Abstract
Widespread use of Pneumococcal Conjugate Vaccines (PCV) has reduced vaccine-type nasopharyngeal colonisation and invasive pneumococcal disease. In a double-blind, randomised controlled trial using the Experimental Human Pneumococcal Challenge (EHPC) model, PCV-13 (Prevenar-13) conferred 78% protection against colonisation acquisition and reduced bacterial intensity (AUC) as measured by classical culture. We used a multiplex qPCR assay targeting lytA and pneumococcal serotype 6A/B cpsA genes to re-assess the colonisation status of the same volunteers. Increase in detection of low-density colonisation resulted in reduced PCV efficacy against colonisation acquisition (29%), compared to classical culture (83%). For experimentally colonised volunteers, PCV had a pronounced effect on decreasing colonisation density. These results obtained in adults suggest that the success of PCV vaccination could primarily be mediated by the control of colonisation density. Studies assessing the impact of pneumococcal vaccines should allow for density measurements in their design.
Keywords: Pneumococcus, PCV, Colonisation, Density, QPCR
1. Introduction
Pneumonia is a leading cause of death in children under 5 years worldwide, causing up to 1.4 million deaths annually [1]. Of these deaths, approximately 38% are caused by Streptococcus pneumoniae (pneumococcus) [2]. Current licensed pneumococcal conjugate vaccines (PCVs) are highly effective in protecting against invasive pneumococcal diseases caused by vaccine-type serotypes in children [3]. Immunization with PCVs also has beneficial indirect effects, conferring herd immunity to unvaccinated adults [4]. Disease and transmission prevention requires interruption of colonisation [3]. Numerous studies have reported a reduction in vaccine-type (VT) pneumococcal colonisation acquisition after PCV vaccination, however, the impact of PCVs on pneumococcal colonisation density is less clear [5], [6], [7], [8]. Studies have shown that vaccination with PCV reduces colonisation densities with VT serotypes when comparing to control vaccines [5]. However, other studies have reported that vaccination with PCV do not affect pneumococcal density in children colonised by VT serotypes [7].
We have previously reported the results of a double-blind, randomised controlled trial investigating the effect of the 13-valent PCV (PCV-13, Prevnar-13, Pfizer) on pneumococcal colonisation using the Experimental Human Pneumococcal Challenge (EHPC) model [9]. PCV showed 78% protection against acquisition of pneumococcus serotype 6B (BHN418) (5/48 became colonised in PCV arm vs 23/48 in the control arm). Moreover, although the number of volunteers who became experimentally colonised following PCV vaccination was limited, we observed a significant reduction in nasal pneumococcal colonisation density (3 log difference in PCV arm compared to control arm at day 2) [9].
Interest in using molecular methods for colonisation detection is increasing because of their high sensitivity and, therefore, their ability to detect pneumococcus at low colonisation densities. Combining the results of classical culture and molecular methods has been suggested in order to improve accuracy of reported colonisation rates [10]. The lytA (autolysin) gene qPCR strategy developed by the Centers for Disease Control (CDC) is currently the WHO-recommended culture-independent method to detect pneumococci [11]. However, given the capacity of pneumococcus to exchange genes with other streptococci [12] a multiplex approach is valuable. Therefore, in this study, we employed a multiplex qPCR assay targeting lytA and pneumococcal serotype 6A/B cpsA genes to re-assess volunteer samples from our PCV study for experimental colonisation of 6B pneumococcus. We compared the results obtained by this molecular method with the results previously obtained by classical culture.
