield of the WT. These results suggested that increased oleosin content might promote lipid synthesis.
Conclusion
In this study, we identified 48 oleosin genes in B. napus and divided these genes into four types, and at least one intron was found in all the oleosin sequences except U oleosins. Over half of the BnaOLE genes are associated with segmental duplications. Synteny analysis showed that most of the oleosin genes in B. napus are relatively conserved, and all of them were faced purifying selection pressure in evolution. In addition, several important oleosins from Arabidopsis and B. napus were cloned, and the functions of these oleosins in Arabidopsis seeds were analyzed. It was shown that overexpression of oleosin genes in A. thaliana has a weak effect on seed oil content but could increase the linoleic acid content (13.3% at most) and decrease the peanut acid content (11% at most) compared to the levels in the WT. The TSWs also increased, which could lead to increased total lipid production. The oil body size in transgenic seeds was larger than that in the WT, except for BnaOLE1. This study provides a foundation for future studies regarding the oleosin superfamily in B. napus.
Methods
Identification of oleosin genes in B. napus and other related species
Arabidopsis genes were searched in the TAIR and Phytozome databases [53] using oleosin as the key word query and then confirmed using the hidden Markov model (HMM) with the Pfam [54], SMART [55] and NCBI Conserved Domain Search databases [56]. The oleosin genes were identified in B. napus based on homologous sequences from A. thaliana using the BLAST search program in the CNS-Genoscope database [57]. The oleosin gene sequences from B. rapa and B. oleracea were obtained from the BRAD database [58, 59]. The oleosin sequences from B. napus, B. rapa, and B. oleracea were also confirmed in the Pfam, SMART and NCBI Conserved Domain Search databases. Redundant sequences and some proteins containing no characteristic oleosin domain were removed manually. According to the gene synteny with oleosins of Arabidopsis and the corresponding chromosomal distribution, a univocal name consisting of two italic letters indicating the source organism and the family was assigned to each oleosin gene (Additional file 5: Table S1). When searching the sequence information, the number of amino acids, coding sequence (CDS) lengths, intron and exon numbers, and chromosomal locations of the BnaOLE genes were obtained from the CNS-Genoscope and BRAD databases. ExPASy was used to calculate the physicochemical parameters, including the molecular weight (kDa), pI and GRAVY, of all the identified oleosin proteins [60].
Gene structure, multiple sequence alignment and phylogenetic analysis of oleosin genes
After obtaining complete oleosin CDS and genomic sequences from the CNS-Genoscope, TAIR and BRAD databases, GSDS [61] was used to illustrate the exon-intron structures of the oleosin genes and the intron phases. If an intron is between two complete codons, then the intron phase was defined as 0; if an intron is after the first or second nucleotide within the codon, the intron phase was defined as phase 1 or phase 2, respectively. Multiple sequence alignment of the oleosin protein sequences of A. thaliana, B. rapa, B. oleracea and B. napus was performed using ClustalW software [62]. Then, the conserved central domain was used to construct an unrooted cladogram using MEGA 6 with the neighbor-joining (NJ) method, and a bootstrap analysis was conducted using 1000 replicates [63]. These conserved portions were also predicted by MEME to show the highly conserved amino acids of oleosin genes [64].
Chromosomal location and gene duplication of the BnaOLE family genes
The chromosomal locations of the BnaOLE genes from the B. napus database and the syntenic relationships between the BnaOLEs and AtOLEs, BraOLEs, and BolOLEs were acquired with the syntenic genes search tool in the BRAD database. Then, Circos was used to show the oleosin distribution in the B. napus genome and the syntenic relationships in the four types of plants [65]. TBtools was used to calculate the nonsynonymous and synonymous substitution rates of every paralogous pair of oleosin genes between A. thaliana and B. napus. The Ka/Ks ratio represents the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks). When the Ka/Ks ratio was less than 1, purifying selection occurred, and otherwise, positive selection occurred. When two genes with sequence distances less than 50 kb belong to the same gene family, we defined these genes as tandemly duplicated genes.
