Abstract
GAS41 is a chromatin-associated protein that belongs to the YEATS family and is involved in the recognition of acetyl-lysine in histone proteins. A unique feature of GAS41 is the presence of a C-terminal coiled-coil domain, which is responsible for protein dimerization. Here, we characterized specificity of the GAS41 YEATS domain, and found that it preferentially binds to acetylated H3K18 and H3K27 peptides. Interestingly, we found that full-length, dimeric GAS41 binds to di-acetylated H3 peptides with an enhanced affinity when compared to mono-acetylated peptides, through a bivalent binding mode. We determined the crystal structure of the GAS41 YEATS domain with H3K23acK27ac to visualize the molecular basis of di-acetylated histone binding. Our results suggest a unique binding mode in which full-length GAS41 is a reader of di-acetylated histones.
INTRODUCTION
GAS41, also known as YEATS4, is a nuclear protein encoded by the GAS41 (glioma-amplified sequence 41) gene that was identified in a glioblastoma cell line1. It is a member of the YEATS family of proteins, and is implicated in chromatin remodeling and transcriptional regulation 2, 3. GAS41 interacts with chromatin-modifying complexes, including TIP60 and SRCAP 4, 5, and promotes deposition of histone H2A.Z 6. Functional studies implicate GAS41 in cell growth and survival 7, 8, and in maintenance of embryonic stem cell identity 9.
Acetylation of histone lysines is abundant in cells, and a high level of acetylated histones typically correlates with actively transcribed genes 10. Proteins involved in transcriptional regulation recognize acetylated lysines utilizing two major families of reader domains: bromodomains 11 and YEATS domains 12. There are four YEATS domain-containing proteins in humans: ENL, YEATS2, AF9, and GAS41. Both AF9 and ENL bind histone H3 peptides with acetylated K9, K18, and K27, with low- to mid-micromolar affinities 13, 14. Recently, GAS41 has been shown to interact with H3K14ac and H3K27ac peptides with affinities ranging from 9.3 to 32.7 μM 6, 9, and with H3K122 succinylated peptide at low pH (pH = 6.0) 15. Notably, YEATS domains have been observed to bind histone peptides with crotonylated lysine with higher affinities 16–19. This is particularly evident for YEATS2, which favors crotonylated lysine with a seven-fold stronger binding affinity when compared to acetylated lysine 16.
In this study, we report that the YEATS domain of GAS41 binds acetylated histone peptides with moderate, mid-micromolar affinities, and preferentially recognizes acetylated H3K18 and H3K27. GAS41 contains a C-terminal coiled-coil domain, and we demonstrate that full-length GAS41 is dimeric in HEK293T cells. We found that full-length GAS41 binds with an enhanced affinity towards di-acetylated over mono-acetylated histone H3, in vitro and in pull-down experiments. We determined the crystal structure of the YEATS domain in complex with H3K23acK27ac, illustrating a bivalent mechanism of di-acetylated histone recognition by GAS41. Our findings suggest a unique recognition mode of acetylated histone by GAS41 through higher-order interactions.
RESULTS and DISCUSSION
The GAS41 YEATS domain preferentially binds H3K18ac and H3K27ac peptides
Previous studies revealed that the human YEATS domain-containing proteins AF9 and ENL bind histone H3 peptides with acetylated K9, K18, and K27, with low- to mid-micromolar affinities 13, 14. To determine the binding affinity and specificity of the GAS41 YEATS domain, we tested a series of peptides derived from histones H3 and H4 with mono-acetylated lysine residues encompassing major sites for lysine acetylation. The recombinant GAS41 YEATS domain yields a well-dispersed 1H-15N HSQC spectrum, which enabled us to perform NMR titration experiments and determine the binding affinity of selected histone peptides (Supplementary figure S1). We found that all acetylated peptides bind to the YEATS domain with relatively modest affinities, and the most potent binding was observed for H3K27ac (KD = 58 μM) and H3K18ac (KD = 106 μM) (Figures 1A, C). All remaining peptides bind to the GAS41 YEATS domain with at least ten-fold lower affinity (Figure 1C). Contradictory to the recent report, we have observed low affinity of GAS41 YEATS toward H3K14ac (KD = 720 μM) (Supplementary figure S2). No binding was detected for the non-acetylated H3K27, indicating that acetylated lysine is essential for the interaction with the GAS41 YEATS domain (Figure 1B). To assess the contribution of acetylated lysine, we tested the tripeptide AKacA, and found that it binds to the GAS41 YEATS domain with a KD of ~1mM, making it comparable to weakly binding acetylated H3 and H4 peptides (Figure 1C). Next, we performed a sequence alignment of all tested peptides, and found that two common features of the two peptides binding with the highest affinities, namely H3K18ac and H3K27ac, are the presence of an arginine residue directly preceding the acetylated lysine and an additional lysine located four residues upstream of the acetyl-lysine (Figure 1D). This result suggests that these positively charged residues are likely important for the recognition of acetylated histone H3 by GAS41.
