Abstract
The mammalian/mechanistic target of rapamycin (mTOR) is a key component of cellular metabolism that integrates nutrient sensing with cellular processes that fuel cell growth and proliferation. Although the involvement of the mTOR pathway in regulating life span and aging has been studied extensively in the last decade, the underpinning mechanisms remain elusive. In this review, we highlight the emerging insights that link mTOR to various processes related to aging, such as nutrient sensing, maintenance of proteostasis, autophagy, mitochondrial dysfunction, cellular senescence, and decline in stem cell function.
Keywords: mTOR, aging, senescence, mitochondria, lifespan, stem cell, proteostasis, nutrient sensing
Introduction
Aging is characterized by the gradual decline in physiological functions occurring in most tissues and organisms 1 . The acceleration of aging in specific tissues leads to a variety of disorders, including neurodegeneration, obesity, diabetes, and cardiovascular and neoplastic diseases 2 . One of the main pharmacological interventions prolonging life span in several model organisms (that is, yeast, nematodes, fruit flies, and mice) is rapamycin 3 . Rapamycin, a natural product isolated from Streptomyces hygroscopicus, was discovered on the island of Rapa Nui in 1972 4 . It possesses anti-fungal, immunosuppressive, and anti-cancer proprieties, which are mediated by the inhibition of its target: mechanistic/mammalian target of rapamycin (mTOR) 5– 7 . Accordingly, mTOR has been implicated in many of the processes that are associated with aging, including cellular senescence, immune responses, cell stem regulation, autophagy, mitochondrial function, and protein homeostasis (proteostasis) 3, 8– 10 . Finally, in some model organisms, interventions expanding life span (for example, caloric restriction, or CR) were shown to involve TOR 3 . This article provides an overview of the role of mTOR signaling in coordinating cellular processes involved in regulation of life span, aging, and age-related pathologies and puts an emphasis on mammals.
Regulation and functions of mTOR pathway
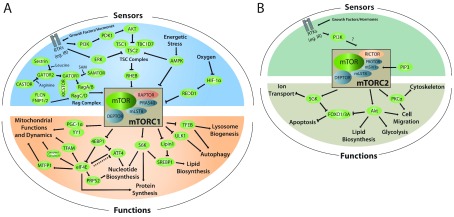
TOR is a serine/threonine kinase that is evolutionary conserved, and homologues are found in yeast, nematodes, flies, plants, and mammals 11 . In higher eukaryotes, including mammals, mTOR is encoded by a single gene and its protein product is a component of two distinct complexes—mTOR complex 1 (mTORC1) and 2 (mTORC2) 3 —which differ functionally and structurally and in their sensitivity to rapamycin 12– 14 ( Figure 1). The two mTOR complexes share the components mLST8 and DEPTOR (DEP domain-containing mTOR-interacting protein), whereas RAPTOR and PRAS40 are present exclusively in mTORC1 3 . In turn, RICTOR, mSIN1, and Protor-1/2 are found within mTORC2 3 . In yeast, TOR1 and TOR2 are encoded by distinct genes; TOR2 engages in both TORC1 and TORC2 complexes, and TOR1 is exclusive for the TORC1 complex 15 . mTORC1 responds to a plethora of extracellular stimuli and intracellular cues, such as amino acids, hormones, growth factors, energetic stress, and oxygen. These factors initiate mTOR-dependent anabolic processes, including nucleotide, lipid, and protein synthesis while inhibiting autophagy, which results in stimulation of cellular growth and proliferation 3, 16 ( Figure 1). Several regulators that signal via the PI3K/PDK1/AKT (phosphoinositide 3-kinase/3-phosphoinositide-dependent protein kinase 1/protein kinase B) pathway (for example, insulin and IGFs) stimulate mTORC1 by inhibiting the tuberous sclerosis complex (TSC) ( Figure 1) which is composed of TSC1 scaffold and TSC2. This complex acts as a Ras homologue enriched in brain (RHEB) GTPase-activating protein 17 . TSC1/2 complex associates with auxiliary factor TBC1D7 (TBC1 domain family member 7) 18 . TSC2 inactivates RHEB, leading to mTORC1 inhibition 19, 20 . One of the major roles of mTORC1 is the regulation of protein synthesis, which is mediated via the phosphorylation of a multitude of substrates 3, 16 , the best characterized of which are the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs) and ribosomal protein S6 kinases (S6Ks) 21 . By affecting protein synthesis (that is, 4E-BPs) or by substrate phosphorylation (that is, S6Ks) or both, these factors also mediate the effects of mTOR on metabolic processes, including nucleotide synthesis, lipid metabolism (via sterol regulatory element-binding protein 1, or SREBP1), and mitochondrial function and dynamics (detailed in the “mTOR and the regulation of mitochondrial function” section) ( Figure 1). In most cell types, mTORC1 inhibition by rapamycin leads to a strong decrease in S6K phosphorylation while only marginally affecting 4E-BP phosphorylation 22, 23 .
Figure 1. mTOR acts as a nutrient sensor coordinating cellular functions linked to proliferation, growth, and survival.
mTOR operates within two functionally and structurally distinct complexes: mTORC1 and mTORC2. The two mTOR complexes share the components mLST8 and DEPTOR, while RAPTOR and PRAS40 are present exclusively in mTORC1. RICTOR, mSIN1, and Protor-1/2 are found exclusively within mTORC2. Growth factors or hormones (for example, insulin through IR) activate the PI3K/PDK1/AKT or ERK signaling pathways, which inactivate the TSC2 subunit of the TSC complex. Inactivation of the TSC complex upregulates the activity of RHEB, which in turn stimulates mTORC1. The activity of mTORC1 is also positively regulated by amino acid–mediated stimulation of the RAG complex of GTPases: Rag A/B and Rag C/D. GATOR1 inhibits RAG GTPases. SAM inhibits the activation of GATOR1 by SAMTOR. GATOR1 is also repressed by GATOR2, which in turn is regulated by Sestrin2 and CASTOR in response to leucine and arginine, respectively. The FLCN FNIP1/2 complex is also thought to stimulate the RAG activity. mTORC1 is suppressed under conditions where energy or glucose is limiting through AMPK signaling, which activates TSC2 and inhibits the mTORC1 subunit RAPTOR, and by hypoxia via the HIF-1α/REDD1 axis. mTORC1 orchestrates several anabolic processes via transcriptional or translational regulation or both. mTORC1 controls protein synthesis in part through its two main effectors: S6K and 4E-BP1. mTORC1 also stimulates mitochondrial function, through 4E-BP1 and PGC-1α/YY1, and mitochondrial dynamics (via MTFP1). The control of mTORC1-mediated nucleotide synthesis is governed by S6K, ATF4, and PRPS2 (which is translationally regulated via 4E-BP/eIF4E), while lipid biosynthesis and adipogenesis are regulated by S6K and Lipin1. mTORC1 controls autophagy by inhibiting the activity of ULK1 and TFEB; the latter also mediates mTORC1-dependent lysosome biogenesis. Conversely, the activity of mTORC2 may be regulated by growth factors through PI3K activation and generation of PIP3. PIP3 has been suggested to bind to mSin1, thereby activating mTORC2. mTORC2 activation promotes AKT signaling involved in glycolysis, lipid biosynthesis, and cell migration, while SGK signaling is involved in ion transport. Both AKT and SGK negatively regulate FOXO1/3A, which is a regulator of key metabolic pathways and apoptosis. mTORC2 also controls cytoskeleton and cell migration through PKCα. 4E-BP, eukaryotic initiation factor 4E-binding protein; AKT, protein kinase B; AMPK, AMP-activated protein kinase; ATF4, activating transcription factor 4; CASTOR, cellular arginine sensor for mTORC1; DEPTOR, DEP domain-containing mTOR-interacting protein; eIF4E, eukaryotic translation initiation factor 4E; ERK, extracellular signal-regulated kinase; FLCN, folliculin; FNIP1/2, folliculin interacting protein 1/2; FOXO1/3, forkhead box protein O1/O3; GATOR1, GTPase-activating proteins toward Rags 1; GATOR2, GTPase activating proteins toward Rags 2; HIF-1α, hypoxia-inducible factor 1 alpha; IGF1R, insulin-like growth factor 1 receptor; IR, insulin receptor; KICSTOR, KPTN-, ITFG2-, C12orf66-, and SZT2-containing regulator of mTORC1; mLST8, mammalian lethal with SEC13 protein 8; mSIN1, mammalian stress-activated protein kinase interacting protein 1; MTFP1, mitochondrial fission process 1; mTOR, mechanistic target of rapamycin kinase; mTORC1, mechanistic target of rapamycin complex 1; mTORC2, mechanistic target of rapamycin complex 2; PDK, 3-phosphoinositide-dependent protein kinase-1; PGC1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PI3K, phosphoinositide 3-kinase; PIP3, phosphatidylinositol (3,4,5)-triphosphate; PKC, protein kinase C alpha; Pras40, Proline-rich AKT1 substrate 1; Protor, protein observed with Rictor-1; PRPS2, phosphoribosyl pyrophosphate synthetase 2; RAG, Ras-related GTP-binding protein, subunits A/B or C/D; RAPTOR, regulatory-associated protein of mTOR; REDD1, regulated in development and DNA damage response 1; RHEB, Ras homolog, mTORC1 binding; RICTOR, rapamycin-insensitive companion of mTOR; S6K, ribosomal protein S6 kinase; SAMTOR, S-adenosylmethionine sensor for the mTORC1 pathway; SGK, serum and glucocorticoid-regulated kinase 1; SREBP1, sterol regulatory element-binding transcription factor 1; TBC1D7, TBC1 domain family member 7; TFAM, mitochondrial transcription factor a; TFEB, transcription factor EB; TSC, tuberous sclerosis complex; ULK1, Unc-51-like autophagy activating kinase; YY1, Yin Yang 1.
In contrast, mTORC2 regulates cytoskeletal organization and the activity of several members of the AGC family of kinases (for example, AKT and SGK1) and has been implicated in the degradation of newly synthesized polypeptides. It is involved in glucose and lipid metabolism through AKT-dependent and independent mechanisms 24– 26 , controls ion transport via SGK1 27 , and affects the cytoskeleton and cell migration through protein kinase C alpha (PKCα) 28 ( Figure 1). In addition, both AKT and SGK negatively regulate FOXO1/3A (forkhead box protein O/O3A), which are transcription factors that regulate metabolism and apoptosis 28 . In most cell lines, mTORC2 is insensitive to short-term (<24 hours) rapamycin treatment 14, 29 , but it has been reported that mTORC2 is downregulated during prolonged rapamycin exposure in several cell lines and tissues (such as hepatocytes, adipose tissues, skeletal muscle, heart, pancreas, liver, lung, and spleen) in vivo 30– 32 . Regulation of mTOR and its functions, including governing mRNA translation, is covered in detail in a number of excellent recent reviews (for example, 3, 16, 21, 33, 34 ).
TOR as a negative regulator of life span
The relationship between TOR and longevity was first shown in non-vertebrates by using genetic manipulations. For example, deletion of S6K homologue SCH9 in Saccharomyces cerevisiae or depletion of TOR ( let-363) 35 or RAPTOR (mTORC1 protein member; daf-15) by RNA interference (RNAi) in Caenorhabditis elegans 36, 37 extends life span in both models. Similar effects were observed in Drosophila melanogaster either through the overexpression of TOR suppressors dTsc1 of dTsc2 or by expressing dominant-negative forms of dTOR or dS6K 38 . Pharmacological inhibition of TOR by rapamycin in S. cerevisiae, C. elegans, D. melanogaster, or Mus musculus confirmed the evolutionary conserved and fundamental role of mTOR as a regulator of longevity 3, 39– 47 .
mTOR and the beneficiary effects of dietary restriction on life span
The central role for the mTOR pathway in regulating life span has been attributed in part to its function as a nutrient sensor ( Figure 2). Nutrient-sensing pathways, including the insulin/insulin-like growth factor 1 (IGF-I) signaling (IIS) network, are thought to act as major determinants of longevity. The importance of IIS, like mTOR pathway inhibition, in regulating life span was firmly established in numerous species ( C. elegans, D. melanogaster, or M. musculus) 48– 52 . IIS activation via binding of insulin or IGF-I to the insulin receptor (IR) or IGF-1 receptor (IGF1R) (or both) leads to activation of PI3K/AKT. Signaling through additional growth factor receptors (for example, epidermal growth factor receptor family, or ERBB) activates the RAS/RAF/MEK/ERK pathways. As AKT, ERK, and its downstream effector RSK1 all phosphorylate and inactivate TSC2 53– 56 , PI3K and RAS/RAF/MEK/ERK pathways largely converge on mTORC1 ( Figure 1) 3 .