Gene cloning, vector construction and gene transformation
All plant materials and plant expression vectors used in this study were stored in the laboratory of Prof. Li Maoteng of Huazhong University of Science and Technology. Arabidopsis was cultured at 21 ± 2 °C with a 16 h light/8 h dark photoperiod at a relative humidity of 60–90%. B. napus cultivar was grown in the experimental field of the Huazhong Agricultural University (30°36′ N, 104°18′ E), Wuhan, China. First, the primers used to clone the OLE genes were designed using Oligo 6.0 based on the gene sequence obtained from the TAIR and CNS-Genoscope databases, and restriction sites were added to the primers. The RNAprep Pure Plant Kit (TIANGEN BIOTECH (BEIJING) CO., LTD) was used to isolate total RNA from the developing siliques of A. thaliana and B. napus, and the PrimeScript™ RT Master Mix Kit (TaKaRa) was used to synthesize first-strand DNA according to the manufacturer’s instructions. This DNA was used as a template for subsequent PCR amplification. The target genes were integrated into the plant expression vector using the In-Fusion® HD Cloning Kit (Clontech). All vectors used in this study contained DsRED, a type of fluorescent protein that can emit red light under excitation by green light.
The recombinant vector was transformed into Col Arabidopsis by the floral dip method by using Agrobacterium tumefaciens. We could identify the transgenic seeds by observing the red light with the help of a filter. In addition, T3-generation transgenic Arabidopsis was used to investigate plant phenotypes. Twenty days after flowering, the siliques of transgenic Arabidopsis were harvested to analyze the expression of target genes. SYBR Green Real-time PCR Master Mix was used to perform RT-qPCR.
Measurement of oil content and fatty acid compositions
The seeds were first weighed using a semi-microanalytical balance and then placed on a glass plate. The seeds were scattered such that there was no overlap among seeds for further analysis. Then, the Seed Count image analysis system (WSeen Detection of Zhejiang Sci-tech University, Hangzhou, Zhejiang, China) was used to scan the seeds and analyze the seed number, seed area, seed length and seed width; the TSW was also determined.
The method for oil content measurement has been previously reported, which we used with some modification [66]. Five milligrams of dry seeds were used for oil content and fatty acid analysis. After placing the seeds into a glass tube, 1 ml of 2.5% sulfuric acid-methanol solution, 0.4 ml of toluene and 0.2 ml of a 2 mg/ml C17:0 solution in toluene were added. The mixture was vortexed and then heated in a 90 °C water bath for 1 h, and then, 1.8 ml of ddH2O and 1 ml of hexane were added after the tube had cooled. After 15 min, the supernatant was filtered using a 0.45-μm microporous membrane. The filtrate was used to determine the fatty acid content by GC using an Agilent 7890A instrument. At least three replicate samples were examined for all of the experiments.
Measurement of oil body sizes
The transgenic seeds that emitted red light under excitation by green light were selected for TEM analysis. The dry seeds were first fixed using 2.5% glutaraldehyde solution, and then the manually isolated cotyledons were sliced and photographed. All the associated procedures have been described previously [11], and all the TEM imaging was completed by the Wuhan Institute of Virology. At least three samples were used to measure oil body size. At least 3 TEM images of the cotyledons of each transgenic line were used to measure oil body size using ImageJ. T-test was used to measure significance differences between lines.
Freezing treatment
The method for the freezing treatment has been previously reported by Shimada et al. [6], which we used with some modifications. The dry Arabidopsis seeds were treated by freezing at − 40 °C for 1 or 3 days. Then, the dry seeds were first washed with 70% ethanol for 1 min and then washed 3 times with ddH2O. Fifty percent disinfectant 84 was used to further eliminate bacteria, and the samples were then washed with ddH2O 3 times. Then, the seeds were sown on MS medium containing 1% agar. We took a picture of the dish every 24 h and determined the germination rate.