Figure 1.
Characterization of the binding affinity and selectivity of the GAS41 YEATS domain. A. 1H-15N HSQC spectra of the 65 μM 15N-labeled YEATS domain (black) titrated with H3K27ac and H3K27cr peptides. The YEATS–peptide ratio is labeled. Graphs show the chemical shift titrations used to determine KD values. B. 1H-15N HSQC spectra of the 65 μM 15N-labeled YEATS domain (black) with 400 μM unmodified H3K27 peptide (red). C. Binding affinities for a series of peptides toward the YEATS domain of GS41 determined from NMR titrations. D. Sequence alignment of the peptides used in the binding experiments. Acetyl-lysine is shown in red, and positively charged residues upstream of acetyl-lysine are labeled in cyan.
Our data demonstrate that the binding affinities of GAS41 YEATS toward acetylated H3 peptides are relatively modest, comparable to ENL and YEATS2 14, 16, and in agreement with recently reported data for GAS41 6. Several recent reports revealed that YEATS domains recognize crotonylated lysine residues with higher affinity than acetylated lysines 16–19. Among these proteins, YEATS2 was shown to have the largest selectivity (approximately seven-fold) toward crotonylated over acetylated lysine 16. To assess whether GAS41 YEATS binds crotonylated lysine, we tested the binding of the H3K27cr peptide and found an approximately two-fold improvement in the binding affinity (KD = 34 μM) compared with H3K27ac (Figures 1A, C). According to the recent studies, the GAS41 YEATS domain binds H3 peptide succinylated on K112 at acidic pH (pH = 6.0) 15. We have tested binding of an H3 peptide with succinylated K27 (H3K27su) at pH = 7.5, and found only modest binding with KD > 1mM (Figures 1C, S3) suggesting that GAS41 is not recognizing K27su at physiological pH.
Full-length GAS41 YEATS is dimeric in HEK293T cells
The sequence of GAS41 reveals the presence of a coiled-coil domain immediately following the YEATS domain (Figure 2A), and previous studies have shown that the GAS41 coiled-coil region is dimeric in solution 20. We expressed the C-terminal fragment of GAS41 encompassing residues 149–227, which includes the coiled-coil domain. Using circular dichroism (CD) spectra, we demonstrate helical secondary structure of the C-terminal coiled-coil in solution (Supplementary figure S4). Thermal denaturation indicates melting transition with Tm = 29°C (Figure 2B), which is consistent with the previously reported stability of GAS41 coiled-coil derived peptides 20.
Figure 2.
GAS41 is dimeric. A. Disorder prediction for GAS41 using PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred). B. Thermal denaturation of C-terminal fragment of GAS41 (residues 149–227) determined from CD spectra by monitoring signal at 208 nm. C. NanoBit assay showing luminescence signal in HEK293T cells transfected with three GAS41 constructs cloned into LgBit and SmBit vectors. Luminescence was measured 48h following the transfection after adding Nano-Glo® Live Cell Reagent at four time points.
To test whether full-length GAS41 forms a dimer in cells, we employed the NanoBit protein-protein interaction assay 21. We expressed GAS41 as fusions with LgBit and SmBit proteins in HEK293T cells, and observed a strong luminescence signal, indicating that full-length GAS41 is indeed dimeric in eukaryotic cells (Figure 2C). To the contrary, we have not observed dimerization of the YEATS domain alone (Figure 2C). Indeed, the dimerization of GAS41 has been suggested in previous studies, and L211G and L218G mutations disrupting the coiled-coil motif impaired GAS41-mediated activation of p53 tumor suppressor pathway 22. We tested GAS41 L211G, L218G double-mutant in NanoBit assay, and found that these mutations disrupted GAS41 dimerization (Figure 2C).