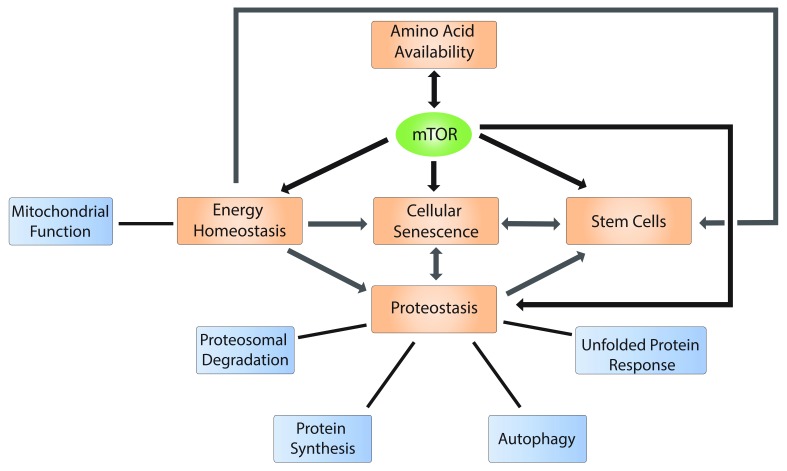
Figure 2. mTOR regulates several hallmarks of aging.
Schematic representation of the role of the mTOR pathway in the regulation of hallmarks of aging (black arrows), such as nutrient availability (represented by amino acid availability), energy homeostasis, cellular senescence, cell stemness, and proteostasis. mTOR activity is regulated in part by amino acid levels, while mTOR in turn stimulates the synthesis of non-essential amino acids (see the “mTOR and the beneficiary effects of dietary restriction on life span” section). The depicted hallmarks of aging are also interconnected (grey arrows), suggesting that aging is a coordinated process in which mTOR plays a significant role. mTOR, mechanistic target of rapamycin kinase.
Suppression of the IIS/mTOR axis is thought to be one of the underpinning mechanisms of the beneficiary effects of dietary restriction (DR), which not only prolongs life span but also delays the onset of age-related pathologies (commonly referred to as health span) across a variety of organisms (yeasts, nematodes, fruit flies, rats, mice, and primates) 57– 59 . DR incorporates both the classic concept of CR (where the total caloric intake is reduced, usually by 20 to 40%) and restriction of specific nutrients or regimens of restriction for intermittent time periods 60 . Consistent with the central role of mTOR in nutrient sensing, CR did not further prolong life span under conditions in which TORC1 was inhibited by TOR1 deletion in S. cerevisiae, TOR RNAi in C. elegans, or overexpression of dTsc2 in D. melanogaster 38, 61, 62 . Conversely, several reports suggested that rapamycin may potentiate the effect of CR in increasing life span in D. melanogaster, indicating potential complexity in the role of mTOR inhibition in mediating the beneficiary effects of CR 40 . Comparison of the effects of rapamycin and CR in liver and white adipose tissues in mice 63, 64 , or experiments carried out in S. cerevisiae 65 , revealed that rapamycin and CR induce discrete changes in transcriptome and metabolome, suggesting that CR may extend life span through mTOR-independent mechanisms. Notably, rapamycin incompletely inhibits some of mTORC1 substrates, including 4E-BPs 22, 23, 66 . This is particularly significant when comparing the impact of mTOR inhibition and CR on metabolome, as 4E-BPs mediate mTORC1-dependent translational regulation of several metabolic processes (for example, cellular energetics, mitochondrial dynamics, and non-essential amino acid synthesis) 67– 69 ( Figure 1). Translatome studies thus are warranted to grasp the full overlap between the effects of CR and mTOR on life span and aging.
As CR is not readily translatable to the clinic, alternative DR strategies based on macronutrient restrictions have been tested. In these approaches, specific macronutrient intake is limited without a reduction in calories 60 . DRs were shown to improve healthy aging in humans; one of the most effective interventions appears to be the reduction of protein and amino acid intake (protein restriction, or PR) 70– 72 . Indeed, PR extends both life span and health span in mice but was linked to reductions in cancer, diabetes, and overall mortality in humans 70 . In some cases, the reduction of a single amino acid, such as methionine or tryptophan, was able to cause these effects 73– 82 . In addition to methionine deprivation, restriction of leucine and other branched-chain amino acids (BCAAs) (isoleucine and valine) improved metabolic health (such as better glucose tolerance and reduced fat mass gain) in both normal and obese mice 83– 85 . Conversely, long-term exposure of mice to an isocaloric yet high BCAA-containing diet led to hyperphagia and obesity and reduced life span 86 . Interestingly, the mechanism was not related to increased total BCAA intake or high mTORC1 but to an amino acid imbalance (shift in the relative quantity of dietary BCAAs versus other amino acids such as tryptophan and threonine) 86 . Metabolites derived from amino acids might also influence life span. Recent evidence indicates that homocysteine, a component of the methionine cycle, can activate mTORC1 87 . Moreover, given that amino acid availability is a major regulator of mTORC1 3 (detailed in the “mTORC1 as a sensor of amino acids” section; Figure 2) and that mTOR inhibition induces increased life span, it is plausible that the beneficial effects of PR on life span and health span are mediated via mTOR. Although this has yet to be determined in the context of aging, a study using a breast cancer xenograft model showed that PR inhibits tumor growth, while reducing mTORC1 (but not mTORC2) activity in both tumor and normal tissues 88 .
mTORC1 as a sensor of amino acids
Nutrient sensing is often dysregulated in aging cells 1 ( Figure 2). Amino acids act as pivotal mTORC1 regulators 3 and indicators of nutrient availability. Indeed, mTORC1 is a central sensor of amino acid availability, which regulates cellular and organismal energetics 89– 91 . In mammals, mTORC1 activation by amino acids is mediated by Ras-related GTP binding (RAG) GTPases 92, 93 which form A/B and C/D heterodimers that associate with lysosomal membranes via the Ragulator complex, also known as LAMTOR (late endosomal/lysosomal adaptor, MAPK and MTOR activator) 94 . Active RAG heterodimers (in which RAG A/B and RAG C/D are bound to GTP and GDP, respectively) recruit mTORC1 to the lysosomal surface, where mTORC1 becomes activated by RHEB 95 . As observed for mTORC1, RHEB localization is also modulated by amino acid signaling, which promotes amino acid–dependent interaction of RHEB with microspherule protein 1 (MCRS1), leading to its maintenance at the lysosome 96 . RAGs are controlled mainly by the GATOR1/2 interplay. GATOR1, a complex containing DEP domain containing 5 (DEPDC5), nitrogen permease regulator 2-like (NPRL2), and nitrogen permease regulator 3-like (NPRL3) proteins, acts as a GTPase-activating protein (GAP) toward RAGs A/B, thereby negatively regulating mTORC1 97 . GATOR1 recruitment to the lysosomal surface in the context of amino acid or glucose deprivation is mediated by KICSTOR, a complex composed of kaptin, actin binding protein (KPTN), integrin alpha FG-GAP repeat containing 2 (ITFG2), chromosome 12 open reading frame 66 (C12orf66), and SZT2 98 . In turn, GATOR2, which is composed of MIOS (meiosis regulator for oocyte development), WD repeat domain 24 (WDR24), WDR59, SEH1-like nucleoporin (SEH1L), and SEC13, suppresses GATOR1, thus activating mTORC1 97 . In addition, the folliculin (FLCN)-folliculin-interacting protein (FNIP) complex was shown to display GAP activity toward RAG C/D, therefore contributing to RAG heterodimer complete activation in response to amino acids 99, 100 . However, it is worth mentioning that FLCN loss does not always result in mTORC1 inhibition, suggesting a context-dependent function for FLCN in TORC1 signaling or the presence of compensatory mechanisms leading to mTORC1 activation in the absence of FLCN (discussed in 100, 101).
It appears that the mechanisms of activation of mTORC1 are amino acid–specific. To this end, lysosomal amino acid transporter SLC38A9 is necessary for the arginine-dependent activation of mTORC1 via its interaction with the RAG-LAMTOR-v-ATPase (vacuolar-type H + ATPase) complex 102– 105 . In parallel, mTORC1 downregulation in response to arginine deprivation was shown to require cellular arginine sensor for mTORC1 (CASTOR1) 106 , which interacts with GATOR2 to inhibit its function 107 . Conversely, arginine binding by CASTOR1 triggers its dissociation from GATOR2 and consequently activates mTORC1 106, 107 . Through a similar mechanism, Sestrin2 functions as a direct leucine sensor 108 by binding to and repressing GATOR2 in the absence of leucine 109, 110 . Upon binding to leucine, Sestrin2 dissociates from GATOR2, which leads to mTORC1 activation 108– 112 . It has been reported that, in addition to Sestrin2, leucyl-tRNA synthetase (LRS) participates in leucine-dependent modulation of mTORC1 signaling 113 . It has been proposed that LRS positively regulates the RAG GTPase cycle by acting as a GAP toward RAG D 113– 115 , although this model has been challenged by other reports 100, 116 . A recent study evidenced LRS-mediated leucylation on lysine residues of RAG A/B, resulting in mTORC1 activation 117 . Methionine availability is indirectly sensed by the SAMTOR protein, which can bind to S-adenosylmethionine (SAM), a metabolite derived from methionine. SAM disrupts the SAMTOR–GATOR1 association and thus relieves the inhibitory effect of this complex on mTORC1 signaling 118 . Finally, glutamine-dependent activation of mTORC1 is dependent on v-ATPase and also requires the adenosine diphosphate ribosylation factor 1 (ARF1) GTPase 119 . Although a number of studies show that alterations of these newly discovered mTOR regulators (for example, Sestrin2) may play a role in aging 120– 122 , their precise effects on life span and aging-related pathologies remain to be established.
mTOR as a regulator of food intake and global energy homeostasis
The mechanisms by which CR and DR expand life span are complex and comprise organismal level regulation, including sensory food perception and modulation of food intake behaviour. For example, recent work in C. elegans showed that activation of a food deprivation signal through chemical manipulation mimics a state of DR, leading to prolonged life span 123 . Food intake behaviour is in part controlled by neurons in the mediobasal hypothalamus (MBH), a key region of the brain implicated in regulation of energy balance 124 . In MBH neurons, mTORC1 activity is induced by food intake and repressed by fasting 125 , thereby suggesting that mTORC1 plays a pivotal role in the hypothalamic regulation of energy balance 126, 127 . Several studies, however, failed to confirm the role of mTOR in the regulation of energy balance or feeding behaviour 91, 128 though confirming mTORC1 function in regulating glucose metabolism 128 . Importantly, overexpression of DEPTOR (either systemic or restricted to MBH neurons) prevents obesity induced by a high-fat diet and improves glucose metabolism 129 . DEPTOR is a component of both mTORC1 and mTORC2 and is able to suppress the activity of both complexes 130 . In addition, mTORC1 in agouti-related protein (AGRP) neurons (part of the MBH) appears to facilitate integration of food-related cues and adaptive energy expenditure 131 . This supports a role for mTOR in regulating systemic energy homeostasis 91 , which may have an impact on metabolic disorders and aging. Moreover, in addition to its activity in MBH neurons, alterations of the mTORC1 pathway in the liver and adipose tissue were shown to impact whole-body metabolism via peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α, a transcriptional coactivator controlling mitochondrial biogenesis 132 ) and fibroblast growth factor 21 (FGF21, a secreted protein that is a key mediator of fatty acid oxidation and lipid metabolism 133 ), respectively 134 . Finally, RAPTOR knockout (KO) in the adipose tissue of mice resulted in lean phenotype which was accompanied by elevated mitochondrial respiration 135 . In conclusion, although these findings position mTOR as a central metabolic regulator at the cellular or whole-organism level or both, the precise relationship between these mTOR functions and life span, health span, and aging is incompletely understood.
The role of mTOR in age-associated processes (hallmarks of aging)
In an attempt to define common denominators of aging, the following nine “hallmarks” have been proposed: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication 1 . Not surprisingly, in keeping with the observations implicating mTOR in increasing life span, a significant proportion of these “hallmarks” are known to be affected by mTOR. In this section, we describe recent advances linking mTOR to these key age-associated processes and explore how aberrant mTOR signaling may orchestrate their coordinated dysregulation.
Loss of proteostasis
Alterations in protein production, degradation, folding, and trafficking—proteostasis—are linked to aging 136 . For instance, the chronic expression and accumulation of misfolded or aggregated proteins lead to the development of various age-associated diseases (for example, Alzheimer’s disease [AD] and Parkinson’s disease and cataracts) 137 . Proteostasis is maintained via orchestration of protein synthesis, protein clearance (via proteasomal degradation or autophagy), and quality control mechanisms (for example, unfolded protein response, or UPR), all of which are thought to be influenced by mTOR. In model organisms, including S. cerevisiae, C. elegans, and D. melanogaster, deletion of different components of the translational machinery and subsequent decrease in protein biosynthesis significantly increase life span 138 . mTORC1 stimulates protein synthesis through several mechanisms that have been highly characterized and that involve two main mTORC1 effectors: S6K and 4E-BPs 21 . mTORC1 also suppresses autophagy, in part by inhibiting Unc-51-like autophagy-activating kinase (ULK1) 139 . It is now emerging that protein synthesis and autophagy, coupled with nutrient sensing, are coordinated by mTORC1 to maintain energy and protein homeostasis 139 ( Figure 2). In addition to autophagy, mTOR may suppress protein degradation by controlling proteasomal activity ( Figure 2). For example, mTORC1, but not mTORC2, inhibition increases protein degradation by the proteasome by enhancing ubiquitination through a mechanism that remains to be elucidated 140 . Moreover, in yeast and mammalian cells, TOR/mTOR inhibition upregulates the levels of the regulatory particle assembly chaperones (RACs) through the activity of MAPKs Mpk1 (in yeast) or ERK5 (in mammalian cells), and increases proteasome abundance 141 . These findings contradict previous reports that mTORC1 inhibition reduces proteolysis by suppressing expression of proteasomal subunits in a nuclear respiratory factor 1 (NRF-1)-dependent manner 142 .