2. Material and methods
2.1. PCV/EHPC clinical trial
A trial investigating the efficacy of the 13-valent PCV vaccine against experimental human pneumococcal challenge was conducted in 2012. Study design and outcomes have been previously reported [9]. Briefly, 96 healthy volunteers aged 18–50 were vaccinated with either PCV-13 (PCV arm, n = 48) or Hepatitis A vaccine (control arm, n = 48). 4–5 weeks post-vaccination, volunteers were inoculated with 80,000 Colony-Forming Units (CFU) per nostril of live 6B pneumococcus (BHN418, sequence type 138) [9]. Nasal wash samples were taken and processed to obtain a nasal wash supernatant and a nasal wash bacterial pellet as described previously [13], before and after pneumococcal inoculation (at days 2, 4, 14 and 21). Samples were stored at −80 °C. This trial was approved by The National Health Service Research and Ethics Committee (REC) (12/NW/0873 Liverpool) and was co-sponsored by the Liverpool School of Tropical Medicine and the Royal Liverpool and Broadgreen University Hospitals Trust. Informed consent was obtained from all volunteers.
2.2. DNA extraction
300 μl of D2, D7 and D14 nasal wash bacterial pellets was centrifuged for 7 min at 20,238g. Following centrifugation, 300 μl of lysis buffer with protease, 100 μl of Zirconium beads (Stratech, Ely, UK) and 300 μl of Phenol (Sigma-Aldrich, St Louis, MO, USA) was added to the pellet and the sample was disrupted twice for 3 min in a tissue homogenizer (Bertin Technologies, Montigny le Bretonneux, France) followed by 3 min on ice. The sample was centrifuged for 10 min at 9391g, and the upper aqueous phase was transferred to a tube pre-filled with 600 μl binding buffer and 10 μl magnetic beads. The samples were incubated at room temperature for 30–120 min then washed twice with 200 μl of wash buffers 1 and 2. Magnetic beads were dried at 55 °C for 10 min, eluted in 63 μl of elution buffer and stored at −20 °C. Lysis buffer, protease, binding buffer, magnetic beads, wash buffers 1 and 2, and elution buffer are part of the Agowa mag Mini DNA isolation kit (LGC Genomics, Berlin, Germany).
2.3. Multiplex qPCR
We developed a novel multiplex qPCR based on methods previously published, using partial amplification of lytA [11] and 6A/B cpsA [14] genes. The sequences of the primers and probes used are: lytA forward primer: 5′-ACG-CAA-TCT-AGC-AGA-TGA-AGC-A-3′; lytA reverse primer 5′-TCG-TGC-GTT-TTA-ATT-CCA-GCT-3′; lytA probe: 5′-(FAM)-TGC-CGA-AAA-CGC-TTG-ATA-CAG-GGA-G-(BHQ-1)-3′; cpsA forward primer: 5′-AAG-TTT-GCA-CTA-GAG-TAT-GGG-AAG-GT-3′; cpsA reverse primer: 5′-ACA-TTA-TGT-CCA-TGT-CTT-CGA-TAC-AAG-3′; cpsA probe: 5′-(HEX)- TGT-TCT-GCC-CTG-AGC-AAC-TGG-(BHQ-1)-3′. The reaction mixture of 25 µl contained 0.6 µM of each lytA primer, 0.3 µM of lytA probe, 0.4 µM of each cpsA primer, 0.2 µM of cpsA probe, 12.5 µM of Taqman Gene Expression Master Mix (Applied Biosystems, USA) and 2.5 µl of extracted DNA. The qPCR reaction was run on a Mx3005P machine (Agilent Technologies, Santa Clara, CA, USA) on the following programme: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. DNA from BHN418 serotype 6B, extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) and serially diluted 1:10 from 4.14 × 106 copies in 2.5 µl, was used as a standard curve. A sample was considered positive if at least one duplicate had a CT value less than 40. The 40 cycle cut-off correlates well with classical culture data where it is known whether or not viable serotype 6B pneumococcus has been isolated.
2.4. Statistical analysis
Risk Ratio, 95% confidence interval (CI) and p-value were calculated. Colonisation densities were determined from number of gene copies per well and analysed in GraphPad Prism v5 (GraphPad Inc.). Densities were log-transformed and means with standard deviations were calculated. Unpaired t-tests were used to compare densities in PCV and control arms.