Full-length GAS41 YEATS domain binds with increased affinity to di-acetylated histone H3
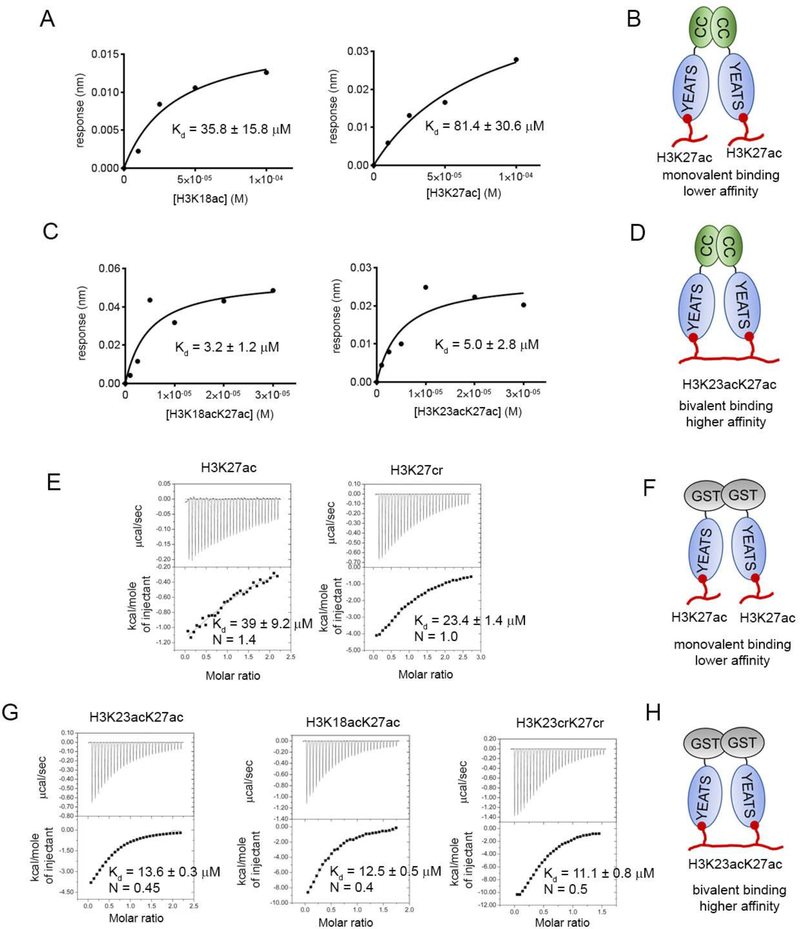
We hypothesized that full-length, dimeric GAS41 may simultaneously recognize di-acetylated histone H3 through a bivalent interaction mode, which could result in enhanced binding affinity. To characterize the interaction of full-length GAS41 with acetylated peptides, we employed bio-layer interferometry using selectively biotinylated GAS41. First, we tested mono acetylated H3K18ac and H3K27ac peptides, and found mid-micromolar KD values (36 μM and 81 μM, respectively, Figures 3A,B) that are consistent with the NMR results for isolated YEATS domain (Figure 1C). Subsequently, we tested the binding of di-acetylated peptides and found low-micromolar affinities for H3K18acK27ac (KD = 3.2 μM) and H3K23acK27ac (KD = 5.0 μM) (Figures 3C, D). This represents an over ten-fold enhancement in the binding affinity of the full-length GAS41 toward di-acetylated H3 peptides.
Figure 3.
Affinity of acylated H3 peptides towards full-length GAS41 and GST-YEATS constructs. A. Binding of H3K18ac and H3K27ac to the full-length GAS41 determined using bio-layer interferometry experiments and corresponding model of studied protein-peptide interaction (B). C. Binding of H3K18acK27ac and H3K23acK27ac to the full-length GAS41 determined using bio-layer interferometry experiments and corresponding model is shown in panel D. E. Determination of the affinity and stoichiometry of binding between GST-YEATS and H3K27ac and H3K27cr peptides using ITC and corresponding model is shown in panel F. G. Characterization of the binding of GST-YEATS and di-acylated H3 peptides using ITC and corresponding model in panel H.
Dimerization of YEATS domain increases the affinity toward the di-acetylated histone H3
To test whether dimerization of the YEATS domain increases the affinity towards di-acetylated H3 peptides in a model system, we purified a dimeric GST-YEATS construct and performed isothermal titration calorimetry (ITC) experiments (Supplementary Table 1). We found that GST-YEATS binds mono-acetylated H3K27ac peptide with 1:1 stoichiometry, and with a similar binding affinity as an isolated YEATS domain (KD = 39 μM, Figures 3E, F). We also confirmed that crotonylated lysine slightly increases the binding affinity of H3K27cr toward GST-YEATS (KD = 23.4 μM, Figure 3E). Subsequently, we tested the di-acetylated H3K23acK27ac peptide, and found that it binds to GST-YEATS with KD = 13.6 μM and 1:2 stoichiometry (single molecule of H3K23acK27ac binds two molecules of YEATS domain, Figure 3G). A very similar binding affinity and stoichiometry was observed for H3K18acK27ac (KD = 12.5 μM and 1:2 stoichiometry, Figure 3G). Therefore, dimerization of the YEATS domain in a model system using GST-fusion leads to two- to three-fold stronger binding of di-acetylated peptides than mono-acetylated H3K27ac. The stoichiometry clearly indicates that the dimerized YEATS domain recognizes di-acetylated H3, and enhanced affinity over mono-acetylated H3 is most likely resulting from a bivalent binding mode (Figure 3H). We also found a very similar effect for a double crotonylated H3K23crK27cr peptide (Figure 3G), which binds to GST-YEATS with KD = 11.1 μM and 1:2 stoichiometry, consistent with an approximately two- to three-fold enhanced affinity compared with that of H3K27cr.