To maintain proteostasis under stress, cells engage quality control mechanisms, such as the UPR, which plays a major role in aging in both cell-autonomous and non-autonomous manners 143 . The activation of the UPR declines with age, as was shown in multiple model organisms ( C. elegans, M. musculus, or Rattus norvegicus) 143 . Conversely, UPR activation can extend life span 143 . The UPR is initiated through the activation of three arms: inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6) 144 . ATF6 and IRE1 increase abundance and splicing of the mRNA encoding X-box binding protein 1 (XBP1), respectively 145– 147 . XBP1 is a transcription factor that controls the expression of genes implicated in protein folding, quality control, and lipid biosynthesis 143 . IRE1 also activates stress signaling, including JNK and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) 148 . Rapamycin suppresses IRE1-dependent JNK induction, XBP1 splicing, and activation of PERK 149, 150 , which suggests significant cross-talk between mTOR and UPR.
The integrated stress response (IRS) is the arm of the UPR which is under governance of PERK 151 . It is centered on the α-subunit of eIF2, which is phosphorylated by four eIF2α kinases—PERK, double-stranded RNA-dependent protein kinase (PKR), heme-regulated eIF2α kinase (HRI), and general control non-derepressible 2 (GCN2)—in response to ER stress, viral infection, heme insufficiency, and amino acid depletion, respectively 7 . eIF2 delivers initiator tRNA (tRNA i Met) as part of the eIF2:GTP:tRNA i Met ternary complex (TC), whereby upon tRNA i Met delivery GTP is hydrolyzed to GDP 33 . A multi-subunit guanine nucleotide exchange factor (GEF) eIF2B exchanges eIF2-bound GDP to GTP, which allows recycling of TC to facilitate the next initiation round 152, 153 . Phosphorylation of eIF2α suppresses the GEF activity of eIF2B, which limits TC recycling 154 . This results in downregulation of global protein synthesis with concomitant translational activation of mRNAs which contain inhibitory upstream open reading frames (uORFs) in their 5′ untranslated regions (5′UTRs) 155 . Transcripts that contain uORFs are enriched in those encoding stress-responsive transcription factors, including activating transcription factor 4 (ATF4) and CCAAT enhancer-binding protein homologous protein (CHOP) 156– 158 . As is the case for UPR, several cross-talk mechanisms between ISR and mTORC1 have been observed. For example, during chronic ER stress, which leads to significant reprogramming of translational machinery that is distinct from the acute ISR 159 , eIF2α phosphorylation is paralleled with the ATF4-dependent transcriptional induction of amino acid transporter expression 160 which increases intracellular amino acid levels, thus elevating mTORC1 activity 161 . However, long-term stress may also decrease protein synthesis via the suppression of mTORC1-mediated translational control, through an AKT-dependent, eIF2α phosphorylation-independent mechanism 161 , highlighting the fine-tuned regulation of translational activity in conditions of stress. In turn, upon nutrient, growth factor, or hormone stimulation (or a combination of these), mTORC1 in concert with casein kinase II (CK2) phosphorylates the eIF2β subunit of eIF2. eIF2β recruits the non-catalytic region of tyrosine kinase adaptor protein 1 (NCK1) that serves as an adaptor for the protein phosphatase 1 (PP1), thereby leading to eIF2α dephosphorylation 162 . mTORC1 inhibition is also paralleled by GCN2 activation and increased eIF2α phosphorylation through a mechanism dependent on protein phosphatase 6 (PP6C) 163 . In addition, in cells characterized by the loss of its negative regulator TSC, mTORC1 downregulates eIF2α phosphorylation by attenuating PERK activity, which is accompanied by increased reactive oxygen species (ROS) production 164 . Finally, mTORC1 can influence ISR via regulation of ATF4 protein levels by stabilizing and promoting translation of ATF4 mRNA, which is mediated by 4E-BP1 165 . Given the central role of UPR and in particular ISR in aging, these data suggest that the aging-related effects of mTOR may be influenced at least in part by its cross-talk with the UPR/ISR machinery.
mTOR and the regulation of mitochondrial function
Aberrant mitochondrial function is a major characteristic of aging cells 1 . For example, genetic models of mitochondrial dysfunction (such as mutation of mitochondrial DNA polymerase in mice) correlate with reduced life span 166 . However, many interventions that extend life span (CR and rapamycin) are associated with reduced energy intake and decreased mitochondrial functions 167 . These apparently contradictory findings revealed complex and antagonistic functions of mitochondria in aging and have been at least partially reconciled by a biphasic modeling of mitochondrial dysregulation 167 . This model proposes that alterations of mitochondrial functions are not linear with the aging of the organism but rather that they increase and peak in middle age, followed by a decline in older age 167 .
Mitochondrial dysfunction in aging cells is characterized by several factors, including elevated ROS production and mitochondrial DNA mutations; decreased electron transport function, membrane potential, and ATP production; altered mitochondrial dynamics; or dysregulated mitophagy 1, 168 . Functionally, altered mitochondrial activity participates in inducing cellular senescence 169 , chronic inflammation, and a decline in stem cell activity associated with aging 1 ( Figure 2).
mTORC1 regulates mitochondrial biogenesis, functions, and dynamics through translational and transcriptional mechanisms ( Figure 1). mTORC1 stimulates translation of nuclear-encoded mitochondria-related mRNAs—for example, components of complex I and V, mitochondrial ribosomal proteins, and mitochondrial transcription factor a (TFAM) 68, 170 —and fission process 1 (MTFP1) mRNA 69 . mTORC1 activity therefore stimulates mitochondrial respiration and ATP production to meet the high energetic requirements of cancer cells 68 . Translational suppression of MTFP1 levels leads to mitochondrial hyperfusion and protection from apoptosis 69 . In addition to these translational mechanisms which are thought to be engaged during acute activation of mTORC1 (12 hours or less), it has been shown that, during prolonged stimulation, mTORC1 regulates transcription of nuclear-encoded mitochondrial genes by engaging PGC-1α and transcription factor Yin-Yang 1 (YY1) 171 . mTORC2 was also shown to play a role in mitochondrial functions in both mouse embryonic fibroblasts and cancer cells, and its suppression was paralleled by increases in mitochondrial membrane potential, ATP production, and calcium uptake 172 .
Mitophagy is the selective degradation of mitochondria by autophagy that serves as a quality control mechanism to ensure recycling and removal of damaged mitochondria 173 . Several studies have shown that mitophagy can extend life span 174, 175 . Interestingly, it was recently revealed that, in addition to having an established role in inhibiting autophagy 139 , mTORC1 is involved in the regulation of mitophagy 176 . In TSC2-null cells, in which mTORC1 is hyperactive, the levels of mitophagy induced by a mitochondrial uncoupling agent were reduced 176 . This was accompanied by an mTORC1-dependent decrease in PTEN-induced kinase 1 (PINK1) levels and a decrease in PARK2 translocation to the outer mitochondrial membrane, which are thought to be essential for the degradation of uncoupled mitochondria by mitophagy 176 .
Collectively, these findings implicate mTOR in many aspects of mitochondrial functions, biogenesis, degradation, and dynamics. However, more work is still needed to establish the extent of mTOR involvement in mitochondrial dysfunction in aging cells and how this is linked to the aging phenotype and orchestrated with other aging hallmarks (for example, cellular senescence).
The role of mTOR in cellular senescence
Cellular senescence is an evolutionary conserved phenomenon characterized by a permanent and stable cell cycle exit 177 . Senescent cells are characterized by an increase in cell size and mitochondrial mass, mitochondrial dysfunction, and the development of a multi-component senescence-associated secretory phenotype (SASP; comprising growth factors, cytokines, metalloproteases, etc) 1, 178– 181 . Cellular senescence can be induced in response to a variety of stresses and signals (oncogenes, DNA damage, telomeric dysfunction, and oxidative stress) 182– 186 and, depending on the context, can be either beneficial (for example, in wound healing) 187 or deleterious (for example, during aging) 188– 190 . The deleterious effects exhibited in aging are strongly linked to the SASP, whose components include a large number of cytokines, chemokines, growth factors, and proteases promoting inflammation, angiogenesis 191 , tissue degeneration, and tumor growth 192 . This explains why the accumulation of only a small percentage (2 to 3%) of senescent cells in tissues can have a significant negative impact 192 . Owing in large part to SASP and the need to produce and secrete large quantities of factors, senescent cells are highly metabolically active and show high levels of protein synthesis 193 . mTOR plays an important role in promoting the secretory phenotype of senescent cells 194 and its inhibition was shown to prevent stem cell senescence 8, 9 . For example, rapamycin was shown to reduce interleukin 1 receptor (IL-1R)-dependent SASP by inhibiting the translation of IL-1α mRNA, which reduces transcription of inflammatory genes regulated by the pro-inflammatory transcription factor NF-κB 195 . Moreover, mTORC1 is thought to interact with MAPK to increase translation of the MK2 kinase (MAPKAPK2), which prevents the degradation of numerous SASP factor transcripts by ZFP36 ring finger protein-like 1 (ZFP36L1) 196 . Inhibition of mTOR by rapamycin was observed to impede increases in mitochondrial DNA, biomass, and ROS associated with genotoxic stress-induced senescence 197 . Interestingly, mTORC1 activation in senescent cells may be the result of defects in amino acid and growth factor sensing 198 . Namely, it was observed that the induction of senescence in human fibroblasts by stress, replicative exhaustion, or oncogene activation renders mTORC1 constitutively active and insensitive to serum and amino acid starvation 198 .
Importantly, several other age-associated hallmarks such as mitochondria dysfunction and dysregulation of proteostasis are directly linked to cellular senescence, as both a cause and a consequence 177 . Given the central role of mTOR in regulating the main aspects of these hallmarks of aging, it is plausible that aberrant mTOR signaling underlies their concerted dysregulation ( Figure 2).
The role of mTOR in stem cell maintenance and stem cell function decline in aging
Adult tissues contain populations of somatic stem cells (also referred to as adult stem cells), which allow their regeneration under normal physiological conditions (for example, due to cellular turnover) and during response to injury 199 . Adult stem cells are able to self-renew and differentiate into multiple cell types and they reside in specialized niches within the tissues 199 . However, they can accumulate mutations or undergo epigenetic changes (or both) which may compromise their functions or lose the ability to divide (a phenomenon referred to as stem cell exhaustion) 199 . To avoid this, adult stem cells are maintained in a quiescent state until activated 199 . Quiescent stem cells are characterized by a reduction in metabolic, transcriptional, and translational activity, which correlates with a suppression of mTORC1 199, 200 . Depending on the tissue type, stem cells accumulate with age (for example, hematopoietic stem cells [HSCs] or intestinal stem cells [ISCs]) or are reduced in number (for example, neural stem cells [NSCs] or germline stem cells) 201– 203 . Importantly, stem cells in aging tissues show impaired stem cell functions because of cell-intrinsic factors (accumulation of DNA damage, epigenetic alterations, and ROS; mitochondrial dysfunction; and loss of proteostasis) and extrinsic factors (disruption of the niche and chronic inflammation) 204 , which ultimately lead to impaired tissue regeneration.
mTOR has been implicated in the self-renewal, proliferation, and differentiation of both embryonic and adult stem cells (reviewed in 205). Although the role of mTORC1 in these processes is better established 205 , the importance of mTORC2 is also observed in promoting osteoblast and inhibiting adipocyte differentiation, reducing neural progenitor cell proliferation, and promoting HSC formation from endothelial cells 205 . Collectively, these studies suggest that increased mTOR activity can be both beneficial and detrimental to stem cells, depending on the cellular context. For example, it was shown that CR in mice causes Paneth cells in the ISC niche to secrete cyclic ADP ribose, which in turn activates SIRT1 in ISCs 206 . Contrary to what is expected under CR, this increases mTORC1 activity toward S6K1 and protein synthesis, thus resulting in higher ISC numbers 206 . mTOR was also shown to be beneficial in NSCs in the hippocampus. Indeed, the mTOR pathway is compromised in aged NSCs, while its activation can revitalize the NSCs by increasing proliferation and promoting neurogenesis 207 . By contrast, in Ercc1 −/Δ mice, in which DNA damage repair is compromised (thus leading to premature aging), mTOR signaling is activated in muscle-derived stem/progenitor cells (MDSPCs) 208 . Rapamycin treatment stimulates autophagy and improves the myogenic differentiation capacity of the Ercc1 −/Δ MDSPCs, suggesting that in this context hyperactive mTOR contributes to stem cell dysfunction 208 . Finally, in S6K1 −/− mice that show extended life span, age-associated decrease in HSC function was improved, as compared with wild-type mice 209 , suggesting that increased mTORC1 signaling may decrease the HSC function during aging.