3. Results
3.1. Colonisation acquisition rates by molecular methods
Samples from 90 of the 98 volunteers enrolled in the EHPC PCV study were available and included in these analyses. The re-assessment of the PCV/EHPC trial samples using the developed multiplex qPCR showed that all volunteers reported as colonisation-positive (acquired the inoculated bacterium) by classical culture were also found to be colonised with pneumococcus by molecular methods. However, the number of colonisation-positive volunteers increased in both PCV and control arms when using molecular methods. Positivity at any day for both lytA and cpsA was found in 22/45 (49%) volunteers in the PCV arm and 31/45 (69%) volunteers in the control arm (Table 1). The risk ratio by classical culture was 0.17 (95% CI, 0.07–0.46; P = 0.0005) and by molecular methods was 0.71 (95% CI, 0.50–1.01; P = 0.06). PCV conferred 83% protection against experimental pneumococcal colonisation by classical culture, and 29% protection by molecular methods.
Table 1.
Comparison of numbers of colonised volunteers by detection method, study day and study arm.
| Classical Culture No. Colonised/Total No. (%) [9] |
lytA/cpsA multiplex qPCR No. Colonised/Total No. (%) |
|||
|---|---|---|---|---|
| PCV | Control | PCV | Control | |
| D2 | 2/38 (5) | 18/39 (46) | 11/38 (29) | 21/39 (54) |
| D7 | 4/43 (9) | 18/42 (43) | 16/43 (37) | 23/41 (56) |
| D14 | 1/9 (11) | 18/23 (79) | 2/9 (22) | 17/23 (74) |
| Any Day | 4/45 (9) | 23/45 (51) | 22/45 (49) | 31/45 (69) |
| Risk Ratio (p-value) | 0.17 (0.0005) | 0.71 (0.06) | ||
Definition of abbreviations: PCV = pneumococcal conjugate vaccine; NW = nasal wash.
When assessing the breakdown of colonisation-positive volunteers per study day, the percentage of experimentally colonised volunteers by lytA/cpsA multiplex qPCR was similar between D2 and D7 regardless of study arm (Table 1).
3.2. Colonisation densities by molecular methods
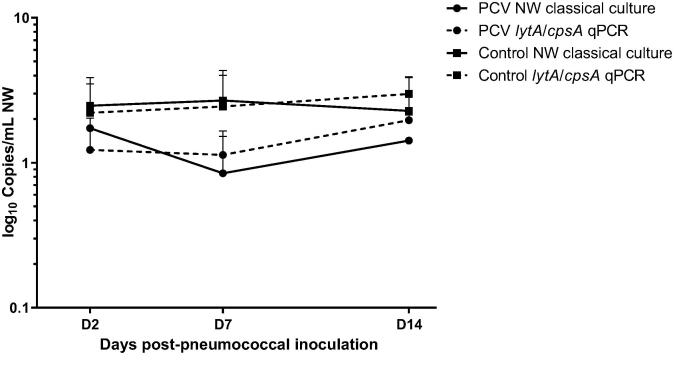
Colonisation densities were significantly lower in volunteers vaccinated with PCV compared to the control arm by both classical culture (P = 0.03) and molecular methods (P < 0.0001) (Fig. 1, Table 2).
Fig. 1.
Colonisation densities by vaccination group, detection method and study time-point. Values shown are the log10 mean copies/mL ± SD. Classical culture data has been previously reported [9].
Table 2.
Comparison of colonisation density of colonised volunteers by detection method, study day and study arm.
| Classical Culture Log10 CFU/ml Mean ± SD [9] |
lytA/cpsA multiplex qPCR Log10 DNA copies/ml Mean ± SD |
|||
|---|---|---|---|---|
| PCV | Control | PCV | Control | |
| D2 | 1.73 ± 0.02 | 2.47 ± 1.39 | 1.22 ± 0.80 | 2.21 ± 1.29 |
| D7 | 0.85 ± 0.67 | 2.70 ± 1.64 | 1.13 ± 0.52 | 2.44 ± 1.56 |
| D14 | 1.42 | 2.28 ± 1.58 | 1.86 ± 0.16 | 2.98 ± 0.93 |
Definition of abbreviations: PCV = pneumococcal conjugate vaccine; CFU = colony forming units; SD = standard deviation; NW = nasal wash.