Di-acetylated H3 peptide pulls-down GAS41 from eukaryotic cells
We also assessed whether di-acetylated H3K23acK27ac peptide can interact with full-length GAS41 expressed in eukaryotic cells. Transfection of HEK293T cells with the construct encoding wild-type GAS41 resulted in robust overexpression of the full-length protein (Figure 4). We then used these cells to perform the pull-down, using biotinylated, di-acetylated H3K23acK27ac and mono-acetylated H3K27ac peptides. While both peptides pulled-down GAS41, the interaction with H3K23acK27ac is considerably stronger when compared to the H3K27ac peptide (Figure 4). To confirm that this effect is specific, we introduced W93A mutation that is known to impair YEATS domain function 6, 9 and found that it leads to the disruption of the binding (Figure 4). Altogether, pull-down experiments validate enhanced affinity of full-length GAS41 towards di-acetylated H3, and are consistent with KD values determined in biochemical experiments.
Figure 4.
Peptide pull-down assay with biotinylated H3K23acK27ac and H3K27ac peptides using 293T cells expressing GAS41. Samples pulled-down using streptavidin beads were immunoblotted with anti-GAS41 antibody. GAS41 and tubulin were detected in input samples (whole cell lysates).
Structural basis for the recognition of di-acetylated histone H3 by the GAS41 YEATS domain
To determine how the GAS41 YEATS domain recognizes acetylated histone peptides, we used X-ray crystallography. First, we determined the structure of the YEATS domain at 2.1 Å resolution (Supplementary figure S5 and Table 1). The GAS41 YEATS domain adopts an immunoglobulin fold with a two-layer β-sandwich consisting of eight antiparallel β strands (Supplementary figure S5), and is similar to the previously reported structures of the YEATS domains from AF9, ENL, and YEATS2 (Supplementary figure S6A). We found a short α-helical region encompassing residues 123–129 as a new structural element not observed in other YEATS domains (Supplementary figure S5).
Table 1.
Data collection and Refinement Statistics
| Crystal Form | GAS41 YEATS | GAS41 YEATS – H3K23acK27ac complex |
|---|---|---|
| Diffraction Data | ||
| X-Ray source | APS 21ID-F | APS 21ID-F |
| Wavelength (Å) | 0.97872 | 0.97872 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 70.16, 79.93, 121.64 | 70.79, 80.31, 121.66 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50 – 2.1 (2.18 – 2.10) | 50 – 2.4 (2.44 – 2.40) |
| Rsym | 0.097 (0.65) | 0.078 (0.492) |
| I / σI | 27.12 (3.0) | 28.0 (3.0) |
| Completeness (%) | 99.6 (99.9) | 97.5 (99.9) |
| Redundancy | 7.2 (6.6) | 7.0 (6.8) |
| Refinement | ||
| Resolutions (Å) | 48.40 – 2.10 | 48.67 – 2.40 |
| No. reflections | 38,396 | 25,753 |
| Rwork /Rfree | 17.38/22.23 | 18.62/24.77 |
| No. atoms | ||
| Protein | 4,210 | 4,198 |
| Ligand/ion | 67 | 127 |
| Water | 394 | 132 |
| B-factors | ||
| Protein | 39.37 | 58.96 |
| Ligand/ion | 81.87 | 81.84 |
| Water | 49.02 | 50.85 |
| RMSD | ||
| Bond length (Å) | 0.02 | 0.015 |
| Bond angle (°) | 1.957 | 1.643 |
| Ramachandran plot (%) | ||
| Favored | 98.59 | 97.42 |
| Allowed | 1.41 | 2.39 |
| Outliers | 0.0 | 0.2 |
| PDB ID | 5VNA | 5VNB |
Values in parentheses indicate the highest-resolution shell.
Analysis of the packing in the crystal structure revealed four YEATS domain molecules in the asymmetric unit, with two molecules showing accessible sites for the binding of acetylated peptides. To determine the crystal structure of a complex with histone peptide, we performed soaking of the YEATS domain crystals with di-acetylated peptides and found the best diffraction (2.4 Å resolution) for H3K23acK27ac complex. Interestingly, we found that di-acetylated H3K23acK27ac binds simultaneously to the two YEATS domains in the crystal structure, and that acetylated K23 and K27 residues occupy the corresponding pockets in two different molecules of the YEATS domain (Figure 5A). Importantly, this binding mode is consistent with a 1:2 binding stoichiometry from the ITC experiment (Figure 3G). Analysis of the crystal structure reveals that only very short segments of the H3 peptide are in contact with the YEATS domains, and the major contacts involve recognition of the acetylated K23 and K27 side chains (Figure 5B). Acetylated lysine fits into the channel comprised of the aromatic side chains of H71, Y74, W93, and F96. The acetyl group is recognized by a network of hydrogen bonds, including interactions with the backbone amides of W93 and G94, as well as the side chain of S73 (Figure 5B). The binding mode of the H3 fragment surrounding K27ac is very similar to the recently reported structure of the GAS41 YEATS domain bound to H3K27ac peptide (Supplementary Figure S7). Both structures show the presence of a hydrophobic interaction between H3P30 and GAS41 F121, which may explain enhanced specificity towards H3K27ac peptide (Supplementary figure S7). Notably, the structure of the YEATS domain with bound H3K23acK27ac reveals reversed direction of polypeptide around H3K23ac, suggesting some plasticity in recognition of acetylated lysines. Overall, the crystal structure of YEATS with bound H3K23acK27ac represents a snapshot of the bivalent recognition mode of di-acetylated histone H3 by full-length dimeric GAS41.