Although there have been great advances in dissecting how the mTOR pathway regulates various aspects of adult stem cell function, it is becoming apparent that this role of mTOR is dependent on the cellular and tissue context. Moreover, the role of mTOR in coordinating stemness with other hallmarks of aging is still largely unknown. For instance, senescence-associated reprogramming was shown to promote cancer stemness 190 , whilst impairment in cell stemness is facilitated by mitochondrial and proteostatic dysfunctions and is accompanied by an altered metabolic state 199 .
The role of mTOR in aging tissues
Although the modulation by mTOR of the hallmarks of aging described above—see the “TOR as a negative regulator of life span” and “The role of mTOR in age-associated processes (hallmarks of aging)” sections—could be generalized to many cellular and tissue types, several studies to date have investigated the role of the mTOR pathway in the aging of specific tissues. Some examples are provided below.
Heart tissue
mTORC1 is necessary for normal cardiovascular development, maintenance, and function at both the embryonic and postnatal states 210 . This was demonstrated in various mouse models carrying constitutive or inducible cardiac tissue-specific deletions of mTOR, RAPTOR, RICTOR, or RHEB 211– 215 . Nonetheless, mTORC1 suppression seems to be beneficial in aging cardiac tissue 210 , as rapamycin treatment in 24-month-old mice showed improved cardiovascular function and a reversal or attenuation of age-related heart pathologies (heart inflammation and cardiac fibrosis) 216 . RNA sequencing analysis suggested that these benefits were linked to changes in inflammatory, metabolic, and anti-hypertrophic profiles 216 . Moreover, both CR and rapamycin treatments significantly reversed age-associated proteomics changes observed in old hearts 217 , which were characterized by reduced abundance of proteins involved in mitochondrial functions, electron transport chain, the citric acid cycle, and fatty acid metabolism 217 .
Central nervous system
Similar to cardiac tissue, mTOR is required for normal neurological development and function (synaptic plasticity, neuroendocrine regulation, and neuronal recovery) and for adequate cognitive function 218– 220 but is dysregulated in older tissue 220, 221 . Although mTOR activity had been found to be downregulated in the aged versus mid-age naked mole rats (as evidenced by the p-mTOR/mTOR ratio) 222 , the mTOR pathway was shown to be hyperactivated in AD in both mouse models and humans 221 . AD is characterized by a reduction in autophagy (and loss of proteostasis in general), impaired glucose metabolism, and decreased mitochondrial functions, all of which are governed by mTOR (as discussed in the “Loss of proteostasis” and “mTOR and the regulation of mitochondrial function” sections and 10). When administered in the early stages of the disease, rapamycin or rapalogs (rapamycin analogs) were shown to prevent cognitive decline in mouse models of AD 223– 226 , which correlated with a decrease in aggregated beta-amyloid plaques, tau tangles, and microglia activation, all main characteristics of AD 224– 226 . In addition, a recent study tested the use of a novel rapamycin intranasal administration protocol (InRapa), aimed at maximizing brain delivery while reducing systemic side effects 227 . In a mouse model of Down syndrome, InRapa administration ameliorated Alzheimer-like cognitive decline, in part by rescuing autophagy and attenuating dysregulated insulin signaling 227 . Intriguingly, in human trials of immunosuppression after heart transplantation, patients taking the rapalog everolimus showed improved memory and concentration compared with the control group 228 . Collectively, these new findings suggesting the use of mTORC1 inhibitors for improving cognitive function and neurogenerative disorders, combined with improved strategies for drug delivery that reduce side effects, represent promising therapeutic perspectives for the future.
Adipose tissue
There are two major adipose tissues: white adipose tissue (WAT), which stores energy in the form of triglyceride droplets and mediates energy status signaling to the hypothalamus 229 , and brown adipose tissue (BAT), which can dissipate energy in response to cold exposure and caloric excess through coupled and uncoupled respiration and heat production 230– 232 . Therefore, BAT confers protection from obesity and other metabolic diseases 231, 233 . BAT also shows increased mitochondrial density and activity compared with WAT 234 . A third population of adipocytes was recently identified, called beige adipocytes 235 , which has characteristics of BAT and can be found in clusters within WAT. With age, there is a decline in both BAT and the “browning” of WAT (the appearance of thermogenic adipocytes within WAT depots) 236 . The importance of BAT in aging was highlighted by recent findings showing that regulator for G protein signaling 14 (RGS14) KO mice have an extended life span that is associated with an increase in BAT, protection against cold exposure, and improved metabolism 237 . Importantly, transplantation of BAT from RGS14 mice exerts the same protective effect in wild-type recipient mice 237 . The TSC1/mTORC1 axis was shown to control the BAT-to-WAT phenotypic switch in adipocytes, whereas rapamycin treatment reverses the adipocyte phenotypic switch 238 . Moreover, adipose-specific depletion of RAPTOR, which leads to a lean phenotype with enhanced mitochondrial respiration 135 , also promotes beige adipogenesis through prostaglandins (PGs) synthesized by cyclooxygenase-2 (COX-2) 239 . Mechanistically, COX-2 is negatively regulated by mTOR via the phosphorylation of CREB-regulated transcription coactivator 2 (CRTC2) 239 . DEPTOR (inhibitor of mTOR and part of mTORC1) is also a regulator of adipogenesis inasmuch as its reduction of mTORC1-mediated feedback inhibition of insulin signaling activates the proadipogenic Akt/PKB-PPAR-γ axis 240 . Indeed, DEPTOR has elevated expression in WAT of obese mice whereas in humans DEPTOR expression in WAT correlates with the degree of obesity 240 .
In light of the protective roles of BAT and adipocyte browning against aging phenotypes, it can be speculated that the positive effects of mTORC1 inhibition on BAT and the stimulation of beige adipogenesis contribute to the anti-aging effects of rapamycin. This is plausibly also linked to the organismal energy status signaling capacity of BAT and the central role that mTOR plays in nutrient and energy sensing (detailed in the “mTORC1 as a sensor of amino acids” and “mTOR as a regulator of food intake and global energy homeostasis” sections). More work is necessary to understand how these complex interplays of networks and tissues are dysregulated in aging organisms.
Skeletal muscle
Aging skeletal muscle is characterized by muscle fiber loss leading to atrophy (sarcopenia). Although mTORC1 signaling is needed for increased muscle mass in response to exercise or during tissue repair 241 , very recent studies showed that mTORC1 signaling is activated in a subset of skeletal muscle fibers in aging mouse and human, which paradoxically was associated with fiber damage 242 . In addition, hyperactivation of mTORC1 (such as in TSC1 KO mice) led to abnormal mitochondria, oxidative stress, and damage and loss of fibers. The mechanisms involved mTORC1 regulation of STAT3 phosphorylation, associated with an increase in the expression of growth differentiation factors (GDF 3, 5, and 15) and of genes involved in oxidative stress and mitochondrial catabolism 242 . These processes were reversed by mTOR inhibition 242 . Moreover, as discussed in the “role of mTOR in stem cell maintenance and stem cell function decline in aging” section, in a model of premature aging, mTOR activity is increased in MDSPCs and causes stem cell dysfunction 208 . This indicates that maintaining a low basal state of the mTOR signaling in aging tissue could be important for maintenance of muscle function.
Future perspectives
Consistent with its role in coordinating protein synthesis, energy metabolism, and autophagy in cancer 10, 139 , emerging evidence suggests that mTOR may act as a central node that orchestrates many aspects of cellular and organismal biology related to aging phenotypes ( Figure 2). Inhibition of the mTOR pathway by rapamycin or genetic means has profound effects on life span and age-associated phenotypes across a wide array of organisms. However, the underlying mechanisms are still unclear as it has been reported that during aging mTOR activity is both increased and decreased, depending on, for example, tissue or sex 202, 243– 251 . It was suggested that, in spite of these variations, overall aging does not result in a generalized increase in mTOR signaling 243 . If this is the case, it is possible that mTOR activity aligns with the antagonistic pleiotropy theory of aging 252 , whereby its levels are beneficial during development but limit the health span in adult life.
Owing to its central role in age-related processes, mTOR represents an appealing target to ameliorate age-related pathologies. Despite its capacity to expand life span, the function of rapamycin (and of rapalogs) as an immunosuppressant 3 might be of concern, as a decline in immune function (immunosenescence) already occurs in the elderly, leading to infection-related morbidity and mortality 253, 254 . Intriguingly, several studies in both mice and humans suggest that mTOR inhibitors could reduce immunosenescence 202, 255 . In mice, rapamycin can restore the self-renewal and hematopoiesis of HSCs and enable effective vaccination against the influenza virus 202 . A randomized trial testing the effects of rapalog RAD001 in a cohort of healthy elderly patients also showed an enhanced response to the influenza vaccination, accompanied by a reduction in programmed death 1 (PD-1) receptor expressing CD4 and CD8 T lymphocytes 255 . PD-1 expression, which increases with age, inhibits T-cell signaling and reduces responses to antigen stimulation 256 . Moreover, a pilot study concluded that short-term rapamycin treatment (8 weeks) in healthy older persons was safe 257 .
Contrary to active-site mTOR inhibitors, allosteric inhibition of mTORC1 by rapamycin has little effect on 4E-BP phosphorylation 22, 23 and thus is expected to incompletely suppress mTORC1-dependent perturbations in translatome, mitochondrial functions, and metabolome 10 . Indeed, studies in D. melanogaster showed that 4E-BP extends life span upon DR by enhancing mitochondrial activity 258 . More recent studies showed that, in D. melanogaster, 4E-BP mediates temperature-induced effects on metabolism and life span 259 but that, in male mice, 4E-BP1 is involved in protecting against diet-induced obesity and insulin resistance 260,261 . Both second generation of mTOR inhibitors that target its active site and third generation (combining allosteric and active-site inhibition; Rapalink-1) potently suppress 4E-BP phosphorylation 7, 262 . Compared with what is known about rapamycin, however, much less is known of the effects of the active-site mTOR inhibitors in the context of aging.
Another limitation of rapamycin is that its chronic exposure in mice leads to mTORC2 inhibition in, for example, hepatocytes 30, 31 . Active-site mTOR inhibitors also inhibit mTORC2 7 . Strikingly, selective suppression of mTORC2 reduces life span 263, 264 and is associated with changes in endocrinology and metabolism (for example, insulin resistance), which have a negative impact on health span 31 . Thus, developing specific inhibitors which effectively suppress all mTORC1 outputs (including 4E-BP phosphorylation), but do not exert a major effect on mTORC2, appears to be warranted as a strategy to target age-related pathologies and improve health span. Interestingly, in a recent trial of healthy elderly patients, the combination of low-dose RAD001 (rapalog) and BEZ235 (dual mTOR/PI3K catalytic inhibitor) was proposed to selectively inhibit mTORC1 and not mTORC2 and led to enhanced immune function and a reduction in infections 265 . However, it is important to note that complete inhibition of mTORC1 can be deleterious. Deficiency in RagA, a GTPase responsible for mTORC1 activation by nutrients, leads to loss of mTORC1 activity and is embryonic lethal in mice 266 . Moreover, conditional deletion of RAPTOR (mTORC1 subunit) causes abnormalities in hematopoietic organs of adult mice 267 . Given these serious side effects of total mTORC1 inhibition and in light of the positive data from the RAD001/BEZ235 low-dose trial, it would be tempting to speculate that incomplete mTORC1 inhibition achieved by intermittent or low-dose treatment with mTOR inhibitors (or both) would carry the benefits of mTORC1 inhibition while limiting the side effects 268, 269 .
Biguanides (for example, metformin) are pharmaceuticals which are thought to have a beneficiary effect (in aging) that indirectly impinges on mTOR 270 . Metformin is a first-line anti-diabetic drug which has been used for more than 60 years in the clinic and has very few side effects. It was shown to modulate life span in model organisms ( C. elegans or M. musculus), to affect several processes dysregulated in aging (for example, cellular senescence, inflammation, autophagy, and protein synthesis), and (with the exception of one study 271 ) to improve cognitive function and neurodegeneration in humans 270 . At the organismal level, metformin reduces gluconeogenesis in the liver, which leads to normalization of glucose levels, decrease in insulinemia, and improvement of insulin resistance in fat, liver, and muscle 272 . By inhibiting mitochondrial complex I, metformin causes energetic stress which results in mTORC1 inhibition through AMPK (5′ AMP-activated protein kinase)-dependent and independent mechanisms 273 . Furthermore, metformin limits the secretion of numerous SASP factors by senescent cells 274 and has been shown to limit the spreading of cellular senescence in vivo 191 . Although many studies have uncovered possible targets of metformin action in the cell in the context of aging, the full extent of metformin’s mechanism of action at the cellular and organismal levels is still incompletely understood. This is complicated by the issues of achievable drug doses in humans compared with the concentration used in cell culture and animal models 272 . Nonetheless, clinical trials in which metformin is used to improve health span or aging-related conditions are being proposed. For instance, in the TAME (targeting aging with metformin) clinical trial, a placebo-controlled multi-center study of about 3000 elderly patients who are 65 to 79 years old, the effects of metformin on the development of age-associated outcomes like cardiovascular events, cancer, dementia, and mortality will be monitored 270 . In addition to providing information about metformin efficacy, the TAME study could provide the basis of establishing aging as an indicator for therapeutic purposes, which may encourage the development of next-generation drugs that target aging and extend healthy life span by modulating mTOR.