There was a significant correlation between densities calculated by classical culture and by lytA/cpsA qPCR for volunteers in both PCV (P < 0.0001; r2 = 0.54) and control arms (P < 0.0001; r2 = 0.85). 91% of samples positive by lytA/cpsA qPCR but not by classical culture had densities < 30 DNA copies/ml. There is no significant difference between mean densities calculated by classical or molecular methods, either in the PCV or in the control arm (Table 2). By either detection method, densities in the PCV arm were significantly lower (P = 0.0010 by classical culture and P < 0.0001 for molecular methods) than in the control arm.
4. Discussion
In this study we re-assessed the experimental colonisation status of all volunteers from our PCV/EHPC trial using a multiplex qPCR. The number of volunteers that became colonised after experimental pneumococcal inoculation by molecular methods was higher than the number previously reported by classical culture [9]. This increase in number is more pronounced in the PCV arm (23 vs 4 volunteers) than in the control arm (31 vs 23 volunteers), which translates into increased risk ratios, and therefore a decrease in the calculated protective efficacy of PCV against pneumococcal colonisation from 83% to 29%.
Molecular methods such as qPCR can detect DNA from dead bacteria. However, it is unlikely that this phenomenon had a major contribution to our findings. If this method were detecting remains of the pneumococcal inoculum administered on Day 0, we would expect to see higher colonisation rates at D2 than D7. However, the colonisation rates on both days are comparable.
The cpsA gene sequence used in our multiplex qPCR is common to both serotypes 6A and 6B. However, epidemiological studies have reported that the incidence of pneumococcal serotype 6A is very low in the UK in the post-PCV era; this was confirmed by screening 795 volunteers for the EHPC studies in Liverpool which found only one volunteer carrying a serotype 6A [15]. Pneumococcal capsular genes have recently been discovered in other streptococci [16], [17], raising the possibility that the additional gene copies detected by qPCR derive from non-pneumococcal streptococci. Nevertheless, in the context of the EHPC model, screening samples are taken before inoculation, carriage is assessed very soon post-inoculation and volunteers have limited contact with children. The most likely explanation for the detection of lytA and cpsA genes in our multiplex qPCR is carriage of the experimental 6B pneumococcus used.
Both classical culture and molecular detection methods demonstrate a lower colonisation density in volunteers vaccinated with PCV. The additional colonised volunteers detected in both arms by molecular methods are mostly colonised at a low density. It has been suggested that colonisation density plays an underestimated but pivotal role in the development of pneumococcal disease and in transmission dynamics [18], [19]. Our results further support the hypothesis that colonisation density is a determining factor for the clinical outcome and spread of S. pneumoniae. It is plausible that a novel vaccine that controlled pneumococcal colonisation density independently of any effect on pneumococcal acquisition would be effective in reducing pneumococcal disease. Measures of colonisation density, by both classical culture and molecular methods, should therefore be accounted for in the design of vaccine efficacy trials, as was demonstrated recently in a Phase 2 trial of a novel pneumococcal vaccine [20].
Findings from previous epidemiological studies have proved inconclusive regarding the impact of PCVs on pneumococcal colonisation density [5], [6], [7], [8]. The advantage of the EHPC model is the ability to study colonisation dynamics in a controlled manner, clarifying causality and eliminating the effect of many confounding factors. Its main disadvantage is the ethical imperative to use adult volunteers rather than children. It is possible that the effect of PCV on colonisation density differs between adults and children, and our findings will need to be validated in younger cohorts.
5. Conclusions
Using molecular methods, we have shown that PCV conferred 29% protection against experimental colonisation acquisition. This may indicate that the main protective mechanism of this vaccine is mediated by reduction of colonisation density, leading to a decreased risk of disease to vaccinated individuals as well as transmission resulting in the observed herd effects in vaccinated populations.