Figure 5.
Structure of GAS41 YEATS in a complex with H3K23acK27ac. A. Structure of the GAS41 YEATS domain with bound H3K23acK27ac determined at 2.4 Å resolution showing Fo - Fc omit electron density maps contoured at 3.0 σ level for H3K23acK27ac (blue mesh). The YEATS domains are shown as yellow and white ribbons, and the H3K23acK27ac peptide is shown in green, with acetylated lysines shown as sticks. B. Details of the binding of K23ac and K27ac to the GAS41 YEATS domains. C. Model of the bivalent recognition of di-acetylated histone protein by full-length GAS41.
CONCLUSIONS
GAS41 belongs to a four-member family of human proteins, characterized by the presence of the conserved YEATS domain. GAS41 is the shortest protein in this family, and is composed of two domains, namely, the N-terminal YEATS and C-terminal coiled-coil (Figure 2A). In this study, we investigated the binding of the GAS41 YEATS domain to a series of histone H3- and H4-derived peptides with acetylated lysine residues, and found that it preferentially recognizes H3K18ac and H3K27ac with modest affinities (KD values ranging from 35 to 106 μM). The presence of the C-terminal coiled-coil suggests GAS41 dimerization 20, 22. Indeed, we have validated that full-length GAS41 is dimeric in HEK293T cells. Disorder prediction for GAS41 sequence indicates that the linker between the YEATS and the coiled-coil domain is relatively short and consists of approximately 20 amino acids (Figure 2A). This implicates the close proximity of the two YEATS domains in dimeric GAS41. In this study, we found that full-length GAS41 preferentially binds to di-acetylated H3 peptides with ~ten-fold enhanced affinity when compared with mono-acetylated H3 (Figures 3A, C). Importantly, we observed a more efficient pull-down of GAS41 using di-acetylated H3K23acK27ac when compared to mono-acetylated H3K27ac (Figure 4), further validating GAS41 as a reader of di-acetylated histone H3. Our results suggest a model in which full-length GAS41 recognizes di-acetylated histone in a unique, bivalent mode (Figure 5C). We further validated this model by testing the binding of dimeric GST-YEATS domain and found enhanced affinity towards di-acylated (including both di-acetylated and di-crotonylated) versus mono-acylated H3 peptides (Figures 3E, G).
Histone acetylation is an epigenetic modification important for regulation of gene expression. Recent ChIP-seq experiments revealed co-localization of GAS41 to gene promoters enriched in H3K14ac and H3K27ac 6. Acetylated lysines of H3K14, H3K18 and H3K27 are enriched at promoters of actively transcribed genes 6, 23, and enhanced affinity of GAS41 towards di-acetylated H3 may explain over 90% of GAS41 bound to highly-acetylated gene promoter regions 6. We also observed approximately two-fold enhanced binding of GAS41 YEATS to peptides with crotonylated versus acetylated lysine. Further studies are required to establish whether GAS41 is a di-acetyl-lysine or di-crotonyl-lysine reader in physiological conditions.
YEATS domain proteins are recently characterized readers of acylated histones, recognizing both acetylated and crotonylated lysines 13, 24. Among human homologs, only AF9 has been reported to bind acetylated histone proteins with relatively high affinity (KD = 2.1 to 3.7 μM) 13, 24, while the KD values for ENL and YEATS2 interactions are weaker (KDs above 30 μM) 14, 16, and are comparable to the affinity of the GAS41 YEATS domain for mono-acetylated H3 6. We found that the affinity of GAS41 toward di-acetylated H3 is significantly improved through a higher-order interaction involving dimerization of the YEATS domain. Whereas a bivalent binding mode has not been proposed for other members of the YEATS domain family, sequence analysis predicts the presence of the coiled-coil domain in YEATS2 (Supplementary figure S6C). This suggests that higher-order interactions might be present in other YEATS protein readers.