Significant progress has been made toward understanding how the mTOR signaling pathway regulates cellular processes relevant to aging (cellular and organismal energetics, proteostasis, cell stemness, cellular senescence, and so on), but as many of these advances were made in the context of cancer, much less is known about how these regulations influence the fate of the aging cell. Notwithstanding that mTOR inhibition clearly extends life span, outstanding questions abound regarding the underpinning mechanisms. For example, is inhibition of mTORC1 (as opposed to mTORC2) the main driver of increased life span and health span? What is the extent to which mTOR inhibition mediates the positive effects of CR/DR? What is the role of the mTORC1-specific mRNA translation program in aging? Much work is still needed to fill many gaps in knowledge related to the function of mTOR in the context of aging. This work may uncover unappreciated regulators or pathways that control the aging process and could lead to the development of drugs aimed at improving the health of the aging population.
Abbreviations
4E-BP, eukaryotic translation initiation factor 4E (eIF4E)-binding protein; AD, Alzheimer’s disease; AKT, protein kinase B; ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; BCAA, branched-chain amino acid; CASTOR, cellular arginine sensor for mTORC1; COX-2, cyclooxygenase-2; CR, caloric restriction; DEPTOR, DEP domain-containing mTOR-interacting protein; DR, dietary restriction; eIF2, eukaryotic translation initiation factor 2; eIF2α , eukaryotic translation initiation factor 2 alpha; eIF2β, eukaryotic translation initiation factor 2 beta; eIF2B, eukaryotic translation initiation factor 2B; ERK, extracellular signal-regulated kinase; FLCN, folliculin; FNIP1, folliculin-interacting protein 1; FNIP2, folliculin interacting protein 2; GATOR1, GTPase activating proteins toward Rags 1; GATOR2, GTPase activating proteins toward Rags 2; GCN2, general control non-derepressible 2; GEF, guanine nucleotide exchange factor; HSC, hematopoietic stem cell; IGF, insulin-like growth factor; IIS, insulin/insulin-like growth factor 1 (IGF-I) signaling; IL, interleukin; IR, insulin receptor; IRE1, inositol-requiring enzyme 1; ISC, intestinal stem cell; KICSTOR, KPTN-, ITFG2-, C12orf66-, and SZT2-containing regulator of mTORC1; KO, knockout; LAMTOR, late endosomal/lysosomal adaptor, MAPK and MTOR activator; LRS, leucyl-tRNA synthetase; MAPK, mitogen-activated protein kinase; MBH, mediobasal hypothalamus; MDSPC, muscle-derived stem/progenitor cell; MTFP1, mitochondrial fission process 1; mTOR, mammalian/mechanistic target of rapamycin; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NSC, neural stem cell; PD-1, programmed death 1; PERK, PKR-like ER kinase; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PR, protein restriction; RAG, Ras-related GTP binding; RHEB, Ras homologue enriched in brain; S6K, ribosomal protein S6 kinase; SAMTOR, S-adenosylmethionine sensor for the mTORC1 pathway; SASP, senescence-associated secretory phenotype; SGK, serum and glucocorticoid-regulated kinase 1; SLC38A9, member 9 of the solute carrier family 38; TAME, targeting aging with metformin; TC, ternary complex; TSC, tuberous sclerosis complex; uORF, upstream open reading frame; UPR, unfolded protein response; v-ATPase, vacuolar-type H + ATPase; XBP1, X-box binding protein 1.
Acknowledgements
We thank Marie-José Blouin of the Lady Davis Institute for Medical Research for critical reading of the manuscript.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Kathrin Thedieck, Institute of Biochemistry and Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innsbruck, Austria; Laboratory of Pediatrics, Section Systems Medicine of Metabolism and Signaling, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands; Faculty VI, School of Medicine and Health Sciences, Department of Neurosciences, Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
Alexander Martin Heberle, Institute of Biochemistry and Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innsbruck, Austria; Laboratory of Pediatrics, Section Systems Medicine of Metabolism and Signaling, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands
Patricia Razquin Navas, Laboratory of Pediatrics, Section Systems Medicine of Metabolism and Signaling, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands; Faculty VI, School of Medicine and Health Sciences, Department of Neurosciences, Carl von Ossietzky University of Oldenburg, Oldenburg, Germany
David M Sabatini, Whitehead Institute for Biomedical Research, Cambridge, MA, USA
Dudley Lamming, Department of Medicine, University of Wisconsin-Madison, Madison, WI, USA
Funding Statement
IT is a scholar of the Fonds de Recherche du Québec-Santé (FRQS; Junior 2). FAM holds the Canada Research Chair in Epigenetics of Aging and Cancer. KB is the recipient of a postdoctoral fellowship from the Cole Foundation. The work in our labs related to these problematics is funded in part by the Terry Fox Research Institute (TFRI-242115 to IT and MP), the CIHR/ISF/IDRC/Azrieli Foundation Joint Canada-Israel Research Program (to IT), and the Canadian Institutes of Health Research (MOP-133442 and PJT-159529 to FAM and LK, respectively).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved]
References
- 1. López-Otín C, Blasco MA, Partridge L, et al. : The hallmarks of aging. Cell. 2013;153:1194–217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Niccoli T, Partridge L: Ageing as a risk factor for disease. Curr Biol. 2012;22(17):R741–52. 10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 3. Saxton RA, Sabatini DM: mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168(6):960–76. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vézina C, Kudelski A, Sehgal SN: Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo). 1975;28(10):721–6. 10.7164/antibiotics.28.721 [DOI] [PubMed] [Google Scholar]
- 5. Heitman J, Movva NR, Hall MN: Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–9. 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- 6. Brown EJ, Albers MW, Shin TB, et al. : A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–8. 10.1038/369756a0 [DOI] [PubMed] [Google Scholar]
- 7. Bhat M, Robichaud N, Hulea L, et al. : Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14(4):261–78. 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- 8. Iglesias-Bartolome R, Patel V, Cotrim A, et al. : mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell. 2012;11(3):401–14. 10.1016/j.stem.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Kolesnichenko M, Hong L, Liao R, et al. : Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell Cycle. 2012;11(2):2391–401. 10.4161/cc.20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uchenunu O, Pollak M, Topisirovic I, et al. : Oncogenic kinases and perturbations in protein synthesis machinery and energetics in neoplasia. J Mol Endocrinol. 2019;62(2):R83–R103. 10.1530/JME-18-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahara T, Maeda T: Evolutionarily conserved regulation of TOR signalling. J Biochem. 2013;154(1):1–10. 10.1093/jb/mvt047 [DOI] [PubMed] [Google Scholar]
- 12. Hara K, Maruki Y, Long X, et al. : Raptor, a Binding Partner of Target of Rapamycin (TOR), Mediates TOR Action. Cell. 2002;110(2):177–89. 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Kim DH, Sarbassov DD, Ali SM, et al. : mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery. Cell. 2002;110(2):163–75. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Sarbassov DD, Ali SM, Kim DH, et al. : Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–302. 10.1016/j.cub.2004.06.054 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Loewith R, Jacinto E, Wullschleger S, et al. : Two TOR Complexes, Only One of which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control. Mol Cell. 2002;10(3):457–68. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- 16. Caron E, Ghosh S, Matsuoka Y, et al. : A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6:453. 10.1038/msb.2010.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwiatkowski DJ, Manning BD: Tuberous sclerosis: A GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14(suppl_2):R251–8. 10.1093/hmg/ddi260 [DOI] [PubMed] [Google Scholar]
- 18. Dibble CC, Manning BD: Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15(6):555–64. 10.1038/ncb2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamagata K, Sanders LK, Kaufmann WE, et al. : rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269(23):16333–9. [PubMed] [Google Scholar]
- 20. Yang H, Jiang X, Li B, et al. : Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552(7685):368–373. 10.1038/nature25023 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Roux PP, Topisirovic I: Signaling Pathways Involved in the Regulation of mRNA Translation. Mol Cell Biol. 2018;38(12): pii: e00070-18. 10.1128/MCB.00070-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang SA, Pacold ME, Cervantes CL, et al. : mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341(6144):1236566. 10.1126/science.1236566 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Yoon SO, Roux PP: Rapamycin Resistance: MTORC1 substrates hold some of the answers. Curr Biol. 2013;23(19):R880–3. 10.1016/j.cub.2013.08.030 [DOI] [PubMed] [Google Scholar]
- 24. Li S, Oh YT, Yue P, et al. : Inhibition of mTOR complex 2 induces GSK3/FBXW7-dependent degradation of sterol regulatory element-binding protein 1 (SREBP1) and suppresses lipogenesis in cancer cells. Oncogene. 2016;35(5):642–50. 10.1038/onc.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hagiwara A, Cornu M, Cybulski N, et al. : Hepatic mTORC2 Activates Glycolysis and Lipogenesis through Akt, Glucokinase, and SREBP1c. Cell Metab. 2012;15(5):725–38. 10.1016/j.cmet.2012.03.015 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Masui K, Cavenee WK, Mischel PS: mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol Metab. 2014;25(7):364–73. 10.1016/j.tem.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lang F, Pearce D: Regulation of the epithelial Na + channel by the mTORC2/SGK1 pathway. Nephrol Dial Transplant. 2016;31(2):200–5. 10.1093/ndt/gfv270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oh WJ, Jacinto E: mTOR complex 2 signaling and functions. Cell Cycle. 2011;10(14):2305–16. 10.4161/cc.10.14.16586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oshiro N, Yoshino K, Hidayat S, et al. : Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9(4):359–66. 10.1111/j.1356-9597.2004.00727.x [DOI] [PubMed] [Google Scholar]
- 30. Sarbassov DD, Ali SM, Sengupta S, et al. : Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68. 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- 31. Lamming DW, Ye L, Katajisto P, et al. : Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–43. 10.1126/science.1215135 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Schreiber KH, Ortiz D, Academia EC, et al. : Rapamycin-mediated mTORC2 inhibition is determined by the relative expression of FK506-binding proteins. Aging Cell. 2015;14(2):265–73. 10.1111/acel.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hinnebusch AG: The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- 34. Hay N, Sonenberg N: Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45. 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- 35. Fabrizio P, Pozza F, Pletcher SD, et al. : Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–90. 10.1126/science.1059497 [DOI] [PubMed] [Google Scholar]
- 36. Jia K, Chen D, Riddle DL: The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131(16):3897–906. 10.1242/dev.01255 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Vellai T, Takacs-Vellai K, Zhang Y, et al. : Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967):620. 10.1038/426620a [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Kapahi P, Zid BM, Harper T, et al. : Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14(10):885–90. 10.1016/j.cub.2004.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anisimov VN, Zabezhinski MA, Popovich IG, et al. : Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10(24):4230–6. 10.4161/cc.10.24.18486 [DOI] [PubMed] [Google Scholar]
- 40. Bjedov I, Toivonen JM, Kerr F, et al. : Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. 10.1016/j.cmet.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harrison DE, Strong R, Sharp ZD, et al. : Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–5. 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Medvedik O, Lamming DW, Kim KD, et al. : MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in saccharomyces cerevisiae. PLoS Biol. 2007;5(10):e261. 10.1371/journal.pbio.0050261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller RA, Harrison DE, Astle CM, et al. : Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66(2):191–201. 10.1093/gerona/glq178 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Powers RW, 3rd, Kaeberlein M, Caldwell SD, et al. : Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20(2):174–84. 10.1101/gad.1381406 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Robida-Stubbs S, Glover-Cutter K, Lamming DW, et al. : TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15(5):713–24. 10.1016/j.cmet.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Antikainen H, Driscoll M, Haspel G, et al. : TOR-mediated regulation of metabolism in aging. Aging Cell. 2017;16(6):1219–1233. 10.1111/acel.12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan H, Finkel T: Key proteins and pathways that regulate lifespan. J Biol Chem. 2017;292(16):6452–6460. 10.1074/jbc.R116.771915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hwangbo DS, Gersham B, Tu MP, et al. : Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–6. 10.1038/nature02549 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Lin K, Dorman JB, Rodan A, et al. : daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–22. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- 50. Selman C, Lingard S, Choudhury AI, et al. : Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22(3):807–18. 10.1096/fj.07-9261com [DOI] [PubMed] [Google Scholar]
- 51. Selman C, Partridge L, Withers DJ: Replication of extended lifespan phenotype in mice with deletion of insulin receptor substrate 1. PLoS One. 2011;6(1):e16144. 10.1371/journal.pone.0016144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tatar M, Kopelman A, Epstein D, et al. : A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–10. 10.1126/science.1057987 [DOI] [PubMed] [Google Scholar]