Funding
This work was supported by the Medical Research Council/FAPESP (grant number MR/M011569/1), Medical Research Council (grant number MR/M011569/1), the Bill & Melinda Gates Foundation, (grant numbers OPP1035281 and OPP1117728); and the National Institute for Health Research (NIHR) Local Clinical Research Network. The funders were not involved in the design, data processing or publication of this work.
Author contributions
ADH-W, AMC, SBG and DMF designed and co-ordinated the PCV clinical trial; JFG, EM and DMF processed clinical samples; ELG, CS, SS, FD, JFG, EM and EN performed DNA extractions and qPCRs; ELG, CS, EM and EN developed the multiplex qPCR; ELG, CS and DMF analysed the data and drafted the manuscript. All authors have read and approved the manuscript.
This work was partially presented as a poster at the 10th International Symposium on Pneumococci and Pneumococcal Diseases (ISPPD-10), 26th-30th June 2016, abstract number 330.
Declaration of Competing Interest
None.
Contributor Information
E.L. German, Email: Esther.German@lstmed.ac.uk.
C. Solórzano, Email: Carla.SolorzanoGonzalez@lstmed.ac.uk.
S. Sunny, Email: syba.sunny@rlbuht.nhs.uk.
F. Dunne, Email: f.j.dunne@bham.ac.uk.
J.F. Gritzfeld, Email: j.gritzfeld@liverpool.ac.uk.
E. Mitsi, Email: Elena.Mitsi@lstmed.ac.uk.
E. Nikolaou, Email: Elissavet.Nikolaou@lstmed.ac.uk.
A.D. Hyder-Wright, Email: Angela.Hyder-Wright@lstmed.ac.uk.
A.M. Collins, Email: Andrea.Collins@lstmed.ac.uk.
S.B. Gordon, Email: sgordon@mlw.mw.
D.M. Ferreira, Email: Daniela.Ferreira@lstmed.ac.uk.
References
- 1.Liu L, Johnson H, Cousens S, Perin J, Scott S, Lawn JE, et al. Child Health Epidemiology Reference Group of WHO and UNICEF. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. The Lancet 2012;379(9832):2151–2161. [DOI] [PubMed]
- 2.Rudan I., O’Brien K., Nair H., Liu L., Theodoratou E., Qazi S., on behalf of Child Health Epidemiology Reference Group (CHERG) Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Global Health. 2013;3(1) doi: 10.7189/jogh.03.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rudan I, O.B.K., Nair H, Liu L, Theodoratou E, Qazi S, Lukšić I, Fischer Walker CL, Black RE, Campbell H, and on behalf of Child Health Epidemiology Reference Group (CHERG), Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. Journal of Global Health, 2013. 3(1). [DOI] [PMC free article] [PubMed]
- 3.Simell B., Auranen K., Käyhty H., Goldblatt D., Dagan R., O’Brien K.L., for the Pneumococcal Carriage Group (PneumoCarr) The fundamental link between pneumococcal carriage and disease. Exp Rev Vacc. 2012;11(7):841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]; Simell B, A.K., Käyhty H, Goldblatt D, Dagan R, O’Brien KL & for the Pneumococcal Carriage Group (PneumoCarr), The fundamental link between pneumococcal carriage and disease. Expert Review of Vaccines, 2012. 11(7): p. 841-855. [DOI] [PubMed]
- 4.Davis S.M., Deloria-Knoll M., Kassaa H.T., O’Brien K.L. Impact of pneumococcal conjugate vaccines on nasopharyngeal carriage and invasive disease among unvaccinated people: review of evidence on indirect effects. Vaccine. 2014;32:133–145. doi: 10.1016/j.vaccine.2013.05.005. [DOI] [PubMed] [Google Scholar]; Davis SM, D.-K.M., Kassaa HT, O’Brien KL, Impact of pneumococcal conjugate vaccines on nasopharyngeal carriage and invasive disease among unvaccinated people: Review of evidence on indirect effects. Vaccine, 2014. 32: p. 133-145. [DOI] [PubMed]