Additional files
Figure S1 Sequence alignment and MEME analysis of B. napus oleosin genes. (A) MEME logo of the conserved Pro knot; (B) sequence alignment of the 72 conserved amino acids in 48 B. napus oleosin proteins. The relatively highly conserved amino acids are marked in different colors. (PNG 3427 kb)
Figure S2 Genetic structure of A. thaliana, B. oleracea, B. rapa and B. napus oleosin genes. The motifs in oleosin are shown on the left, and the genetic structure is shown on the right. The numbers near the introns indicate the intron phase. (PNG 579 kb)
Figure S3. Subcellular localization of AtOLE1, AtOLE2, AtOLE3 and AtOLE4. A, B, C, and D represent AtOLE1; E, F, G, and H represent AtOLE2; I, J, K, and L represent AtOLE3; and M, N, O, and P represent AtOLE4. A, E, I, M: EGFP fluorescence; B, F, G, N: chloroplast autofluorescence; C, G, K, N: background; D, H, L, P: merged EGFP and chloroplast fluorescence. (PNG 3259 kb)
Figure S4 Distribution of different sizes of oil bodies in transgenic seeds. Different sizes of oil bodies are marked with different colors. (TIF 12720 kb)
Table S1 Gene names and the corresponding gene IDs. (XLS 34 kb)
Table S2 Ka/Ks ratios between all of the oleosins from A. thaliana, B. oleracea, B. rapa, and B. napus. (XLS 1511 kb)
Acknowledgments
Not applicable
Abbreviations
- CDS
Coding sequence
- DGAT
Diacylgycerol acyltransferase
- HMM
Hidden Markov model
- MEME
Multiple Expectation Maximization for Motif Elicitation
- OBAP1
Oil body-associated protein 1
- QTL
Quantitative trait locus
- TAG
Triacylglycerol
- TAIR
the Arabidopsis Information Resource
- TEM
Transmission electron microscopy
- TSW
Thousand-seed weight
- WGD
Whole-genome duplication
- WT
Wild type
Authors’ contributions
KC performed overall data analysis and wrote the manuscript. YY and SL constructed the vectors and performed the plant transformation experiment. YY provided guidance for gene transformation and seed lipid analysis, led the study and modified the manuscript. ZG and KZ helped determined the oil content and fatty acid composition. YL provided guidance for gene family analysis. LZ, HC, and WZ helped modify some Figs. ML designed, led and coordinated the overall study. All the authors read and approved the final manuscript. KC and YY contributed equally to this work.
Funding
The work was supported by the National Natural Science Foundation of China (31871656), the National Program on Key Basic Research Project (2015CB150205) and the New Century Talents Support Program of the Ministry of Education of China (NCET110172). The funding bodies were not involved in the design of the study, data collection, interpretation of data, or in writing the manuscript.
Availability of data and materials
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kang Chen and Yongtai Yin contributed equally to this work.
Contributor Information
Kang Chen, Email: ace.ckang@foxmail.com.
Yongtai Yin, Email: yinyongtaishuai@163.com.
Si Liu, Email: 1040295512@qq.com.
Zhenyi Guo, Email: 2712158874@qq.com.
Kai Zhang, Email: kaizhanghb@163.com.
Yu Liang, Email: 262621874@qq.com.
Lina Zhang, Email: zhangln0217@163.com.
Weiguo Zhao, Email: zhaoweiguo0517@126.com.
Hongbo Chao, Email: chaohongbo@yeah.net.
Maoteng Li, Email: limaoteng426@hust.edu.cn.