- 53. Ma L, Chen Z, Erdjument-Bromage H, et al. : Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121(2):179–93. 10.1016/j.cell.2005.02.031 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Manning BD, Tee AR, Logsdon MN, et al. : Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10(1):151–62. 10.1016/S1097-2765(02)00568-3 [DOI] [PubMed] [Google Scholar]
- 55. Inoki K, Li Y, Zhu T, et al. : TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–57. 10.1038/ncb839 [DOI] [PubMed] [Google Scholar]
- 56. Roux PP, Ballif BA, Anjum R, et al. : Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101(37):13489–94. 10.1073/pnas.0405659101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anderson RM, Le Couteur DG, de Cabo R: Caloric Restriction Research: New Perspectives on the Biology of Aging. J Gerontol A Biol Sci Med Sci. 2017;73(1):1–3. 10.1093/gerona/glx212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gems D, Partridge L: Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol. 2013;75:621–44. 10.1146/annurev-physiol-030212-183712 [DOI] [PubMed] [Google Scholar]
- 59. Mattison JA, Colman RJ, Beasley TM, et al. : Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. 10.1038/ncomms14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee C, Longo V: Dietary restriction with and without caloric restriction for healthy aging [version 1; peer review: 3 approved]. F1000Res. 2016;5: pii: F1000 Faculty Rev-117. 10.12688/f1000research.7136.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hansen M, Taubert S, Crawford D, et al. : Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6(1):95–110. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- 62. Kaeberlein M, Powers RW, 3rd, Steffen KK, et al. : Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193–6. 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Fok WC, Livi C, Bokov A, et al. : Short-term rapamycin treatment in mice has few effects on the transcriptome of white adipose tissue compared to dietary restriction. Mech Ageing Dev. 2014;140:23–9. 10.1016/j.mad.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu Z, Wang R, Fok WC, et al. : Rapamycin and dietary restriction induce metabolically distinctive changes in mouse liver. J Gerontol A Biol Sci Med Sci. 2015;70(4):410–20. 10.1093/gerona/glu053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choi KM, Hong SJ, van Deursen JM, et al. : Caloric Restriction and Rapamycin Differentially Alter Energy Metabolism in Yeast. J Gerontol A Biol Sci Med Sci. 2018;73(1):29–38. 10.1093/gerona/glx024 [DOI] [PubMed] [Google Scholar]
- 66. Thoreen CC, Kang SA, Chang JW, et al. : An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–32. 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hulea L, Gravel SP, Morita M, et al. : Translational and HIF-1α-Dependent Metabolic Reprogramming Underpin Metabolic Plasticity and Responses to Kinase Inhibitors and Biguanides. Cell Metab. 2018;28(6):817–832.e8. 10.1016/j.cmet.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morita M, Gravel SP, Chénard V, et al. : mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18(5):698–711. 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Morita M, Prudent J, Basu K, et al. : mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell. 2017;67(6):922–935.e5. 10.1016/j.molcel.2017.08.013 [DOI] [PubMed] [Google Scholar]
- 70. Mirzaei H, Suarez JA, Longo VD: Protein and amino acid restriction, aging and disease: from yeast to humans. Trends Endocrinol Metab. 2014;25(11):558–66. 10.1016/j.tem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Solon-Biet SM, McMahon AC, Ballard JW, et al. : The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19(3):418–30. 10.1016/j.cmet.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Levine ME, Suarez JA, Brandhorst S, et al. : Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab. 2014;19(3):407–17. 10.1016/j.cmet.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun L, Sadighi Akha AA, Miller RA, et al. : Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64(7):711–22. 10.1093/gerona/glp051 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Segall PE, Timiras PS: Patho-physiologic findings after chronic tryptophan deficiency in rats: a model for delayed growth and aging. Mech Ageing Dev. 1976;5(2):109–24. 10.1016/0047-6374(76)90012-9 [DOI] [PubMed] [Google Scholar]
- 75. Richie JP, Jr, Leutzinger Y, Parthasarathy S, et al. : Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8(15):1302–7. 10.1096/fasebj.8.15.8001743 [DOI] [PubMed] [Google Scholar]
- 76. Orentreich N, Matias JR, DeFelice A, et al. : Low methionine ingestion by rats extends life span. J Nutr. 1993;123(2):269–74. [DOI] [PubMed] [Google Scholar]
- 77. Ooka H, Segall PE, Timiras PS: Histology and survival in age-delayed low-tryptophan-fed rats. Mech Ageing Dev. 1988;43(1):79–98. 10.1016/0047-6374(88)90099-1 [DOI] [PubMed] [Google Scholar]
- 78. Miller RA, Buehner G, Chang Y, et al. : Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4(3):119–25. 10.1111/j.1474-9726.2005.00152.x [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Malloy VL, Krajcik RA, Bailey SJ, et al. : Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell. 2006;5(4):305–14. 10.1111/j.1474-9726.2006.00220.x [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. De Marte ML, Enesco HE: Influence of low tryptophan diet on survival and organ growth in mice. Mech Ageing Dev. 1986;36(2):161–71. 10.1016/0047-6374(86)90017-5 [DOI] [PubMed] [Google Scholar]
- 81. Lees EK, Król E, Grant L, et al. : Methionine restriction restores a younger metabolic phenotype in adult mice with alterations in fibroblast growth factor 21. Aging Cell. 2014;13(5):817–27. 10.1111/acel.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yu D, Yang SE, Miller BR, et al. : Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J. 2018;32(6):3471–82. 10.1096/fj.201701211R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cummings NE, Williams EM, Kasza I, et al. : Restoration of metabolic health by decreased consumption of branched-chain amino acids. J Physiol. 2018;596(4):623–45. 10.1113/JP275075 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Fontana L, Cummings NE, Arriola Apelo SI, et al. : Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell Rep. 2016;16(2):520–30. 10.1016/j.celrep.2016.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lees EK, Banks R, Cook C, et al. : Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci Rep. 2017;7(1):9977. 10.1038/s41598-017-10381-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Solon-Biet SM, Cogger VC, Pulpitel T, et al. : Branched-chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat Metab. 2019;1:532–45. 10.1038/s42255-019-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Khayati K, Antikainen H, Bonder EM, et al. : The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. 2017;31(2):598–609. 10.1096/fj.201600915R [DOI] [PubMed] [Google Scholar]
- 88. Lamming DW, Cummings NE, Rastelli AL, et al. : Restriction of dietary protein decreases mTORC1 in tumors and somatic tissues of a tumor-bearing mouse xenograft model. Oncotarget. 2015;6(31):31233–40. 10.18632/oncotarget.5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hardie DG, Ross FA, Hawley SA: AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–62. 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Efeyan A, Comb WC, Sabatini DM: Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–10. 10.1038/nature14190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Albert V, Hall MN: mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. 10.1016/j.ceb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 92. Kim E, Goraksha-Hicks P, Li L, et al. : Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–45. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sancak Y, Peterson TR, Shaul YD, et al. : The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 94. Sancak Y, Bar-Peled L, Zoncu R, et al. : Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Bar-Peled L, Schweitzer LD, Zoncu R, et al. : Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Fawal MA, Brandt M, Djouder N: MCRS1 binds and couples Rheb to amino acid-dependent mTORC1 activation. Dev Cell. 2015;33(1):67–81. 10.1016/j.devcel.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 97. Bar-Peled L, Chantranupong L, Cherniack AD, et al. : A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–6. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 98. Wolfson RL, Chantranupong L, Wyant GA, et al. : KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543(7645):438–42. 10.1038/nature21423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Petit CS, Roczniak-Ferguson A, Ferguson SM: Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202(7):1107–22. 10.1083/jcb.201307084 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Tsun ZY, Bar-Peled L, Chantranupong L, et al. : The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52(4):495–505. 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Paquette M, El-Houjeiri L, Pause A: mTOR Pathways in Cancer and Autophagy. Cancers (Basel). 2018;10(1): pii: E18. 10.3390/cancers10010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jung J, Genau HM, Behrends C: Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–94. 10.1128/MCB.00125-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rebsamen M, Pochini L, Stasyk T, et al. : SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–81. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Wang S, Tsun ZY, Wolfson RL, et al. : Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188–94. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Wyant GA, Abu-Remaileh M, Wolfson RL, et al. : mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell. 2017;171(3):642–654.e12. 10.1016/j.cell.2017.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Saxton RA, Chantranupong L, Knockenhauer KE, et al. : Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536(7615):229–33. 10.1038/nature19079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chantranupong L, Scaria SM, Saxton RA, et al. : The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165(1):153–64. 10.1016/j.cell.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Saxton RA, Knockenhauer KE, Wolfson RL, et al. : Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2015;351(6268):53–8. 10.1126/science.aad2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kim JS, Ro SH, Kim M, et al. : Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci Rep. 2015;5:9502. 10.1038/srep09502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Parmigiani A, Nourbakhsh A, Ding B, et al. : Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9(4):1281–91. 10.1016/j.celrep.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wolfson RL, Chantranupong L, Saxton RA, et al. : Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351(6268):43–8. 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Peng M, Yin N, Li MO: Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159(1):122–33. 10.1016/j.cell.2014.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Han JM, Jeong SJ, Park MC, et al. : Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–24. 10.1016/j.cell.2012.02.044 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Kim JH, Lee C, Lee M, et al. : Control of leucine-dependent mTORC1 pathway through chemical intervention of leucyl-tRNA synthetase and RagD interaction. Nat Commun. 2017;8(1):732. 10.1038/s41467-017-00785-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lee M, Kim JH, Yoon I, et al. : Coordination of the leucine-sensing Rag GTPase cycle by leucyl-tRNA synthetase in the mTORC1 signaling pathway. Proc Natl Acad Sci U S A. 2018;115(23):E5279–E5288. 10.1073/pnas.1801287115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yoon MS, Son K, Arauz E, et al. : Leucyl-tRNA Synthetase Activates Vps34 in Amino Acid-Sensing mTORC1 Signaling. Cell Rep. 2016;16(6):1510–7. 10.1016/j.celrep.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. He XD, Gong W, Zhang JN, et al. : Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations. Cell Metab. 2018;27(1):151–166.e6. 10.1016/j.cmet.2017.10.015 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 118. Gu X, Orozco JM, Saxton RA, et al. : SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358(6364):813–8. 10.1126/science.aao3265 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 119. Jewell JL, Kim YC, Russell RC, et al. : Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347(6218):194–8. 10.1126/science.1259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Quan N, Sun W, Wang L, et al. : Sestrin2 prevents age-related intolerance to ischemia and reperfusion injury by modulating substrate metabolism. FASEB J. 2017;31(9):4153–67. 10.1096/fj.201700063R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Quan N, Wang L, Chen X, et al. : Sestrin2 prevents age-related intolerance to post myocardial infarction via AMPK/PGC-1α pathway. J Mol Cell Cardiol. 2018;115:170–8. 10.1016/j.yjmcc.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang C, Sun W, Li J, et al. : Loss of sestrin 2 potentiates the early onset of age-related sensory cell degeneration in the cochlea. Neuroscience. 2017;361:179–91. 10.1016/j.neuroscience.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lucanic M, Garrett T, Yu I, et al. : Chemical activation of a food deprivation signal extends lifespan. Aging Cell. 2016;15(5):832–41. 10.1111/acel.12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gao Q, Horvath TL: Neuronal control of energy homeostasis. FEBS Lett. 2008;582(1):132–41. 10.1016/j.febslet.2007.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cota D, Proulx K, Smith KA, et al. : Hypothalamic mTOR signaling regulates food intake. Science. 2006;312(5775):927–30. 10.1126/science.1124147 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 126. Blouet C, Ono H, Schwartz GJ: Mediobasal hypothalamic p70 S6 kinase 1 modulates the control of energy homeostasis. Cell Metab. 2008;8(6):459–67. 10.1016/j.cmet.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cota D, Matter EK, Woods SC, et al. : The role of hypothalamic mammalian target of rapamycin complex 1 signaling in diet-induced obesity. J Neurosci. 2008;28(28):7202–8. 10.1523/JNEUROSCI.1389-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Smith MA, Katsouri L, Irvine EE, et al. : Ribosomal S6K1 in POMC and AgRP Neurons Regulates Glucose Homeostasis but Not Feeding Behavior in Mice. Cell Rep. 2015;11(3):335–43. 10.1016/j.celrep.2015.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Caron A, Labbé SM, Lanfray D, et al. : Mediobasal hypothalamic overexpression of DEPTOR protects against high-fat diet-induced obesity. Mol Metab. 2016;5(2):102–12. 10.1016/j.molmet.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Catena V, Fanciulli M: Deptor: not only a mTOR inhibitor. J Exp Clin Cancer Res. 2017;36(1):12. 10.1186/s13046-016-0484-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Burke LK, Darwish T, Cavanaugh AR, et al. : mTORC1 in AGRP neurons integrates exteroceptive and interoceptive food-related cues in the modulation of adaptive energy expenditure in mice. eLife. 2017;6: pii: e22848. 10.7554/eLife.22848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lin J, Handschin C, Spiegelman BM: Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–70. 10.1016/j.cmet.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 133. Fisher FM, Maratos-Flier E: Understanding the Physiology of FGF21. Annu Rev Physiol. 2016;78:223–41. 10.1146/annurev-physiol-021115-105339 [DOI] [PubMed] [Google Scholar]