- 5.O'Brien K.L., Millar E.V., Zell E.R., Bronsdon M., Weatherholtz R., Reid R. Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J Infect Dis. 2007;196(8):1211–1220. doi: 10.1086/521833. [DOI] [PubMed] [Google Scholar]; O'Brien KL, Millar EV, Zell ER, Bronsdon M, Weatherholtz R, Reid R, Bencenti J, Kvamme S, Whitney CG & Santosham M. Effect of Pneumococcal Conjugate Vaccine on Nasopharyngeal Colonization among Immunized and Unimmunized Children in a Community-Randomized Trial. The Journal of Infectious Diseases 2007. 196(8): p. 1211-1220. [DOI] [PubMed]
- 6.Dunne E.M., Satzke C., Ratu F.T., Neal E.F.G., Boelsen L.K., Matanitobua S. Effect of ten-valent pneumococcal conjugate vaccine introduction on pneumococcal carriage in Fiji: results from four annual cross-sectional carriage surveys. Lancet Global Health. 2018;6:e1375–e1385. doi: 10.1016/S2214-109X(18)30383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dunne EM, Satzke C, Ratu FT, Neal EFG, Boelsen LK, Matanitobua S, Pell CL, Nation ML, Ortika BD, Reyburn R, Jenkins K, Nguyen C, Gould K, Hinds J, Tikoduadua L, Kado J, Rafai E, Kama M, Mulholland EK & Russell FM. Effect of ten-valent pneumococcal conjugate vaccine introduction on pneumococcal carriage in Fiji: results from four annual cross-sectional carriage surveys. Lancet Global Health 2018. 6: e1375-1385 [DOI] [PMC free article] [PubMed]
- 7.Hanke C.R., Grijalva G.C., Chochua A., Pletz M.W., Hornberg C., Edwards K.M. Bacterial density, serotype distribution and antibiotic resistance of pneumococcal strains from the nasopharynx of peruvian children before and after pneumococcal conjugate vaccine 7. Pediatric Infect Dis J. 2016;35(4):432–439. doi: 10.1097/INF.0000000000001030. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hanke CR, Grijalva GC, Chochua A, Pletz MW, Hornberg C, Edwards KM, Griffin MR, Verastegui H, Gil AI, Lanata CF, Klugman KP & Vidal JE. Bacterial Density, Serotype Distribution and Antibiotic Resistance of Pneumococcal Strains from the Nasopharynx of Peruvian Children Before and After Pneumococcal Conjugate Vaccine 7. Pediatric Infectious Disease Journal 2016. 35(4): p. 432-439. [DOI] [PMC free article] [PubMed]
- 8.Satzke C., Dunne E.M., Choummanivong M., Ortika B.D., Neal E.F.G., Pell C.L. Pneumococcal carriage in vaccine-eligible children and unvaccinated infants in Lao PDR two years following the introduction of the 13-valent pneumococcal conjugate vaccine. Vaccine. 2019;37:296–305. doi: 10.1016/j.vaccine.2018.10.077. [DOI] [PubMed] [Google Scholar]; Satzke C, Dunne EM, Choummanivong M, Ortika BD, Neal EFG, Pell CL, Nation ML, Fox KK, Nguyen CD, Gould KA, Hinds J, Chanthongthip A, Xeuatvongsa A, Mulholland EK, Sychareun V & Russelll FM. Pneumococcal carriage in vaccine-eligible children and unvaccinated infants in Lao PDR two years following the introduction of the 13-valent pneumococcal conjugate vaccine. Vaccine 2019. 37: p. 296-305 [DOI] [PubMed]
- 9.Collins A.M., Wright A.D., Mitsi E., Gritzfeld J.F., Hancock C.A., Pennington S.H. First human challenge testing of a pneumococcal vaccine: double-blind randomized controlled trial. Am J Respiratory Critical Care Med. 2015;192(7):853–858. doi: 10.1164/rccm.201503-0542OC. [DOI] [PubMed] [Google Scholar]; Collins AM, W.A., Mitsi E, Gritzfeld JF, Hancock CA, Pennington SH, Wang D, Morton B, Ferreira DM and Gordon SB., First Human Challenge Testing of a Pneumococcal Vaccine: Double-Blind Randomized Controlled Trial. American Journal of Respiratory Critical Care Medicine, 2015. 192(7): p. 853-858. [DOI] [PubMed]