References
- 1.Liu S, Fan C, Li J, Cai G, Yang Q, Wu J, Yi X, Zhang C, Zhou Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet. 2016;129(6):1203–1215. doi: 10.1007/s00122-016-2697-z. [DOI] [PubMed] [Google Scholar]
- 2.Lu S, Sturtevant D, Aziz M, Jin C, Li Q, Chapman KD, Guo L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018;94(6):915–932. doi: 10.1111/tpj.13959. [DOI] [PubMed] [Google Scholar]
- 3.Niu Y, Wu GZ, Ye R, Lin WH, Shi QM, Xue LJ, Xu XD, Li Y, Du YG, Xue HW. Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana. Mol Plant. 2009;2(5):1107–1122. doi: 10.1093/mp/ssp042. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Huang J, Chen F, Xu F, Ni X, Xu H, Wang Y, Jiang C, Wang H, Xu A, et al. Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus. Theor Appl Genet. 2012;124(2):407–421. doi: 10.1007/s00122-011-1716-3. [DOI] [PubMed] [Google Scholar]
- 5.Huang AHC. Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 2018;176(3):1894–1918. doi: 10.1104/pp.17.01677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 2008;55(5):798–809. doi: 10.1111/j.1365-313X.2008.03553.x. [DOI] [PubMed] [Google Scholar]
- 7.Huang AH. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110(4):1055–1061. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40(5):325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 9.Frandsen GI, Mundy J, Tzen JT. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant. 2001;112(3):301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 10.Miquel M, Trigui G, D'Andrea S, Kelemen Z, Baud S, Berger A, Deruyffelaere C, Trubuil A, Lepiniec L, Dubreucq B. Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol. 2014;164(4):1866–1878. doi: 10.1104/pp.113.233262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siloto RMP. The accumulation of Oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell Online. 2006;18(8):1961–1974. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatla SC, Kaushik V, Yadav MK. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol Adv. 2010;28(3):293–300. doi: 10.1016/j.biotechadv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Taurino M, Costantini S, De Domenico S, Stefanelli F, Ruano G, Delgadillo MO, Sanchez-Serrano JJ, Sanmartin M, Santino A, Rojo E. SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 2018;176(2):1531–1546. doi: 10.1104/pp.17.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Ribera I, La Paz JL, Repiso C, Garcia N, Miquel M, Hernandez ML, Martinez-Rivas JM, Vicient CM. The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol. 2014;164(3):1237–1249. doi: 10.1104/pp.113.233221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vance VB, Huang AH. The major protein from lipid bodies of maize. Characterization and structure based on cDNA cloning. J Biol Chem. 1987;262(23):11275–11279. [PubMed] [Google Scholar]
- 16.Huang MD, Huang AH. Bioinformatics reveal five lineages of Oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol. 2015;169(1):453–470. doi: 10.1104/pp.15.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JC, Lin RH, Huang HC, Tzen JT. Cloning, expression and isoform classification of a minor oleosin in sesame oil bodies. J Biochem. 1997;122(4):819–824. doi: 10.1093/oxfordjournals.jbchem.a021828. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Bih FY, Learn GH, Ting JT, Sellers C, Huang AH. Oleosins in the gametophytes of Pinus and Brassica and their phylogenetic relationship with those in the sporophytes of various species. Planta. 1994;193(3):461–469. doi: 10.1007/BF00201827. [DOI] [PubMed] [Google Scholar]
- 19.Keddie JS, Hubner G, Slocombe SP, Jarvis RP, Cummins I, Edwards EW, Shaw CH, Murphy DJ. Cloning and characterisation of an oleosin gene from Brassica napus. Plant Mol Biol. 1992;19(3):443–453. doi: 10.1007/BF00023392. [DOI] [PubMed] [Google Scholar]