- 134. Cornu M, Oppliger W, Albert V, et al. : Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci U S A. 2014;111(32):11592–9. 10.1073/pnas.1412047111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Polak P, Cybulski N, Feige JN, et al. : Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8(5):399–410. 10.1016/j.cmet.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 136. Koga H, Kaushik S, Cuervo AM: Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–15. 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Powers ET, Morimoto RI, Dillin A, et al. : Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- 138. Gonskikh Y, Polacek N: Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev. 2017;168:30–6. 10.1016/j.mad.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 139. Lindqvist LM, Tandoc K, Topisirovic I, et al. : Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr Opin Genet Dev. 2018;48:104–11. 10.1016/j.gde.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 140. Zhao J, Zhai B, Gygi SP, et al. : mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112(52):15790–7. 10.1073/pnas.1521919112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Rousseau A, Bertolotti A: An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536(7615):184–9. 10.1038/nature18943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang Y, Nicholatos J, Dreier JR, et al. : Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513(7518):440–3. 10.1038/nature13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Taylor RC: Aging and the UPR(ER). Brain Res. 2016;1648(Pt B):588–93. 10.1016/j.brainres.2016.04.017 [DOI] [PubMed] [Google Scholar]
- 144. Hetz C: The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. 10.1038/nrm3270 [DOI] [PubMed] [Google Scholar]
- 145. Calfon M, Zeng H, Urano F, et al. : IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–6. 10.1038/415092a [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 146. Lee K, Tirasophon W, Shen X, et al. : IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16(4):452–66. 10.1101/gad.964702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Yoshida H, Matsui T, Yamamoto A, et al. : XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–91. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 148. Hetz C, Chevet E, Oakes SA: Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17(7):829–38. 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bachar E, Ariav Y, Ketzinel-Gilad M, et al. : Glucose amplifies fatty acid-induced endoplasmic reticulum stress in pancreatic beta-cells via activation of mTORC1. PLoS One. 2009;4(3):e4954. 10.1371/journal.pone.0004954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pfaffenbach KT, Nivala AM, Reese L, et al. : Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J Nutr. 2010;140(5):879–84. 10.3945/jn.109.119883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Walter P, Ron D: The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 152. Konieczny A, Safer B: Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983;258(5):3402–8. [PubMed] [Google Scholar]
- 153. Krishnamoorthy T, Pavitt GD, Zhang F, et al. : Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol. 2001;21(15):5018–30. 10.1128/MCB.21.15.5018-5030.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Clemens MJ, Pain VM, Wong ST, et al. : Phosphorylation inhibits guanine nucleotide exchange on eukaryotic initiation factor 2. Nature. 1982;296(5852):93–5. 10.1038/296093a0 [DOI] [PubMed] [Google Scholar]
- 155. Dever TE, Feng L, Wek RC, et al. : Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68(3):585–96. 10.1016/0092-8674(92)90193-G [DOI] [PubMed] [Google Scholar]
- 156. B'chir W, Chaveroux C, Carraro V, et al. : Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal. 2014;26(7):1385–91. 10.1016/j.cellsig.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 157. Harding HP, Zhang Y, Zeng H, et al. : An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–33. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 158. Ye J, Koumenis C: ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med. 2009;9(4):411–6. 10.2174/156652409788167096 [DOI] [PubMed] [Google Scholar]
- 159. Guan BJ, van Hoef V, Jobava R, et al. : A Unique ISR Program Determines Cellular Responses to Chronic Stress. Mol Cell. 2017;68(5):885–900.e6. 10.1016/j.molcel.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Krokowski D, Han J, Saikia M, et al. : A self-defeating anabolic program leads to β-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. J Biol Chem. 2013;288(24):17202–13. 10.1074/jbc.M113.466920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Guan BJ, Krokowski D, Majumder M, et al. : Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. J Biol Chem. 2014;289(18):12593–611. 10.1074/jbc.M113.543215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gandin V, Masvidal L, Cargnello M, et al. : mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat Commun. 2016;7: 11127. 10.1038/ncomms11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wengrod J, Wang D, Weiss S, et al. : Phosphorylation of eIF2α triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci Signal. 2015;8(367):ra27. 10.1126/scisignal.aaa0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Krishnamoorthy J, Tenkerian C, Gupta J, et al. : Downregulation of PERK activity and eIF2α serine 51 phosphorylation by mTOR complex 1 elicits pro-oxidant and pro-death effects in tuberous sclerosis-deficient cells. Cell Death Dis. 2018;9(3): 254. 10.1038/s41419-018-0326-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Park Y, Reyna-Neyra A, Philippe L, et al. : mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 2017;19(6):1083–90. 10.1016/j.celrep.2017.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Trifunovic A, Wredenberg A, Falkenberg M, et al. : Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 167. Baker DJ, Peleg S: Biphasic Modeling of Mitochondrial Metabolism Dysregulation during Aging. Trends Biochem Sci. 2017;42(9):702–11. 10.1016/j.tibs.2017.06.005 [DOI] [PubMed] [Google Scholar]
- 168. Dhillon RS, Denu JM: Using comparative biology to understand how aging affects mitochondrial metabolism. Mol Cell Endocrinol. 2017;455:54–61. 10.1016/j.mce.2016.12.020 [DOI] [PubMed] [Google Scholar]
- 169. Moiseeva O, Bourdeau V, Roux A, et al. : Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol. 2009;29(16):4495–507. 10.1128/MCB.01868-08 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 170. Gandin V, Masvidal L, Hulea L, et al. : nanoCAGE reveals 5' UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26(5):636–48. 10.1101/gr.197566.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Cunningham JT, Rodgers JT, Arlow DH, et al. : mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–40. 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 172. Betz C, Stracka D, Prescianotto-Baschong C, et al. : Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A. 2013;110(31):12526–34. 10.1073/pnas.1302455110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 173. Kim I, Rodriguez-Enriquez S, Lemasters JJ: Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–53. 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ryu D, Mouchiroud L, Andreux PA, et al. : Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22(8):879–88. 10.1038/nm.4132 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 175. Schiavi A, Maglioni S, Palikaras K, et al. : Iron-Starvation-Induced Mitophagy Mediates Lifespan Extension upon Mitochondrial Stress in C. elegans. Curr Biol. 2015;25(14):1810–22. 10.1016/j.cub.2015.05.059 [DOI] [PubMed] [Google Scholar]
- 176. Bartolomé A, García-Aguilar A, Asahara SI, et al. : MTORC1 Regulates both General Autophagy and Mitophagy Induction after Oxidative Phosphorylation Uncoupling. Mol Cell Biol. 2017;37(23): pii: e00441-17. 10.1128/MCB.00441-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wiley CD, Campisi J: From Ancient Pathways to Aging Cells-Connecting Metabolism and Cellular Senescence. Cell Metab. 2016;23(6):1013–21. 10.1016/j.cmet.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 178. Passos JF, Nelson G, Wang C, et al. : Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6(1):347. 10.1038/msb.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Passos JF, Saretzki G, Ahmed S, et al. : Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5(5):e110. 10.1371/journal.pbio.0050110 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 180. Rodier F, Coppé JP, Patil CK, et al. : Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–9. 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 181. Sapieha P, Mallette FA: Cellular Senescence in Postmitotic Cells: Beyond Growth Arrest. Trends Cell Biol. 2018;28(8):595–607. 10.1016/j.tcb.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 182. Serrano M, Lin AW, McCurrach ME, et al. : Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16 INK4a. Cell. 1997;88(5):593–602. 10.1016/S0092-8674(00)81902-9 [DOI] [PubMed] [Google Scholar]
- 183. Frippiat C, Chen QM, Zdanov S, et al. : Subcytotoxic H 2O 2 stress triggers a release of transforming growth factor-beta 1, which induces biomarkers of cellular senescence of human diploid fibroblasts. J Biol Chem. 2001;276(4):2531–7. 10.1074/jbc.M006809200 [DOI] [PubMed] [Google Scholar]
- 184. Karlseder J, Smogorzewska A, de Lange T: Senescence induced by altered telomere state, not telomere loss. Science. 2002;295(5564):2446–9. 10.1126/science.1069523 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 185. Mallette FA, Gaumont-Leclerc MF, Ferbeyre G: The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21(1):43–8. 10.1101/gad.1487307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Mallette FA, Gaumont-Leclerc MF, Huot G, et al. : Myc down-regulation as a mechanism to activate the Rb pathway in STAT5A-induced senescence. J Biol Chem. 2007;282(48):34938–44. 10.1074/jbc.M707074200 [DOI] [PubMed] [Google Scholar]
- 187. Demaria M, Ohtani N, Youssef SA, et al. : An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–33. 10.1016/j.devcel.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Baker DJ, Childs BG, Durik M, et al. : Naturally occurring p16 Ink4a-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–9. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 189. Baker DJ, Wijshake T, Tchkonia T, et al. : Clearance of p16 Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–6. 10.1038/nature10600 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 190. Milanovic M, Fan DNY, Belenki D, et al. : Senescence-associated reprogramming promotes cancer stemness. Nature. 2018;553(7686):96–100. 10.1038/nature25167 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 191. Oubaha M, Miloudi K, Dejda A, et al. : Senescence-associated secretory phenotype contributes to pathological angiogenesis in retinopathy. Sci Transl Med. 2016;8(362):362ra144. 10.1126/scitranslmed.aaf9440 [DOI] [PubMed] [Google Scholar]
- 192. Campisi J: Aging, Cellular Senescence, and Cancer. Annu Rev Physiol. 2013;75:685–705. 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Dörr JR, Yu Y, Milanovic M, et al. : Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501(7467):421–5. 10.1038/nature12437 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 194. Narita M, Young AR, Arakawa S, et al. : Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332(6032):966–70. 10.1126/science.1205407 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 195. Laberge RM, Sun Y, Orjalo AV, et al. : MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17(8):1049–61. 10.1038/ncb3195 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 196. Herranz N, Gallage S, Mellone M, et al. : mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17(9):1205–17. 10.1038/ncb3225 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 197. Correia-Melo C, Marques FDM, Anderson R, et al. : Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016;35(7):724–42. 10.15252/embj.201592862 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 198. Carroll B, Nelson G, Rabanal-Ruiz Y, et al. : Persistent mTORC1 signaling in cell senescence results from defects in amino acid and growth factor sensing. J Cell Biol. 2017;216(7):1949–57. 10.1083/jcb.201610113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 199. Ermolaeva M, Neri F, Ori A, et al. : Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol. 2018;19(9):594–610. 10.1038/s41580-018-0020-3 [DOI] [PubMed] [Google Scholar]
- 200. Sampath P, Pritchard DK, Pabon L, et al. : A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–60. 10.1016/j.stem.2008.03.013 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 201. Schultz MB, Sinclair DA: When stem cells grow old: phenotypes and mechanisms of stem cell aging. Development. 2016;143(1):3–14. 10.1242/dev.130633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Chen C, Liu Y, Liu Y, et al. : mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2(98):ra75. 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Yilmaz ÖH, Katajisto P, Lamming DW, et al. : mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486(7404):490–5. 10.1038/nature11163 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 204. Oh J, Lee YD, Wagers AJ: Stem cell aging: Mechanisms, regulators and therapeutic opportunities. Nat Med. 2014;20(8):870–80. 10.1038/nm.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Meng D, Frank AR, Jewell JL: mTOR signaling in stem and progenitor cells. Development. 2018;145(1): pii: dev152595. 10.1242/dev.152595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Igarashi M, Guarente L: mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell. 2016;166(2):436–50. 10.1016/j.cell.2016.05.044 [DOI] [PubMed] [Google Scholar]