- 10.Gritzfeld J.F., Cremers A.J.H., Ferwerda G., Ferreira G.M., Kadioglu A., Hermans P.W.M. Density and duration of experimental human pneumococcal carriage. Clin Microbiol Infect. 2014;20(12):O1145–O1151. doi: 10.1111/1469-0691.12752. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gritzfeld JF, C.A., Ferwerda G, Ferreira GM, Kadioglu A, Hermans PWM and Gordon SB., Density and duration of experimental human pneumococcal carriage. Clinical Microbiology and Infection, 2014. 20(12): p. O1145–O1151. [DOI] [PMC free article] [PubMed]
- 11.Carvalho M.S., Tondella M.L., McCaustland K., Weidlich L., McGee L., Mayer L.W. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45(8):2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carvalho MS, T.M., McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R and Sampson JS., Evaluation and Improvement of Real-Time PCR Assays Targeting lytA, ply, and psaA Genes for Detection of Pneumococcal DNA. Journal of Clinical Microbiology, 2007. 45(8): p. 2460-2466. [DOI] [PMC free article] [PubMed]
- 12.Johnston C., Hinds J., Smith A., van der Linden M., Van Eldere J., Mitchell T.J. Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J Clin Microbiol. 2010;48(8):2762–2769. doi: 10.1128/JCM.01746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johnston C, H.J., Smith A, van der Linden M, Van Eldere J, Mitchell TJ., Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. Journal of Clinical Microbiology, 2010. 48(8): p. 2762-2769. [DOI] [PMC free article] [PubMed]
- 13.Gritzfeld J.F., Wright A.D., Collins A.M., Pennington S.H., Wright A.K., Kadioglu A. Experimental human pneumococcal carriage. J Visual Exp. 2013;72 doi: 10.3791/50115. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gritzfeld JF, W.A., Collins AM, Pennington SH, Wright AK, Kadioglu A, Ferreira DM and Gordon SB., Experimental human pneumococcal carriage. Journal of Visual Experiments, 2013. 72. [DOI] [PMC free article] [PubMed]
- 14.Azzari C., Moriondo M., Indolfi G., Cortimiglia M., Canessa C., Becciolini L. Realtime PCR is more sensitive than multiplex PCR for diagnosis and serotyping in children with culture negative pneumococcal invasive disease. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009282. [DOI] [PMC free article] [PubMed] [Google Scholar]; Azzari C, M.M., Indolfi G, Cortimiglia M, Canessa C, Becciolini L, Lippi F, de Martino M and Resti M., Realtime PCR Is More Sensitive than Multiplex PCR for Diagnosis and Serotyping in Children with Culture Negative Pneumococcal Invasive Disease. PLoS ONE, 2010. 5(2). [DOI] [PMC free article] [PubMed]
- 15.Adler H., Nikolaou E., Gould K., Hinds J., Collins A.M., Connor V., Hales C., Hill H., Hyder-Wright A.D., Zaidi S.R., German E.L., Gritzfeld J.F., Mitsi E., Pojar S., Gordon S.B., Roberts A.P., Rylance J., Ferreira D.M. Pneumococcal colonization in healthy adult research participants in the conjugate vaccine era, United Kingdom, 2010–2017. J Infect Dis. 2019 doi: 10.1093/infdis/jiz034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adler H, Nikolaou E, Gould K, Hinds J, Collins AM, Connor V, Hales C, Hill H, Hyder-Wright AD, Zaidi SR, German EL, Gritzfeld JF, Mitsi E, Pojar S, Gordon SB, Roberts AP, Rylance J & Ferreira DM. Pneumococcal Colonization in Healthy Adult Research Participants in the Conjugate Vaccine Era, United Kingdom, 2010–2017. The Journal of Infectious Diseases 2019. [DOI] [PMC free article] [PubMed]