METHODS
Cloning, Expression and Protein Purification
The synthetic gene encoding Human GAS41 YEATS (residues 13–158) was ordered from Life Technologies and subcloned using the BamHI and HindIII restriction sites into pQE-80L expression vector (Qiagen) with an N-terminal hexahistidine (His6) tag. The recombinant plasmid pQE80L-GAS41 (residues 13–158) was transformed into E. coli strain BL21(DE3). Transformants were grown in 15N- labeled M9 minimal medium containing ampicillin at 37 °C until reaching an OD600 between 0.6 and 0.8. After induction with 0.25 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG), cultures were grown for an additional 16 h at 18 °C. Harvested cells were resuspended in lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl and 1 mM TCEP) and lysed using a cell disrupter. Clarified lysate was applied to Ni–NTA (Qiagen) affinity column. The column was extensively washed with lysis buffer containing 35 mM imidazole and eluted with lysis buffer containing 200 mM imidazole. The eluted pure fractions were pooled and dialyzed against 50 mM Tris, pH 7.5, and 200 mM NaCl buffer and then concentrated to ~60 μM. The final purified 15N-labeled GAS41(13–158) was used in NMR studies.
Codon-optimized cDNAs of full-length human GAS41 was synthesized by Life Technologies and amplified GAS41 YEATS (residues 1–148) by the polymerase chain reaction (PCR). The PCR product was subcloned using the BamHI and EcoRI restriction sites into pGST-parallel vector 25 with an N-terminal GST tag followed by a TEV cleavage site. The resulting plasmid pGST-GAS41(1–148) was transformed into E. coli strain BL21(DE3). Transformed cells were grown in Luria broth medium with ampicillin selection. After 18 h induction with 0.2 mM IPTG at 18°C, cells were harvested by centrifugation and resuspended in lysis buffer containing 50 mM Tris, pH 7.5, 300 mM NaCl and 1 mM TCEP and lysed using a cell disruptor. The soluble fraction of the cell lysate was then loaded onto a glutathione-Sepharose 4B (GE Healthcare) affinity column. The column was thoroughly washed with buffer containing 50 mM Tris, pH 8.0 and 500 mM NaCl and eluted with 10 mM reduced glutathione. GST-tagged GAS41(1–148) protein was used in ITC experiment. The eluted proteins were proteolytically cleaved with TEV protease, followed by S-75 size exclusion chromatography purification into buffer containing 20 mM Tris, pH 7.5, 300 mM NaCl. The final purified GAS41(1–148) was used in crystallization trial.
GAS41 coiled coil region (residues 149–227) was cloned from the synthetic human GAS41 gene by PCR. The PCR product was subcloned using the BamHI and HindIII restriction sites into pMocr vector with an N-terminal hexahistidine (His6) tag followed by a TEV cleavage site. GAS41 149–227 protein was expressed as inclusion bodies and solubilized in buffer with 6 M Guanidine HCl. Re-folding was performed using dialysis to 50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT buffer following the cleavage with TEV protease. N-terminal Mocr-His6 was extracted by re-application to Ni-NTA resin. Protein was dialyzed extensively against Storage Buffer (50 mM tris pH 7.5, 150 mM NaCl, 1 mM TCEP) and concentrated for storage at –80 °C.
NMR Titration
All NMR spectra were collected at 30°C on a 600 MHz Bruker Advance III spectrometer equipped with cryogenic probe, running Topspin version 2.1. Binding of histone peptides to GAS41 YEATS (residues 13–158) were characterized by measuring chemical shift perturbations of selected amide resonances on the 1H-15N HSQC spectra of 60 μM 15N-labeled GAS4113–158 titrated with peptides at molar ratios of 1.0, 2.7, 4.3 and 6.0. Dissociation constants were determined from least-squares fitting of chemical shift perturbations as a function of ligand concentration
with a = (KA/δb) × [Pt], b = 1 + KA([Lti] + [Pt]), and c = δb * KA * [Lti], where δi is the absolute change in chemical shift for each titration point, [Lti] is the total ligand concentration at each titration point, [Pt] is the total protein concentration, KA = 1/KD is the binding constant, and δb is the chemical shift of the resonance in question in the complex. KD and δb were used as fitting parameters in analysis 26.
Isothermal Titration Calorimetry
The measurements were performed using a VP-ITC titration calorimetric system (MicroCal) at 25°C. GST-tagged GAS41 YEATS (residues 1–148) was dialyzed extensively against ITC buffer consisting of 50 mM phosphate, pH 7.5, 150 mM NaCl and 2 mM β-mercaptoethanol. Histone derived peptides were directly dissolved in the ITC buffer at 500 μM concentration. The titration curve was obtained by injecting 10 μL aliquots of histone derived peptides into the cell containing 50 μM GST-GAS41(1–148), at a time interval of 200 s. All samples were degassed by vacuum aspiration for 20 min prior to measurements. ITC titration data were analyzed with a single-site fitting model using Origin 7.0 software.