- 207. Romine J, Gao X, Xu XM, et al. : The proliferation of amplifying neural progenitor cells is impaired in the aging brain and restored by the mTOR pathway activation. Neurobiol Aging. 2015;36(4):1716–26. 10.1016/j.neurobiolaging.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 208. Takayama K, Kawakami Y, Lavasani M, et al. : mTOR signaling plays a critical role in the defects observed in muscle-derived stem/progenitor cells isolated from a murine model of accelerated aging. J Orthop Res. 2017;35(7):1375–82. 10.1002/jor.23409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Selman C, Sinclair A, Pedroni SM, et al. : Evidence that hematopoietic stem cell function is preserved during aging in long-lived S6K1 mutant mice. Oncotarget. 2016;7(21):29937–43. 10.18632/oncotarget.8729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Sciarretta S, Forte M, Frati G, et al. : New Insights Into the Role of mTOR Signaling in the Cardiovascular System. Circ Res. 2018;122(3):489–505. 10.1161/CIRCRESAHA.117.311147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Shende P, Plaisance I, Morandi C, et al. : Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123(10):1073–82. 10.1161/CIRCULATIONAHA.110.977066 [DOI] [PubMed] [Google Scholar]
- 212. Zhang D, Contu R, Latronico MV, et al. : MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120(8):2805–16. 10.1172/JCI43008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Tamai T, Yamaguchi O, Hikoso S, et al. : Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem. 2013;288(14):10176–87. 10.1074/jbc.M112.423640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Zhu Y, Pires KM, Whitehead KJ, et al. : Mechanistic target of rapamycin ( Mtor) is essential for murine embryonic heart development and growth. PLoS One. 2013;8(1):e54221. 10.1371/journal.pone.0054221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Mazelin L, Panthu B, Nicot AS, et al. : mTOR inactivation in myocardium from infant mice rapidly leads to dilated cardiomyopathy due to translation defects and p53/JNK-mediated apoptosis. J Mol Cell Cardiol. 2016;97:213–25. 10.1016/j.yjmcc.2016.04.011 [DOI] [PubMed] [Google Scholar]
- 216. Flynn JM, O'Leary MN, Zambataro CA, et al. : Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12(5):851–62. 10.1111/acel.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Dai DF, Karunadharma PP, Chiao YA, et al. : Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13(3):529–39. 10.1111/acel.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 218. Laplante M, Sabatini DM: mTOR signaling in growth control and disease. Cell. 2012;149(2):274–93. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Hoeffer CA, Klann E: mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33(2):67–75. 10.1016/j.tins.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Garza-Lombó C, Gonsebatt ME: Mammalian Target of Rapamycin: Its Role in Early Neural Development and in Adult and Aged Brain Function. Front Cell Neurosci. 2016;10:157. 10.3389/fncel.2016.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Perluigi M, Di Domenico F, Butterfield DA: mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol Dis. 2015;84:39–49. 10.1016/j.nbd.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 222. Triplett JC, Tramutola A, Swomley A, et al. : Age-related changes in the proteostasis network in the brain of the naked mole-rat: Implications promoting healthy longevity. Biochim Biophys Acta. 2015;1852(10 Pt A):2213–24. 10.1016/j.bbadis.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Caccamo A, Majumder S, Richardson A, et al. : Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: Effects on cognitive impairments. J Biol Chem. 2010;285(17):13107–20. 10.1074/jbc.M110.100420 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 224. Spilman P, Podlutskaya N, Hart MJ, et al. : Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One. 2010;5(4):e9979. 10.1371/journal.pone.0009979 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 225. Lin AL, Zheng W, Halloran JJ, et al. : Chronic rapamycin restores brain vascular integrity and function through NO synthase activation and improves memory in symptomatic mice modeling Alzheimer's disease. J Cereb Blood Flow Metab. 2013;33(9):1412–21. 10.1038/jcbfm.2013.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Majumder S, Richardson A, Strong R, et al. : Inducing Autophagy by Rapamycin Before, but Not After, the Formation of Plaques and Tangles Ameliorates Cognitive Deficits. PLoS One. 2011;6(9):e25416. 10.1371/journal.pone.0025416 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 227. Tramutola A, Lanzillotta C, Barone E, et al. : Intranasal rapamycin ameliorates Alzheimer-like cognitive decline in a mouse model of Down syndrome. Transl Neurodegener. 2018;7:28. 10.1186/s40035-018-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Lang UE, Heger J, Willbring M, et al. : Immunosuppression using the mammalian target of rapamycin (mTOR) inhibitor everolimus: pilot study shows significant cognitive and affective improvement. Transplant Proc. 2009;41(10):4285–8. 10.1016/j.transproceed.2009.08.050 [DOI] [PubMed] [Google Scholar]
- 229. Ghaben AL, Scherer PE: Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20(4):242–58. 10.1038/s41580-018-0093-z [DOI] [PubMed] [Google Scholar]
- 230. Chouchani ET, Kazak L, Spiegelman BM: New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019;29(1):27–37. 10.1016/j.cmet.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 231. Kazak L, Chouchani ET, Lu GZ, et al. : Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 2017;26(4):660–671.e3. 10.1016/j.cmet.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Rothwell NJ, Stock MJ: A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281(5726):31–5. 10.1038/281031a0 [DOI] [PubMed] [Google Scholar]
- 233. Kazak L, Rahbani JF, Samborska B, et al. : Ablation of adipocyte creatine transport impairs thermogenesis and causes diet-induced obesity. Nat Metab. 2019;1(3):360–370. 10.1038/s42255-019-0035-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Fawcett DW: Differences in Physiological Activity in Brown and White Fat as Revealed by Histochemical Reactions. Science. 1947;105(2718):123. 10.1126/science.105.2718.123 [DOI] [PubMed] [Google Scholar]
- 235. Wu J, Boström P, Sparks LM, et al. : Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–76. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 236. Gonçalves LF, Machado TQ, Castro-Pinheiro C, et al. : Ageing is associated with brown adipose tissue remodelling and loss of white fat browning in female C57BL/6 mice. Int J Exp Pathol. 2017;98(2):100–108. 10.1111/iep.12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Vatner DE, Zhang J, Oydanich M, et al. : Enhanced longevity and metabolism by brown adipose tissue with disruption of the regulator of G protein signaling 14. Aging Cell. 2018;17(4):e12751. 10.1111/acel.12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Xiang X, Lan H, Tang H, et al. : Tuberous Sclerosis Complex 1–Mechanistic Target of Rapamycin Complex 1 Signaling Determines Brown-to-White Adipocyte Phenotypic Switch. Diabetes. 2015;64(2):519–28. 10.2337/db14-0427 [DOI] [PubMed] [Google Scholar]
- 239. Zhang X, Luo Y, Wang C, et al. : Adipose mTORC1 Suppresses Prostaglandin Signaling and Beige Adipogenesis via the CRTC2-COX-2 Pathway. Cell Rep. 2018;24(12):3180–3193. 10.1016/j.celrep.2018.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Laplante M, Horvat S, Festuccia WT, et al. : DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metab. 2012;16(2):202–12. 10.1016/j.cmet.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Frey JW, Jacobs BL, Goodman CA, et al. : A role for Raptor phosphorylation in the mechanical activation of mTOR signaling. Cell Signal. 2014;26(2):313–22. 10.1016/j.cellsig.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Tang H, Inoki K, Brooks SV, et al. : mTORC1 underlies age-related muscle fiber damage and loss by inducing oxidative stress and catabolism. Aging Cell. 2019;18(3):e12943. 10.1111/acel.12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Baar EL, Carbajal KA, Ong IM, et al. : Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice. Aging Cell. 2016;15(1):155–66. 10.1111/acel.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Calhoun C, Shivshankar P, Saker M, et al. : Senescent Cells Contribute to the Physiological Remodeling of Aged Lungs. J Gerontol A Biol Sci Med Sci. 2016;71(2):153–60. 10.1093/gerona/glu241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Harries LW, Fellows AD, Pilling LC, et al. : Advancing age is associated with gene expression changes resembling mTOR inhibition: Evidence from two human populations. Mech Ageing Dev. 2012;133(8):556–62. 10.1016/j.mad.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Houtkooper RH, Argmann C, Houten SM, et al. : The metabolic footprint of aging in mice. Sci Rep. 2011;1:134. 10.1038/srep00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Leontieva OV, Paszkiewicz GM, Blagosklonny MV: Fasting levels of hepatic p-S6 are increased in old mice. Cell Cycle. 2014;13(17):2656–9. 10.4161/15384101.2014.949150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Linford NJ, Beyer RP, Gollahon K, et al. : Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell. 2007;6(5):673–88. 10.1111/j.1474-9726.2007.00319.x [DOI] [PubMed] [Google Scholar]
- 249. Sengupta S, Peterson TR, Laplante M, et al. : mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468(7327):1100–4. 10.1038/nature09584 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 250. Xie K, Ryan DP, Pearson BL, et al. : Epigenetic alterations in longevity regulators, reduced life span, and exacerbated aging-related pathology in old father offspring mice. Proc Natl Acad Sci U S A. 2018;115(10):E2348–E2357. 10.1073/pnas.1707337115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 251. Yang SB, Tien AC, Boddupalli G, et al. : Rapamycin ameliorates age-dependent obesity associated with increased mTOR signaling in hypothalamic POMC neurons. Neuron. 2012;75(3):425–36. 10.1016/j.neuron.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Blagosklonny MV: Revisiting the antagonistic pleiotropy theory of aging: TOR-driven program and quasi-program. Cell Cycle. 2010;9(16):3151–6. 10.4161/cc.9.16.13120 [DOI] [PubMed] [Google Scholar]
- 253. Murphy SL, Xu J, Kochanek KD: Deaths: Final data for 2010. Natl Vital Stat Rep. 2013;61(4):1–117. [PubMed] [Google Scholar]
- 254. National Center for Health Statistics (US): Health, United States, 2015: With Special Feature on Racial and Ethnic Health Disparities. Health, United States,2016. [PubMed] [Google Scholar]
- 255. Mannick JB, Del Giudice G, Lattanzi M, et al. : mTOR inhibition improves immune function in the elderly. Sci Transl Med. 2014;6(268):268ra179. 10.1126/scitranslmed.3009892 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 256. Lages CS, Lewkowich I, Sproles A, et al. : Partial restoration of T-cell function in aged mice by in vitro blockade of the PD-1/PD-L1 pathway. Aging Cell. 2010;9(5):785–98. 10.1111/j.1474-9726.2010.00611.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Kraig E, Linehan LA, Liang H, et al. : A randomized control trial to establish the feasibility and safety of rapamycin treatment in an older human cohort: Immunological, physical performance, and cognitive effects. Exp Gerontol. 2018;105:53–69. 10.1016/j.exger.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Zid BM, Rogers AN, Katewa SD, et al. : 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139(1):149–60. 10.1016/j.cell.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 259. Carvalho GB, Drago I, Hoxha S, et al. : The 4E-BP growth pathway regulates the effect of ambient temperature on Drosophila metabolism and lifespan. Proc Natl Acad Sci U S A. 2017;114(36):9737–42. 10.1073/pnas.1618994114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Tsai SY, Rodriguez AA, Dastidar SG, et al. : Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice. Cell Rep. 2016;16(7):1903–14. 10.1016/j.celrep.2016.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Le Bacquer O, Petroulakis E, Paglialunga S, et al. : Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117(2):387–96. 10.1172/JCI29528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Fan Q, Aksoy O, Wong RA, et al. : A Kinase Inhibitor Targeted to mTORC1 Drives Regression in Glioblastoma. Cancer Cell. 2017;31(3):424–35. 10.1016/j.ccell.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Lamming DW, Mihaylova MM, Katajisto P, et al. : Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell. 2014;13(5):911–7. 10.1111/acel.12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Soukas AA, Kane EA, Carr CE, et al. : Rictor/TORC 2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23(4):496–511. 10.1101/gad.1775409 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 265. Mannick JB, Morris M, Hockey HP, et al. : TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci Transl Med. 2018;10(449): pii: eaaq1564. 10.1126/scitranslmed.aaq1564 [DOI] [PubMed] [Google Scholar]
- 266. Efeyan A, Schweitzer LD, Bilate AM, et al. : RagA, but Not RagB, Is Essential for Embryonic Development and Adult Mice. Dev Cell. 2014;29(3):321–9. 10.1016/j.devcel.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Hoshii T, Tadokoro Y, Naka K, et al. : mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Invest. 2012;122(6):2114–29. 10.1172/JCI62279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Arriola Apelo SI, Neuman JC, Baar EL, et al. : Alternative rapamycin treatment regimens mitigate the impact of rapamycin on glucose homeostasis and the immune system. Aging Cell. 2016;15(1):28–38. 10.1111/acel.12405 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 269. Arriola Apelo SI, Pumper CP, Baar EL, et al. : Intermittent Administration of Rapamycin Extends the Life Span of Female C57BL/6J Mice. J Gerontol A Biol Sci Med Sci. 2016;71(7):876–81. 10.1093/gerona/glw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Barzilai N, Crandall JP, Kritchevsky SB, et al. : Metformin as a Tool to Target Aging. Cell Metab. 2016;23(6):1060–5. 10.1016/j.cmet.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Kuan YC, Huang KW, Lin CL, et al. : Effects of metformin exposure on neurodegenerative diseases in elderly patients with type 2 diabetes mellitus. Prog Neuropsychopharmacol Biol Psychiatry. 2017;79(Pt B):77–83. 10.1016/j.pnpbp.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 272. Pollak MN: Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov. 2012;2(9):778–90. 10.1158/2159-8290.CD-12-0263 [DOI] [PubMed] [Google Scholar]
- 273. Dowling RJ, Goodwin PJ, Stambolic V: Understanding the benefit of metformin use in cancer treatment. BMC Med. 2011;9:33. 10.1186/1741-7015-9-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Moiseeva O, Deschênes-Simard X, St-Germain E, et al. : Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK/NF-κB activation. Aging Cell. 2013;12(3):489–98. 10.1111/acel.12075 [DOI] [PubMed] [Google Scholar]