- 16.Lessa F.C., Milucky J., Rouphael N.G., Bennett N.M., Keipp Talbot H., Harrison L.H. Streptococcus mitis expressing pneumococcal serotype 1 capsule. Sci Rep. 2018;8:17959. doi: 10.1038/s41598-018-35921-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lessa FC, Milucky J, Rouphael NG, Bennett NM, Keipp Talbot H, Harrison LH, Farley MM, Walston J, Pimenta F, Gertz RE, Gowrisankar R, daGloria Carvalho M, Beall B & Whitney CG. Streptococcus mitis Expressing Pneumococcal Serotype 1 Capsule. Scientific Reports 2018. 8:17959 [DOI] [PMC free article] [PubMed]
- 17.Pimenta J., Gertz R.E., Park S.H., Kim E., Moura I., Milucky J. Streptococcus infantis, Streptococcus mitis, and Streptococcus oralis Strains With Highly Similar cps5 Loci and antigenic relatedness to serotype 5 pneumococci. Front Microbiol. 2019;9:3199. doi: 10.3389/fmicb.2018.03199. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pimenta J, Gertz RE, Park SH, Kim E, Moura I, Milucky J, Rouphael N, Farley MM, Harrison LH, Bennett NM, Bigogo G, Feikin DR, Breiman R, Lessa FC, Whitney CG, Rajam G, Schiffer J, daGloria Carvalho M & Beall B.Streptococcus infantis, Streptococcus mitis, andStreptococcus oralisStrains With Highly Similarcps5Loci and Antigenic Relatedness to Serotype 5 Pneumococci. Frontiers in Microbiology 2019. 9:3199 [DOI] [PMC free article] [PubMed]
- 18.Thors V., Morales-Aza B., Pidwill G., Vipond I., Muir P., Finn A. Population density profiles of nasopharyngeal carriage of 5 bacterial species in pre-school children measured using quantitative PCR offer potential insights into the dynamics of transmission. Human Vacc Immunotherap. 2016;12(2):375–382. doi: 10.1080/21645515.2015.1090069. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thors V, M.-A.B., Pidwill G, Vipond I, Muir P, and Finn A, Population density profiles of nasopharyngeal carriage of 5 bacterial species in pre-school children measured using quantitative PCR offer potential insights into the dynamics of transmission. Human Vaccines & Immunotherapeutics, 2016. 12(2): p. 375--382. [DOI] [PMC free article] [PubMed]
- 19.Vu H.T., Yoshida L.M., Suzuki M., Nguyen H.A., Nguyen C.D., Nguyen A.T. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatric Infect Dis J. 2011;30(1):11–18. doi: 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]; Vu HT, Yoshida LM, Suzuki M, Nguyen HA, Nguyen CD, Nguyen AT, Oishi K, Yamamoto T, Watanabe K & Vu TD. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatric Infectious Disease Journal 2011. 30(1): p.11-18 [DOI] [PubMed]
- 20.Odutola A., Ota M.O.C., Ezra M.A., Ogundare O., Saidu Y., Foster-Nyarko E. Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: a phase 2, randomized, controlled, observer-blind study. Vaccine. 2017;35(19):2531–2542. doi: 10.1016/j.vaccine.2017.03.071. [DOI] [PubMed] [Google Scholar]; Odutola A, Ota MOC, Ezra MA, Ogundare O, Saidu Y, Foster-Nyarko E, Owiafe PK, Ceesay F, Worwui A, Idoko OT, Owolavi O, Bojang A, Jarju S, Drammeh I, Kampmann B, Greenwood BM, Alderson M, Traskine M, Devos N, Schoonbroodt S, Swinnen K, Verlant V, Dobbelaere K & Borys D. Efficacy of a novel, protein-based pneumococcal vaccine against nasopharyngeal carriage of Streptococcus pneumoniae in infants: A phase 2, randomized, controlled, observer-blind study. Vaccine 2017; 35(19): 2531-2542. [DOI] [PubMed]