Determination of binding affinity using bio-layer interferometry
To determine the affinity of full-length GAS41 towards H3 peptides we employed Bio-Layer Interferometry experiments using an Octet Red 96 instrument (ForteBio, Inc.). Mono-biotinylated GAS41 was obtained by co-expression of Avi-tagged GAS41 with BirA enzyme in E. coli BL21(DE3) cells as previously described 27. The purification procedure for biotinylated GAS41 was identical to that for the His tagged GAS41 YEATS (residues 13–158) protein. Prior to protein immobilization, streptavidin biosensor tips were incubated in 50 mM Tris, pH 7.5, and 200 mM NaCl buffer for 600 s. Subsequently, protein was loaded onto tips for 600 s, followed by 1200 s equilibration step. Peptide binding experiments were performed in following order: 600 s equilibration, 300 s association, and 600 s dissociation. Experimental set-up was performed using Octet Data Acquisition Software, and data were analyzed by Octet Data Analysis Software (Pall ForteBio, LLC.). Signal was plotted as a function of ligand concentration to determine KD, using Prism software (GraphPad Software, Inc.). To correct for drift during association, the slope during the drift period was fit by linear regression, and the product of slope and time (in seconds) was subtracted from the signal.
Circular Dichroism Spectroscopy
Spectra and thermal denaturation experiments were measured at 50 μM concentration of GAS41 (149–227) in 20 mM sodium phosphate, pH 6.5, and 250 mM sodium fluoride using a Jasco CD-810 spectropolarimeter with constant N2 flushing. Rectangular cells with 1 mm path length were used, and a circular water bath was used to control temperature of the optic cell chamber. Protein spectra were averaged from three wavelength-scans collected at 0.1 nm-intervals from 178 – 260 nm. Baseline spectrum of buffer was recorded prior to recording protein spectra. Mean residue ellipticity was determined by the equation
where Θobs is observed ellipticity (in millidegrees), mrw is mean residue molecular weight, l is optical path length of the cell (in cm), and c is peptide concentration. For thermal denaturation experiments, we recorded CD spectra over temperature gradient from 5 – 95°C at 1 degree increments.
X-Ray Crystallography
Protein crystals of GAS41 YEATS (residues 1–148) were obtained by sitting drop method using 6–8.5 mg/ml protein mixed in a 1:1 volume ratio with precipitation solution containing 100 mM CHES, pH 9.4 and 1.24 –1.36 M ammonium sulfate. The crystals were cryoprotected in reservoir solution supplemented with 25% ethylene glycol. X-ray diffraction data were collected on LS-CAT beam line 21-ID-F at the Advanced Photon Source. The data were then indexed, integrated, and scaled using the HKL2000 suite 28. The structure was determined by molecular replacement method with the CCP4 version of MOLREP 29 using the structure of yeast Taf14 YEATS domain (PDB code: 3QRL) as a search model. To obtain a histone peptide bound complex structure, native GAS41 crystals were soaked in a 1:2 mixture of protein buffer and reservoir solution containing saturating amounts of peptide. Model building and structure refinement were carried out using WinCOOT 30 and Phenix.refine 31. The data statistics are summarized in Table 1.
GAS41 pull-down assay
Full-length GAS41 and GAS41 W93A mutant was cloned into pCMV vector and used to transfect HEK293T cells using FuGENE6 (Promega). Cell lysates were prepared in lysis buffer (PBS with 1 % Triton X-100 and protease inhibitor cocktail) and sonicated. Biotinylated H3K23acK27ac and H3K27ac peptides were incubated with streptavidin-magnetic beads for 6 h at 4 °C. Cell lysates were precleared with streptavidin-magnetic beads for 2 h at 4 °C and incubated with the biotinylated peptides immobilized on streptavidin-magnetic beads overnight at 4 °C. The beads were washed 10 times with wash buffer (PBS with 0.2 % Triton X-100). Pulled-down proteins were analyzed using western blot with GAS41 antibody (sc-393708 from Santa Cruz).
NanoBiT protein-protein interaction assay
The NanoBiT protein-protein interaction assays (Promega) were conducted according to the manufacturer’s instructions. Briefly, full-length GAS41, YEATS (residues 1–158), and GAS41 L211G/L218G mutant were cloned into pBiT1.1-C[TK/LgBiT] and pBiT2.1-C[TK/SmBiT] vectors for NanoBiT system. All plasmids were verified by sequencing. HEK293T cells were plated into 96-well white plates at 1×104 cells per well. Each Lg-BiT and Sm-BiT plasmids were co-transfected using FuGENE HD (Promega) the next day and incubated for 48 h. After we added the Nano-Glo Live Cell Reagent to each well, the luminescence was measured on 0 min, 10 min, 30 min, and 60 min using the PHERAstar FS (BMG Labtech). The positive control (Lg-PRKAR2A and Sm-PRKACA) and the negative control vectors were provided by the manufacture.