- 20.Huang CY, Huang A. Unique motifs and length of hairpin in Oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol. 2017;174(4):2248–2260. doi: 10.1104/pp.17.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang NL, Huang MD, Chen TL, Huang AH. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 2013;161(4):1862–1874. doi: 10.1104/pp.112.212514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9(8):1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Y, Zhu RL, Mishler BD. Evolution of oleosin in land plants. PLoS One. 2014;9(8):e103806. doi: 10.1371/journal.pone.0103806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baud S, Lepiniec L. Physiological and developmental regulation of seed oil production. Prog Lipid Res. 2010;49(3):235–249. doi: 10.1016/j.plipres.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14(4):504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu YY, Chou YR, Wang CS, Tseng TH, Chen LJ, Tzen JT. Different effects on triacylglycerol packaging to oil bodies in transgenic rice seeds by specifically eliminating one of their two oleosin isoforms. Plant Physiol Biochem. 2010;48(2–3):81–89. doi: 10.1016/j.plaphy.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Tzen J, Cao Y, Laurent P, Ratnayake C, Huang A. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 1993;101(1):267–276. doi: 10.1104/pp.101.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Wang X, Ren K, Li K, Wei M, Wang W, Sheng X. Light deprivation-induced inhibition of chloroplast biogenesis does not arrest embryo morphogenesis but strongly reduces the accumulation of storage reserves during embryo maturation in Arabidopsis. Front Plant Sci. 2017;8:1287. doi: 10.3389/fpls.2017.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Fulda M, Wallis JG, Browse J. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 2006;45(5):847–856. doi: 10.1111/j.1365-313X.2005.02636.x. [DOI] [PubMed] [Google Scholar]
- 30.Levesque-Lemay M, Chabot D, Hubbard K, Chan JK, Miller S, Robert LS. Tapetal oleosins play an essential role in tapetosome formation and protein relocation to the pollen coat. New Phytol. 2016;209(2):691–704. doi: 10.1111/nph.13611. [DOI] [PubMed] [Google Scholar]
- 31.Hu Z, Wang X, Zhan G, Liu G, Hua W, Wang H. Unusually large oilbodies are highly correlated with lower oil content in Brassica napus. Plant Cell Rep. 2009;28(4):541–549. doi: 10.1007/s00299-008-0654-2. [DOI] [PubMed] [Google Scholar]
- 32.Gu J, Chao H, Wang H, Li Y, Li D, Xiang J, Gan J, Lu G, Zhang X, Long Y, et al. Identification of the relationship between oil body morphology and oil content by microstructure comparison combining with QTL analysis in Brassica napus. Front Plant Sci. 2017;7:1989. doi: 10.3389/fpls.2016.01989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. CELL. 1997;91(7):875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 34.Bartschat S, Samuelsson T. U12 type introns were lost at multiple occasions during evolution. BMC Genomics. 2010;11:106. doi: 10.1186/1471-2164-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng F, Wu J, Wang X. Genome triplication drove the diversification of Brassica plants. Hortic Res. 2014;1:14024. doi: 10.1038/hortres.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng F, Wu J, Fang L, Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci. 2012;3:198. doi: 10.3389/fpls.2012.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao H, Wang H, Wang X, Guo L, Gu J, Zhao W, Li B, Chen D, Raboanatahiry N, Li M. Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci Rep. 2017;7:46295. doi: 10.1038/srep46295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Sun Y, Su W, Yang J, Liu X, Wang Y, Wang F, Li H, Li X. Species-specific size expansion and molecular evolution of the oleosins in angiosperms. GENE. 2012;509(2):247–257. doi: 10.1016/j.gene.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Rana D, van den Boogaart T, O'Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004;40(5):725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- 40.Cummins I, Hills MJ, Ross JH, Hobbs DH, Watson MD, Murphy DJ. Differential, temporal and spatial expression of genes involved in storage oil and oleosin accumulation in developing rapeseed embryos: implications for the role of oleosins and the mechanisms of oil-body formation. Plant Mol Biol. 1993;23(5):1015–1027. doi: 10.1007/BF00021816. [DOI] [PubMed] [Google Scholar]