Supplementary Material
Acknowledgements
This work was funded by the National Institute of Health (NIH) R01 grant (CA207272) to T.C., funding from The Forbes Institute for Cancer Discovery (University of Michigan) and LLS Scholar grants (1340-17) to T.C. and (1215-14) to J.G. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Supporting Information
Supporting information, consisting of seven figures and one table, is available free of charge via the internet at http://pubs.acs.org
References
- 1.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, and Meese E (1997) Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I, Hum Mol Genet 6, 1817–1822. [DOI] [PubMed] [Google Scholar]
- 2.Schulze JM, Wang AY, and Kobor MS (2009) YEATS domain proteins: a diverse family with many links to chromatin modification and transcription, Biochem Cell Biol 87, 65–75. [DOI] [PubMed] [Google Scholar]
- 3.Schulze JM, Wang AY, and Kobor MS (2010) Reading chromatin: insights from yeast into YEATS domain structure and function, Epigenetics 5, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyon Y, Selleck W, Lane WS, Tan S, and Cote J (2004) Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans, Mol Cell Biol 24, 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, and Conaway JW (2005) The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes, J Biol Chem 280, 13665–13670. [DOI] [PubMed] [Google Scholar]
- 6.Hsu CC, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, Wang X, Li H, Wen H, Li W, and Shi X (2018) Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer, Genes Dev 32, 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harborth J, Elbashir SM, Bechert K, Tuschl T, and Weber K (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs, J Cell Sci 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann K, Ahrens K, Matthes S, Buerstedde JM, Stratling WH, and Phi-van L (2002) Targeted disruption of the GAS41 gene encoding a putative transcription factor indicates that GAS41 is essential for cell viability, J Biol Chem 277, 18626–18631. [DOI] [PubMed] [Google Scholar]
- 9.Hsu CC, Zhao D, Shi J, Peng D, Guan H, Li Y, Huang Y, Wen H, Li W, Li H, and Shi X (2018) Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity, Cell Discov 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bannister AJ, and Kouzarides T (2011) Regulation of chromatin by histone modifications, Cell Res 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, and Knapp S (2012) Histone recognition and large-scale structural analysis of the human bromodomain family, Cell 149, 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao D, Li Y, Xiong X, Chen Z, and Li H (2017) YEATS Domain-A Histone Acylation Reader in Health and Disease, J Mol Biol 429, 1994–2002. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, Li H, and Shi X (2014) AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation, Cell 159, 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan L, Wen H, Li Y, Lyu J, Xi Y, Hoshii T, Joseph JK, Wang X, Loh YE, Erb MA, Souza AL, Bradner JE, Shen L, Li W, Li H, Allis CD, Armstrong SA, and Shi X (2017) ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia, Nature 543, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Jin J, Chung MWH, Feng L, Sun H, and Hao Q (2018) Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation, Proc Natl Acad Sci U S A 115, 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, Zhao Y, Allis CD, Shi X, and Li H (2016) YEATS2 is a selective histone crotonylation reader, Cell Res 26, 629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Zhao D, Chen Z, and Li H (2016) YEATS domain: Linking histone crotonylation to gene regulation, Transcription, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Zeng L, Zhao C, Ju Y, Konuma T, and Zhou MM (2016) Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain, Structure 24, 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, and Kutateladze TG (2016) The Taf14 YEATS domain is a reader of histone crotonylation, Nat Chem Biol 12, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen S, Mattay D, Rauber C, Muller KM, and Arndt KM (2014) Characterization and inhibition of AF10-mediated interaction, J Pept Sci 20, 385–397. [DOI] [PubMed] [Google Scholar]
- 21.Oh-Hashi K, Hirata Y, and Kiuchi K (2016) SOD1 dimerization monitoring using a novel split NanoLuc, NanoBit, Cell Biochem Funct 34, 497–504. [DOI] [PubMed] [Google Scholar]
- 22.Park JH, and Roeder RG (2006) GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation, Mol Cell Biol 26, 4006–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, and Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome, Nat Genet 40, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, Roeder RG, Shi X, Allis CD, and Li H (2016) Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain, Mol Cell 62, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheffield P, Garrard S, and Derewenda Z (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors, Protein Expr Purif 15, 34–39. [DOI] [PubMed] [Google Scholar]
- 26.Tugarinov V, and Kay LE (2003) Quantitative NMR studies of high molecular weight proteins: application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G, J Mol Biol 327, 1121–1133. [DOI] [PubMed] [Google Scholar]
- 27.Gray F, Cho HJ, Shukla S, He S, Harris A, Boytsov B, Jaremko L, Jaremko M, Demeler B, Lawlor ER, Grembecka J, and Cierpicki T (2016) BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization, Nat Commun 7, 13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode, Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A, and Teplyakov A (2010) Molecular replacement with MOLREP, Acta Crystallogr D Biol Crystallogr 66, 22–25. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.