- 41.Zale J, Jung JH, Kim JY, Pathak B, Karan R, Liu H, Chen X, Wu H, Candreva J, Zhai Z, et al. Metabolic engineering of sugarcane to accumulate energy-dense triacylglycerols in vegetative biomass. Plant Biotechnol J. 2016;14(2):661–669. doi: 10.1111/pbi.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zulu NN, Popko J, Zienkiewicz K, Tarazona P, Herrfurth C, Feussner I. Heterologous co-expression of a yeast diacylglycerol acyltransferase (ScDGA1) and a plant oleosin (AtOLEO3) as an efficient tool for enhancing triacylglycerol accumulation in the marine diatom Phaeodactylum tricornutum. Biotechnol Biofuels. 2017;10:187. doi: 10.1186/s13068-017-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El TA, Reynolds KB, Petrie JR, Singh SP, Vanhercke T. Thioesterase overexpression in Nicotiana benthamiana leaf increases the fatty acid flux into triacylgycerol. FEBS Lett. 2017;591(2):448–456. doi: 10.1002/1873-3468.12539. [DOI] [PubMed] [Google Scholar]
- 44.Yurchenko O, Shockey JM, Gidda SK, Silver MI, Chapman KD, Mullen RT, Dyer JM. Engineering the production of conjugated fatty acids in Arabidopsis thaliana leaves. Plant Biotechnol J. 2017;15(8):1010–1023. doi: 10.1111/pbi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh K, Huang AH. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 2004;136(3):3427–3434. doi: 10.1104/pp.104.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Long Y, Yin Y, Zhang C, Gan L, Liu L, Yu L, Meng J, Li M. New insights into the genetic networks affecting seed fatty acid concentrations in Brassica napus. BMC Plant Biol. 2015;15:91. doi: 10.1186/s12870-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Chi M, Li L, Li H, Noman M, Yang Y, Ji K, Lan X, Qiang W, Du L, et al. Genome-wide identification, expression profiling, and functional validation of Oleosin gene family in Carthamus tinctorius L. Front Plant Sci. 2018;9:1393. doi: 10.3389/fpls.2018.01393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu WX, Liu HL, Qu LQ. Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds. Theor Appl Genet. 2013;126(9):2289–2297. doi: 10.1007/s00122-013-2135-4. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem. 2010;48(1):9–15. doi: 10.1016/j.plaphy.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001;126(2):861–874. doi: 10.1104/pp.126.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian Y, Lv X, Xie G, Zhang J, Xu Y, Chen F. Seed-specific overexpression of AtFAX1 increases seed oil content in Arabidopsis. Biochem Biophys Res Commun. 2018;500(2):370–375. doi: 10.1016/j.bbrc.2018.04.081. [DOI] [PubMed] [Google Scholar]
- 53.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32(Database issue):D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 59.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. BIOINFORMATICS. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. GENE. 1988;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 63.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Iskandarov U, Klotz ET, Stevens RL, Cahoon RE, Nazarenus TJ, Pereira SL, Cahoon EB. A thraustochytrid diacylglycerol acyltransferase 2 with broad substrate specificity strongly increases oleic acid content in engineered Arabidopsis thaliana seeds. J Exp Bot. 2013;64(11):3189–3200. doi: 10.1093/jxb/ert156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequence alignment and MEME analysis of B. napus oleosin genes. (A) MEME logo of the conserved Pro knot; (B) sequence alignment of the 72 conserved amino acids in 48 B. napus oleosin proteins. The relatively highly conserved amino acids are marked in different colors. (PNG 3427 kb)
Figure S2 Genetic structure of A. thaliana, B. oleracea, B. rapa and B. napus oleosin genes. The motifs in oleosin are shown on the left, and the genetic structure is shown on the right. The numbers near the introns indicate the intron phase. (PNG 579 kb)
Figure S3. Subcellular localization of AtOLE1, AtOLE2, AtOLE3 and AtOLE4. A, B, C, and D represent AtOLE1; E, F, G, and H represent AtOLE2; I, J, K, and L represent AtOLE3; and M, N, O, and P represent AtOLE4. A, E, I, M: EGFP fluorescence; B, F, G, N: chloroplast autofluorescence; C, G, K, N: background; D, H, L, P: merged EGFP and chloroplast fluorescence. (PNG 3259 kb)
Figure S4 Distribution of different sizes of oil bodies in transgenic seeds. Different sizes of oil bodies are marked with different colors. (TIF 12720 kb)
Table S1 Gene names and the corresponding gene IDs. (XLS 34 kb)
Table S2 Ka/Ks ratios between all of the oleosins from A. thaliana, B. oleracea, B. rapa, and B. napus. (XLS 1511 kb)
Data Availability Statement
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.